Abstract
Although many Escherichia coli strains are considered commensals in mammals, strains encoding the cyclomodulin genotoxins are associated with clinical and subclinical disease in the urogenital and gastrointestinal tracts, meningitis, and inflammatory disorders. These genotoxins include the polyketide synthase (pks) pathogenicity island, cytolethal distending toxin (cdt), and hemolysin-associated cytotoxic necrotizing factor (cnf). E. coli strains are not excluded from rodents housed under SPF conditions in academic or vendor facilities. This study isolated and characterized genotoxin-encoding E. coli from laboratory rats obtained from 4 academic institutions and 3 vendors. A total of 69 distinct E. coli isolates were cultured from feces, rectal swab, nares, or vaginal swab of 52 rats and characterized biochemically. PCR analysis for cyclomodulin genes and phylogroup was performed on all 69 isolates. Of the 69 isolates, 45 (65%) were positive for pks, 20/69 (29%) were positive for cdt, and 4 (6%) were positive for cnf. Colibactin was the sole genotoxin identified in 21 of 45 pks+ isolates (47%), whereas cdt or cnf was also present in the remaining 24 isolates (53%); cdt and cnf were never present together or without pks. All genotoxin-associated strains were members of pathogen-associated phylogroup B2. Fisher exact and χ2 tests demonstrated significant differences in genotoxin prevalence and API code distribution with regard to vendor. Select E. coli isolates were characterized by HeLa cell in vitro cytotoxicity assays, serotyped, and whole-genome sequenced. All isolates encoding cyclomodulins induced megalocytosis. Serotypes corresponded with vendor origin and cyclomodulin composition, with the cnf+ serotype representing a known human uropathogen. Whole-genome sequencing confirmed the presence of complete pks, cdt, and hemolysin-cnf pathogenicity islands. These findings indicate that genotoxin-encoding E. coli colonize laboratory rats from multiple commercial vendors and academic institutions and suggest the potential to contribute to clinical disease and introduce confounding variables into experimental rat models.
Abbreviations: cdt, cytolethal distending toxin; cnf, cytotoxic necrotizing factor; EMEM, Eagle minimal essential medium; pks, polyketide synthase
Escherichia coli is a gram-negative bacillus that colonizes the gastrointestinal tract of humans and animals.46 Although some strains are considered commensals, various intestinal and extraintestinal pathogenic E. coli pathotypes are associated with a wide range of clinical disease states in the host;16,41 these strains are responsible for the deaths of more than 2 million humans annually.65 Specific pathotypes often harbor similar virulence factors and correspond to distinct clinical and histologic lesions. Intestinal pathotypes include enteropathogenic E. coli, enterohemorrhagic E. coli, enteroinvasive E. coli, enterotoxigenic E. coli, enteroaggregative E. coli, diffusely adhering E. coli, and adherent-invasive E. coli.65 Extraintestinal pathotypes include uropathogenic E. coli and neonatal meningitis E. coli, both of which have an enhanced ability to translocate through the intestinal epithelium and cause severe clinical disease.
E. coli strains typically are classified into 1 of the 4 major phylogenetic groups: A, B1, B2, and D.10,14,60 Groups B2 and D are often associated with pathogenicity, whereas fecal strains belonging to groups A and B1 generally lack virulence factors.22,60 Strains belonging to pathogroup B2 have been isolated from the feces of persons from developed countries with increasing frequency.52,70
These pathogenic strains encode various combinations of virulence genes and pathogenicity islands which promote invasion and colonization, evasion of host defenses, and damage to host tissues. Associated virulence factors include cytotoxins such as genotoxic cyclomodulins, cytotoxic necrotizing factors (cnf), cytolethal distending toxin (cdt), and the genotoxin colibactin (pks). These virulence factors modulate host cellular differentiation, proliferation, and apoptosis and promote cytopathic effects.7,21,69
CNF is a 115-kDa cyclomodulin protein that induces cell-cycle alterations and cytoskeletal changes by activating rho GTPases, which leads to a variety of aberrant phenotypic effects including micropinocytosis, megalocytosis, and multinucleation.62 cnf1 is chromosomally encoded,23 whereas cnf2 is plasmid-encoded.20 cnf-producing E. coli are considered necrotoxigenic and are associated with intestinal, urinary,23 and meningeal infection of humans.41 cnf+ E. coli have previously been isolated from clinically normal and clinically ill ferrets,49 cats,25 dogs,37,67 pigs,73 birds,42 and macaques.24
Cytolethal distending toxins (CDT) are encoded by 3 adjacent genes—cdtA, cdtB, and cdtC—that can be either chromosomally or plasmid-encoded.72 All 3 genes are required for the production of this heat-stable exotoxin, which bears considerable homology to DNAse I and causes DNA breaks.13 CDT have been classified into subgroups I through V21 according to variations in amino-acid sequences and genomic locations.34 Various enteropathogenic E. coli serotypes carry this cyclomodulin, which, like colibactin, induces irreversible megalocytosis and G1 or G2 cell-cycle arrest. cdt+ E. coli have been isolated from healthy and diseased humans as well as cattle, swine, and birds.34,58
A 54-kb polyketide synthase (pks) pathogenicity island encodes multiple clb genes (nonribosome peptide synthases) that are collectively responsible for colibactin synthesis. The pks island was first identified in 2006 in a case of neonatal meningitis induced by extraintestinal pathogenic E. coli57 and is associated with a variety of extraintestinal infections in humans, including bacterial meningitis, septicemia, and infections of the genitourinary tract.28,53 In addition, the pks island is associated with increased persistence in the gastrointestinal tract. Colibactin induces double-stranded DNA breaks, which lead to chromosomal instability and subsequent promotion of carcinogenesis.53 In human studies, colibactin-producing E. coli are isolated from human colorectal tumors with significantly increased frequency.7 Furthermore, pks+ E. coli promoted tumor survival by inducing cellular senescence through growth factor secretion.15 This association is recapitulated in laboratory animal models. Monoassociation of the pks+ E. coli strain NC101 caused typhlitis44 and promoted invasive carcinoma in azoxymethane-treated IL10 knockout (C57BLIL10−/−) mice;1 these effects were dependent on the presence of the pks island. In vitro studies have confirmed these findings by demonstrating that colibactin-encoding E. coli strains induce significant megalocytosis, double-stranded DNA breaks, phosphorylated γ-H2AX foci,1 and G2 cell-cycle arrest in eukaryotic cells.69
The presence of these genotoxins in human E. coli isolates is variable; prevalence is dependent on geographic location. In Puerto Rico, 8 of 41 (20%) fecal isolates tested positive for pks. However, only 1 isolate encoded cnf, and none encoded cdt. The cnf-encoding isolate was also pks+.30 Similarly, a group in France found that 26% of their 81 patients harbored pks+ E. coli strains, 18% were cnf+, and 11% were cdt+; cnf and cdt were often associated with pks, with a minority of genotoxin+ strains encoding either cnf or cdt only.61 Only 2 isolates (originating from patients with colon cancer) were positive for all 3 genotoxins. All cnf+ strains demonstrated a hemolytic phenotype.61 In Mexico, a single uropathogenic strain among 108 tested encoded both cnf and cdt; pks was not evaluated.47
Our laboratory has demonstrated that 88% of E. coli isolates from laboratory mice were pks+ and belonged to pathogen-associated phylogroup B2.28 Genotoxic E. coli strains have been identified in several other species of laboratory animals; pks+ E. coli has also been identified in laboratory macaques, and cnf+ E. coli in ferrets and NHP.24,49
Rats provide valuable models of both neonatal meningitis and uropathogenic E. coli infection. Young rats are commonly used to study systemic dissemination of neonatal meningitis E. coli K1 infection through the gastrointestinal tract17,79 and methods of prevention,82 intestinal barrier permeability,33 and sequelae of bacterial neonatal meningitis.29 Neonatal rats have recently been used to model maternal to neonatal transmission of pks+ E. coli, which resulted in increased rates of intestinal epithelial cell proliferation, apoptosis, and permeability that was transmissible through generations.59 In addition, numerous studies have used rats experimentally infected with cnf+ uropathogenic E. coli to study the dissemination and pathogenesis of E. coli associated recurrent urinary tract infections, pyelonephritis, and acute kidney injury.63,68,77 In addition, potential novel treatments for these conditions, such as photodynamic therapy,35 and novel drug delivery methods are investigated in these experimentally infected rat models.
The prevalence of pks and other cyclomodulin+ E.coli strains in SPF laboratory rats is currently unknown; vendors typically do not include E. coli on their health surveillance reports. Therefore, this study focused on determining the comparative prevalence of pks-, cdt-, and cnf+ isolates from the gastrointestinal tract and several other sites from rats obtained from multiple institutions and vendors. Given previous work regarding prevalence in laboratory mice and its association with urosepsis and meningitis in immunocompromised mice,28 we hypothesized that the majority of isolates from rats encoded the pks genomic island regardless of institution or vendor; we then asked whether these isolates also encoded cdt or cnf.
Materials and Methods
Animals.
The study population comprised 52 rats from 3 vendors originating from multiple barriers within each vendor facility and ultimately residing at 4 academic institutions. Vendor A rats were housed in institutions W, X, and Z; vendor B supplied rats for institutions Y and Z; and vendor C supplied rats only to institution Z. The most commonly represented strain was Sprague–Dawley; 4 rats were Long Evans, and 3 were c-fos–lacZ transgenic rats. There was an even distribution of male and female rats. According to health surveillance reports, all animals were considered SPF. E. coli was absent from vendor surveillance reports. Samples were collected from 2015 through 2017, and rats ranged in age from 8 wk to 2 y. Animals were group-housed at both the vendors and academic institutions; 3 of 4 academic institutions maintained AAALAC-accredited facilities. Rodent chow and water were provided free-choice and housed in polycarbonate cages. All animals were on IACUC-approved studies.
Culture and isolation.
E. coli was isolated from fecal contents, vagina, or nares of clinically normal rats immediately upon delivery to the academic institutions or after being housed in academic facilities. In total, 69 E.coli isolates were cultured from feces or rectal swabs (n = 49), vaginal swabs (n = 1), or nares (n =33). Fecal or rectal samples were collected directly from the rectum of the animals in shipping crates prior to their entrance into the institutional facilities. Fecal pellets or rectal swabs were placed into tubes containing sterile Gram Negative broth (Becton Dickenson, Franklin Lakes, NJ) and incubated at 37 °C overnight. A broth swab was plated onto MacConkey lactose agar plates (Remel, San Diego, CA), and lactose+ colonies were then plated onto sheep blood agar plates (Remel) according to distinct colony morphologies. The presence or absence of β-hemolysis was noted and recorded; suspect E. coli isolates were biochemically characterized by using API 20 E (Biomérieux, Marcy l'Etoile, France).
DNA extraction and PCR amplification.
A loop of each of the 69 E. coli isolates grown overnight on sheep's blood agar plates was placed in 500 μL of sterile PBS in a microfuge tube and swirled until thoroughly dissolved. Samples were boiled for 10 min, followed by 10 min of centrifugation at 12,000 × g. The supernatants were used in the PCR reactions. Two sets of primers (clbA, clbQ) were used to identify pks genes.24 Multiplex PCR analysis was used to amplify cnf and cdt genes. Primers for viaA, TSPE4.C2, chuA, svg, and uidA were used in multiplex PCR analysis to determine the phylogroup of each isolate.5,14 The phylogenetic groups were determined according to the PCR gel pattern.
Statistical analysis
Webtool (http://www.physics.csbsju.edu/stats/) was used to calculate expected contingency tables for each genotoxin, phylogroup, and API code according to results of PCR and microbiologic characterization. Prism version 6.01 (GraphPad Software, San Diego, CA) was used to perform either 2-tailed χ2 or Fisher exact tests to evaluate significant differences between the distribution of pks, cdt, and cnf encoding isolates among the 3 vendors and correlation between genotoxin-encoding ability, phylogroup, and API code. Statistical significance was set at a P value of less than 0.05.
Serotyping.
Nine E. coli isolates chosen from different vendors, barriers, and institutions and representing pks–cdt–cnf−, pks+cdt–cnf−, pks+cdt–cnf+, and pks+cdt+cnf– and genotypes were submitted to the E. coli Reference Center (Penn State University) for serotype testing. This testing included O and H typing and PCR analyses for heat-labile enterotoxin (elt), heat-stabile enterotoxin (estA and estB), Shiga-type toxin 1 and 2 (stx1 and stx2), intimin γ (eae), cnf1, and cnf2.
Cytotoxicity assay.
Control strains included NC101 (pks+cdt– cnf−) and NC101Δpks (pks– mutant), which were gifts from Dr Christian Jobin. Other control strains included V27 (positive control; pks+cdt+cnf–, acquired from the E. coli Reference Center), and K12 (triple-negative control). Eleven isolates representing all possible combinations of genotype, vendors, and institution were evaluated; these isolates included pks+, cdt+, and cnf+ isolates; triple-negative isolates; and isolates from all anatomic locations sampled.
Cell culture assay for colibactin cytotoxicity.
The cytotoxicity assay was performed as described previously with modifications.28,57 HeLa S3 cells (CCL2.2, ATCC, Manassas, VA) were grown and maintained in Eagle minimal essential medium (EMEM, ATCC) containing 10% FCS (Sigma, St Louis, MO) and 1% antibiotic–antimycotic (Gibco, Gaithersburg, MD) at 37 °C with 5% CO2. Cells (5 × 103 per well) were seeded onto 96-well cell culture plates and incubated at 37 °C with 5% CO2 for 24 h. Overnight liquid cultures of E. coli strains were grown for 2 h at 37 °C and then adjusted to OD600 in 1% FCS EMEM to concentrations corresponding to a multiplicity of infection (the number of bacteria per cell at the onset of infection) of 100. After inoculation, plates were centrifuged at 200 × g for 10 min to facilitate bacterial interaction and then incubated at 37 °C with 5% CO2 for 4 h. Cells were then washed with EMEM and resuspended in EMEM containing 10% FCS and 200 µg/mL gentamicin (Gibco). After a 72-h incubation, plates were stained with Diff-quick stain (Thermo Fisher Scientific, Waltham, MA), after which cells were inspected for confluence and morphologic changes and imaged at 20× magnification under a microscope (Axiovert-10, Zeiss, Jena, Germany) by using Image Pro-Plus software (version 7.0, Media Cybernetics, Rockville, MD).
Cell culture assay for sonicate cytotoxicity.
Overnight cultures of E. coli strains were pelleted by centrifugation at 13,500 × g for 5 min. The pelleted cells were washed in 1 mL of PBS and pelleted again by centrifugation at 13,500 × g for 5 min. Pellets were resuspended in 2 mL of PBS and then sonicated on ice (amplitude, 35; power, 7 W; 30-s intervals) for a total of 5 min with 1-min breaks between intervals. Sonicated samples were centrifuged at 13,500 × g for 10 min at 4 °C to remove large debris. Supernatant was collected and then filter-sterilized through 0.2 μm filters. Total protein was quantified by using the BCA assay (Thermo Fisher Scientific). HeLa cells (5 × 103 per well) were seeded onto 96-well cell culture plates and incubated at 37 °C with 5% CO2 for 24 h. Cells were treated with 1 or 40 μg/mL total protein of crude bacterial sonicate for 72 h. Cells were stained and microscopically analyzed for confluence and morphologic changes as described earlier.
Draft genome sequencing and comparative analysis.
Genomic DNA was isolated from 7 representative isolates by using the MasterPure Complete DNA and RNA Purification Kit (Epicentre, Madison, WI) according to the manufacturer's protocol for bacterial cell samples. DNA libraries were prepared by the Sequencing Core at the Forsyth Institute (Cambridge, MA) by using NextraXT technology for sequencing of 2 × 150 paired-end reads by Illumina MiSeq. Raw sequencing reads were decontaminated of adapter sequences and quality trimmed to a Phred quality score (Q) ≥ 10 by using BBDuk from the BBMap package version 37.17 (http://sourceforge.net/projects/bbmap/). Decontaminated reads were then assembled into contigs with SPAdes3 and scaffolds with Ragout43 followed by genome annotation with RAST hosted by PATRIC.2,6,78 Sequences encoding putative virulence factor and antibiotic resistance genes were identified by using VirulenceFinder 1.536 and ResFinder 2.1,81 hosted by Center for Genomic Epidemiology. Syntenic relationships of pks, cdt, and hemolysin–cnf operon genes between genomes were determined by using SimpleSynteny.76
Accession numbers.
GenBank accession numbers are available in Table 4.
Table 4.
Novel rat E. coli genomes have similar statistics as pathogenic, PKS-encoding E. coli strains IHE3034 and NC101
| Strain | Isolation source | Genome length (bp) | Contigs | G+C% content | Protein-coding sequences | tRNA | rRNA | Virulence factor genes | GenBank accession no. |
| S11 | Research rat | 5201802 | 32 | 49.72 | 5149 | 81 | 10 | cdtABC, gad, iroN, iss, mchB, mchC, mchF, mcmA, pic, PKS, vat | NHYT00000000 |
| S14 | 5208467 | 37 | 49.64 | 5153 | 81 | 10 | cdtABC, gad, iroN, iss, mchB, mchC, mchF, mcmA, pic, PKS, vat | NHYQ00000000 | |
| S12 | 5092914 | 14 | 50.04 | 4995 | 77 | 11 | iroN, iss, PKS, sfaS, vat | NHYS00000000 | |
| S13 | 5296109 | 47 | 48.57 | 5101 | 73 | 9 | astA, cba, cma, gad, lpfA, pic, PKS | NHYR00000000 | |
| S15 | 5248403 | 58 | 47.71 | 5078 | 79 | 8 | celb, gad, iss, PKS | NHYP00000000 | |
| S16 | 5623575 | 140 | 45.33 | 5509 | 70 | 3 | cdtABC, celb, gad, iss, PKS | NHYO00000000 | |
| S17 | 5139109 | 40 | 49.47 | 5022 | 76 | 9 | cnf1, gad, iroN, mchB, mchC, mchF, mcmA, PKS, vat | QLVH00000000 | |
| IHE3034 | Human neonatal meningitis | 5108383 | 1 (complete genome) | 50.70 | 5045 | 97 | 22 | gad, iroN, iss, PKS, sfaS, vat, cdtABC | CP001969.1 |
| NC101 | Research mouse | 5021144 | 27 | 50.57 | 4917 | 72 | 4 | gad, iroN, iss, PKS, sfaS, vat | AEFA00000000.1 |
| K12 substrain DH10B | Human nonpathogenic | 4686137 | 1 (complete genome) | 50.80 | 4606 | 87 | 14 | gad, iss | CP000948.1 |
astA, EAST-1 heat-stable toxin; cba, colicin B; cdtABC, cytolethal distending toxin subunits A, B, and C; celb, endonuclease colicin E2; cma, colicin M; gad, glutamate decarboxylase; iroN, enterobactin siderophore receptor protein; iss, increased serum survival; lpfA, long polar fimbriae; mchB, microcin H47 part of colicin H; mchC, MchC protein; mchF, ABC transporter protein MchF; mcmA, microcin M part of colicin H; pic, serine protease autotransporters of Enterobacteriaceae (SPATE); PKS, polyketide synthetase (colibactin); sfaS, S-fimbriae minor subunit; vat, vacuolating autotransporter toxin
Virulence factor genes for toxins, survival factors, and adhesions were identified in the rat E. coli genomes.
Results
Microbiologic characterization.
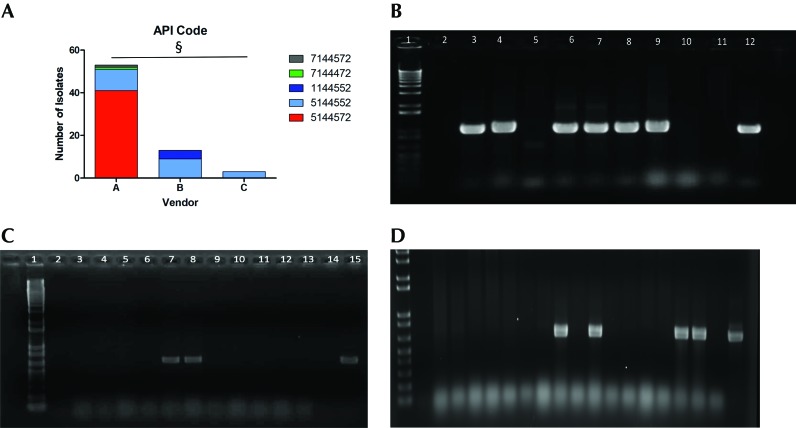
E. coli was isolated from all 52 rats sampled, with all biologic sampling locations (rectum, nares, vagina) yielding E. coli isolates. In total, 69 E. coli isolates were cultured; some animals harbored multiple E. coli isolates as determined by distinct API codes and colony morphology. None of the isolates demonstrated hemolysis. Across all vendors and institutions, there was no correlation between API code and genotoxin genotype. Differences in API codes indicated the ability of the isolates to ferment certain sugars and metabolize specific amino acids. The most common API code, 5144572, was observed in 41 of the 69 isolates, whereas the second most common was 5144552, observed in 23 isolates. The major metabolic difference between these codes is that strains with the most common code (5144572) have the ability to ferment sucrose whereas those with 5144552 cannot. The API code 1144552 appeared in 4 isolates, and codes 7144472 and 7144572 were observed in single isolates from vendor A; codes beginning with 1 lack lysine and arginine decarboxylase activity. χ2 testing revealed significant (P < 0.0001) correlation between API code and vendor, thus suggesting a degree of clonality among isolates from each origin (Figure 1 A). However, there was no statistical correlation between genotype and API code (data not shown). For example, all 9 isolates from vendor B with the API code 5144552 harbored both pks and cdt, whereas all 11 isolates with the same API code from vendor A were negative for all genotoxins. The 4 isolates with API code 1144552 originated from vendor B rats cultured directly from the shipping crate after arriving at the institution. This API code occurred only in isolates that were cnf+.
Figure 1.
(A) API code acccording to vendor. §, P < 0.0001. Amplification of (B) clbQ, (C) cdt, and (D) cnf genes in E. coli isolates from rats. Lane 1, 1-kb ladder; lane 2, negative control; lanes 3 through 11, E. coli isolates from rats; lane 12, positive control.
Identification of pks, cdt, and cnf genes.
Conventional PCR analysis for pks genes clbA and clbQ (Figure 1 B) and multiplex PCR assays for cdt (Figure 1 C) and cnf (Figure 1 D) were performed on all isolates to identify the presence of genotoxin genetic elements. Overall, 45 of 69 (65%) of the total isolates were positive for both pks genes; there were no isolates that tested positive for one gene without the other; 20 of 69 (29%) isolates were positive for cdt, and 4 of 69 (6%) isolates were positive for cnf. pks was the sole genotoxin identified in 21 of the 45 pks+ isolates (47%), whereas cdt or cnf was also present in the remaining 24 isolates (53%). Neither cdt nor cnf was present without pks, and cnf and cdt were never present together (Table 1 and 2). The Fisher exact test determined that the prevalence of all 3 genotoxins was significantly different between the 3 vendors (P < 0.001 in all 3 cases). Roughly half (55%) of the isolates from vendor A rats were positive for pks, with a 15% minority encoding cdt in addition. No vendor A animals tested positive for cnf. Conversely, all isolates from vendor B animals were pks+, and 69% of them also encoded cdt. Isolates that did not encode cdt had cnf instead. Therefore, all isolates from vendor B were positive for multiple genotoxins (Table 2). Overall 51% to 80% of isolates from animals arriving at institutions W and X were pks+, with a minority of isolates (15% to 17%) carrying cdt in addition. All isolates from institutions Y and Z were pks+, with the majority of isolates also harboring cdt (64% to 100%). All rats from vendor C encoded pks and cdt. Those E. coli isolates from institution Y that did not have cdt encoded for cnf instead (36%; Table 2).
Table 1.
Distribution of genotoxin prevalence from Vendor A according to institutional destination
| Total | Institution W | Institution X | Institution Z | |
| Total pks+ E.coli | 29/53 (55%) | 4/5 (80%) | 24/47 (51%) | 1/1 |
| Total cdt+ E.coli | 8/53 (15%) | 0/5 (0%) | 8/47 (17%) | 0/1 |
| Total cnf+ E. coli | 0/53 (0%) | 0/5 (0%) | 0/47 (0%) | 0/1 |
| pks–/ cdt–/ cnf– | 24 | 1 | 23 | 0 |
| pks–/ cdt+/cnf– | 0 | 0 | 0 | 0 |
| pks–/cdt–/cnf+ | 0 | 0 | 0 | 0 |
| pks+/ cdt–/cnf– | 21 | 4 | 16 | 1 |
| pks+/ cdt+/cnf– | 8 | 0 | 8 | 0 |
| pks+/ cdt–/cnf+ | 0 | 0 | 0 | 0 |
Table 2.
Distribution of genotoxin prevalence according to vendor origin and institutional destination
| Vendor B |
Vendor C |
||||
| Total | Institution Y | Institution Z | Institution Z | All isolates | |
| Total pks+ E.coli | 13/13(100%) | 11/11 (100%) | 2/2 (100%) | 3/3(100%) | 45/69 (65%) |
| Total cdt+ E.coli | 9/13 (69%) | 7/11 (64%) | 2/2 (100%) | 3/3 (100%) | 20/69 (29%) |
| Total cnf+ E. coli | 4/13 (31%) | 4/11 (36%) | 0/2 (0%) | 0/3 (0%) | 4/69 (6%) |
| pks–/ cdt–/ cnf– | 0 | 0 | 0 | 0 | 24 |
| pks–/ cdt+/cnf– | 0 | 0 | 0 | 0 | 0 |
| pks–/cdt–/cnf+ | 0 | 0 | 0 | 0 | 0 |
| pks+/ cdt–/cnf– | 0 | 0 | 0 | 0 | 21 |
| pks+/ cdt+/cnf– | 9 | 7 | 2 | 3 | 20 |
| pks+/ cdt–/cnf+ | 4 | 4 | 0 | 0 | 4 |
Phylogenetic analysis.
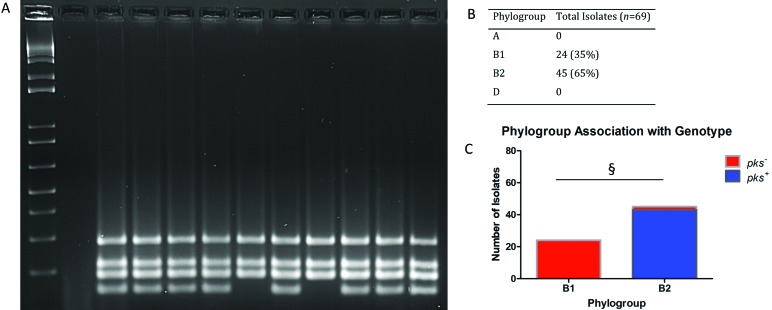
Phylogroup was determined according to the amplification pattern of multiplex PCR analysis of viaA, TSPE4.C2, chuA, svg, and uidA (Figure 2 A); the presence of 3 or more bands identifies the isolate as a member of phylogroup B2. All isolates were members of phylogroup B, with 24 of the 69 (35%) isolates belonging to group B1 and 45 of the 69 (65%) isolates belonging to pathogen-associated phylogroup B2. Only 2 isolates that were members of phylogroup B2 did not test positive for any of the cyclomodulins under evaluation. All genotoxin-positive isolates belonged to group B2 (Figure 2 B). A Fisher exact test demonstrated significant (P < 0.001) association between phylotype and genotype (Figure 2 C).
Figure 2.
Phylogenetic analysis. Sets of primers for viaA, TSPE4.C2, chuA, svg, and uidA genes were used in multiplex PCR assays to determine the phylogroup of each isolate.7,48 The phylogenetic groups were determined according to the PCR gel pattern, with the presence of 3 or more bands indicating membership in phylogroup B2. (A) Phylogroup multiplex PCR gel: lane 1,1-kb ladder; lane 2, negative control; lanes 3 through 12, E. coli isolates from rats. (B) Overall prevalence. (C) Overall distribution; §, P < 0.001.
Serotyping.
The most common serotype among isolates was O7:H7; all originated at vendor A, but each isolate originated from rats housed at a different institution (Table 3). Two of these isolates were pks+ only, and the third encoded both pks and cdt. The next 2 most common serotypes were found in duplicate. The 2 pks+cdt+ isolates from vendor B (rats housed at different institutions) were serotype O166:H6. Two triple-negative E. coli isolates from vendor A were O179:H8. The pks+ cdt+ E. coli isolate from vendor C was OM:H6, and the pks+cnf+ isolate from vendor B was O4:H5, a known uropathogen in humans.19,38 None of the E. coli isolates serotyped were positive for elt, estA, estB, stx1, stx2, eae, or cnf2.
Table 3.
Results from serotyping and virulence factor testing of E. coli isolates from rats
| Sample no. | O-type | H-type | Vendor | Institution | elt | estA | estB | stx1 | stx2 | eae | cnf1 | cnf2 |
| S1: pks– cdt– cnf– | 179 | 8 | A | W | neg | neg | neg | neg | neg | neg | neg | neg |
| S16: pks– cdt– cnf– | 179 | 8 | A | X | neg | neg | neg | neg | neg | neg | neg | neg |
| S5: pks+ cdt– cnf– | 7 | 7 | A | Z | neg | neg | neg | neg | neg | neg | neg | neg |
| S2: pks+ cdt– cnf– | 7 | 7 | A | W | neg | neg | neg | neg | neg | neg | neg | neg |
| S14: pks+ cdt+ cnf– | 7 | 7 | A | X | neg | neg | neg | neg | neg | neg | neg | neg |
| S4: pks+ cdt+ cnf– | 166 | 6 | B | Z | neg | neg | neg | neg | neg | neg | neg | neg |
| S8: pks+ cdt+ cnf– | 166 | 6 | B | Y | neg | neg | neg | neg | neg | neg | neg | neg |
| S7: pks+ cdt+ cnf– | M | 6 | C | Z | neg | neg | neg | neg | neg | neg | neg | neg |
| S9: pks+ cdt– cnf+ | 4 | 5 | B | Y | neg | neg | neg | neg | neg | neg | pos | neg |
neg, negative; pos, positive
In vitro cytotoxicity of E. coli isolates.
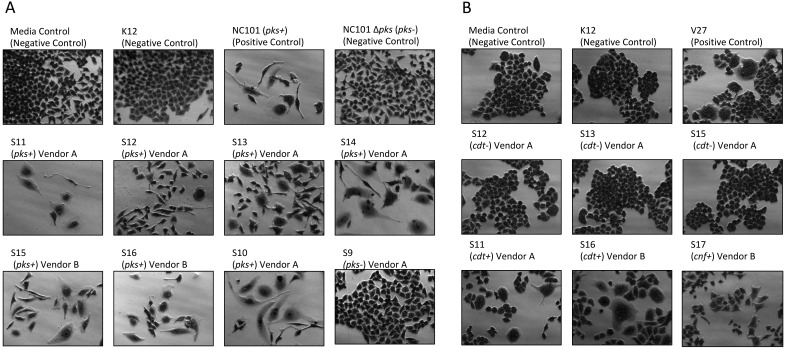
Cell culture assays were performed to determine whether in vitro infection or sonicates of representative rat E. coli isolates were cytotoxic to HeLa cells. A total of 17 isolates encompassing representatives from all institutions, vendors and barriers, anatomic areas of isolation, genotoxin status, and phylogroup were evaluated. Live bacteria were used rather than sonicates, because whole cells are required for the complete expression of colibactin.7 Conversely, CDT and CNF cytotoxicity are detectable only by using sonicate preparations. Viable pks+ E. coli isolates induced megalocytic cytotoxicity to HeLa cells, indicating contact-dependent colibactin expression (Figure 3 A). HeLa cells treated with sonicate from cdt+ or cnf+ E. coli isolates also displayed cell body and nuclear enlargement, which are characteristic of these cytotoxins (Figure 3 B). E. coli isolates that were PCR-negative for pks or cdt lacked cytotoxicity in their respective sonicate-based cell culture assays. These results indicate that rat E. coli isolates exhibit cytotoxic pks, cdt, and cnf activity in vitro, as their genotypes suggest.
Figure 3.
(A) Cell culture assay for cytotoxicity. HeLa cells were treated with E. coli at a multiplicity of infection (MOI) 100 for 4 h followed by a 72 h incubation in gentamicin-containing media. Cells infected with the novel rat isolates encoding pks displayed megalocytosis (enlargement of the cell body and nucleus) similar to the pks+ E. coli control (NC101 mouse isolate). No cytotoxicity was observed for cells treated with novel negative pks, the E. coli negative controls (media control and K12 nonpathogenic laboratory strain), or the NC101 isogenic pks knockout mutant (NC101 Δ pks). (B) Cell culture assay for cytotoxicity. HeLa cells were treated with 4 μg total protein of E. coli sonicate for 72 h. Cells treated with sonicate from cdt+ or cnf+ rat isolates display megalocytosis (enlargement of the cell body and nucleus) similar to the E. coli positive control V27 human urosepsis isolate. No cytotoxicity was observed for cells treated with cdt– or negative-control strains of E. coli. Magnification, 20×.
Draft genome sequencing and comparative analysis.
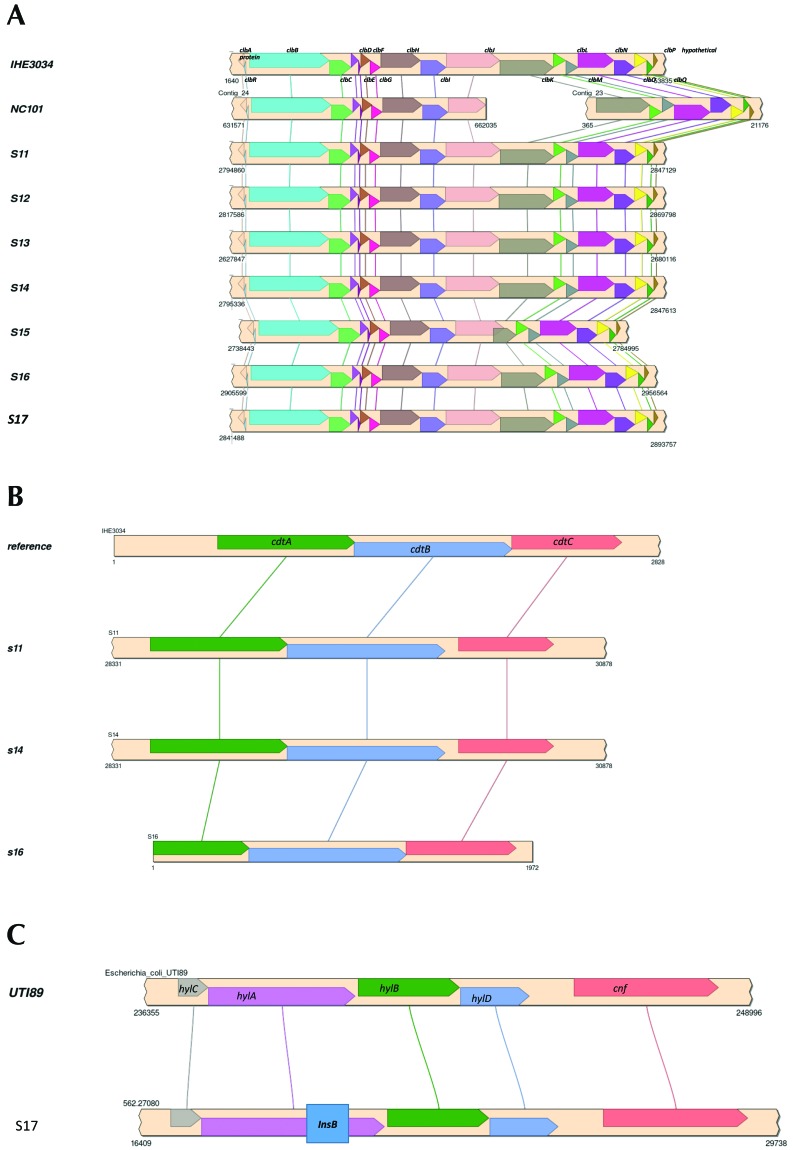
The draft genome sequences of 7 representative rat E. coli isolates were obtained for comparative analysis of the pks, cdt, and cnf genes as well as for identification of other virulence factor and antibiotic resistance genes. The genome sizes, G+C% contents, and protein and RNA genes of the rat E. coli isolates were similar to those of the representative pks+ E. coli strains IHE3034 and NC101 (Table 4). Homologous genes for all pks genes were identified in the rat E. coli isolates and showed identical synteny to IHE3034 and NC101. Compared with IHE3034, all pks genes from the rat E. coli isolates had at least 98% sequence coverage and identity, except the clbJ and clbK genes from isolate S15, which had approximately 90% and approximately 45% sequence coverage, respectively. Further analysis of the clbJ and clbK genes from isolate S15 suggested that they might be expressed as a hybridized gene. The clbJ gene appears to be missing 624 bp at the 3′ end, including the stop codon, but retains 2 nonribosomal peptide synthetase modules. clbK appears to lack 3540 bp at the 5′ end, including a start codon and the pks module, but retains the nonribosomal peptide synthetase module and the oxidase domain. Further analysis of the putative clbJ and clbK genes shows that their sequences overlap by 1480 bp in the genome, suggesting they are not expressed as separate genes but instead form a single, continuous gene sequence. When the clbJ start codon is used as the position of the open reading frame, the predicted sequence is translated into a 2240-amino–acid product (7323 bp) that includes the clbJ and clbK sequences and terminates at the clbK stop codon. This pattern suggests that the putative clbJ and clbK sequences may be transcribed and translated into a hybridized protein (designated clbJK-hybrid). The predicted clbJK-hybrid protein would contain 2 NPRS modules as well as an oxidase domain (Figure 4 A). A BLAST search found identical clbJK-hybrid gene sequences in the 3 other genomes: neonatal meningitis-causing E. coli strain NMEC O18 (GenBank accession no. CP007275), Klebsiella pneumoniae strain Kp52.145 (FO834906), and K. pneumoniae subsp. pneumoniae strain KPNIH32 (CP009775). This finding indicates that other E. coli and K. pneumoniae strains have a putative clbJK-hybrid sequence instead of separate clbJ and clbK genes in their pks islands. Isolate S15 still induced megalocytosis in HeLa cells, indicating cytotoxic colibactin expression despite having a putative clbJK-hybrid gene. Whether E. coli strain NMEC O18 and the 2 K. pneumoniae strains also exhibit colibactin cytotoxicity has not been reported.
Figure 4.
Syntenic alignment of (A) pks island genes between IHE3034, NC101, and the novel rat E. coli isolates. (B) cdt tripartite holotoxin syntenic alignment between reference genome and novel rat E. coli isolates. (C) Hemolysin–cnf operon syntenic alignment between cnf+ UTI89 uropathogen and a representative novel rat E. coli isolate. A 500-bp insertion (box within the InsB gene) is present in the hylA gene.
cdt genes were detected in 3 of the 7 genomes. All 3 genomes had complete tripartite cdt holotoxin islands including cdtA, cdtB, and cdtC (Figure 4 B). The cnf gene was intact, but the adjacent hemolysin operon demonstrated an insertional event that interrupted the hlyA gene (Figure 4 C). None of these cnf+ isolates were hemolytic.. Additional virulence factor genes were identified in the rat E. coli isolate genomes and included toxins (astA, cdtABC, pic, vat), bacteriocin synthesis genes (cba, celb, cma, mchB, mchC, mchF, mcmA), nutrient or survival factors (gad, iroN, iss), and adherence factors (lpfA, sfaS; Table 4). Neither gene sequences for cell cycle inhibiting factor (cif) nor antibiotic resistance genes were detected in any of the rat E. coli isolates. The genomic results suggest the rat E. coli isolates encode pks gene islands, cdt, cnf, and other virulence genes that endow them with pathogenic potential.
Discussion
Because E. coli is a major commensal organism of the human and animal intestinal tracts, a thorough understanding of this organism is warranted in both humans and animals. A shift in the genetic makeup of these E. coli colonizing the gut from phylogroups A and B1 to pathogen-associated phylogroups B2 and D has occurred in recent years in industrialized countries; this shift affects both humans and animals.52,70 Colibactin production induces double-stranded DNA breaks, activation of the DNA damage response, and subsequent genomic instability in the mammalian host. Senescence, cell death, and carcinogenesis are associated with colonization of pks+ E. coli strains. Similarly, cdt encodes a DNAse genotoxin that causes single- and double-stranded DNA breaks and results in increased mutagenesis; this cyclomodulin has been detected in E. coli isolated from proximal and distal colon cancer tissues from human patients.7,61 Cytotoxic necrotizing factor is a third cyclomodulin that is known to induce cell cycle disturbances and abnormal cytoskeletal effects.
Little information is available regarding the E. coli status of laboratory rats and the variability of genotoxin expressing E. coli in animals from different vendors and institutions. To our knowledge, this report is the first to characterize the presence of colibactin, cdt, and cnf in unmanipulated laboratory rats. In this study, we demonstrated statistically significant differences in the distribution of pks+, cdt+, and cnf+ E. coli across 3 popular vendors used by 4 academic institutions; this difference has the potential to affect study outcomes according to rat vendor origin. Overall, the majority of isolates (65%) were pks+ and members of phylogroup B2 (65%). There was a strong association of genotoxin-positive strains with phylogroup B2 in rats, as is the case in human isolates.19 Surprisingly, whereas cdt and cnf were not identified in E. coli-colonizing mice,28 these cyclomodulins were present in laboratory rats: 29% of rat E. coli isolates carried cdt, whereas only 6% carried cnf. This pattern is in contrast to our hypothesis and available human surveys, where cnf+ E. coli (39.5%) is isolated much more commonly than cdt+ E. coli (1% to 6%).7 In addition, cdt and cnf were never present in the E. coli strains without colibactin or in strains with each other. The coassociation of pks and cdt in some E. coli strains suggests mechanisms that potentiate genotoxicity, although pks and cdt are not commonly identified within the same human E. coli isolate.27,30 Double-positive isolates (pks+ cnf+) have been characterized from both healthy humans and patients with urosepsis;21 this is in contrast to surveys in humans and other laboratory animals, where cnf is occasionally present in colibactin-negative isolates.24,49,61 Many previous studies have shown a correlation between cnf and hemolysis,45,49,51 which is consistent with the proximity of the hemolysin gene to the cnf gene. Interestingly, none of the cnf+ isolates from laboratory rats demonstrated hemolysis due to an insertion event in the hylA gene. All cnf+ E. coli strains isolates were isolated from vendor B rats.
The results of both the whole cell and sonicate cytotoxicity assays correlated with the presence or absence of pks, cdt, and cnf. Because cell contact is required for colibactin cytotoxicity,69 HeLa cell death and megalocytosis was due to cdt or cnf in the sonicate assay. Genotoxin-negative E. coli isolates produced results that were indistinguishable from those of the nonpathogenic strain K12, thus suggesting attenuated pathogenicity due to lack of genotoxins. Whereas only 55% of E. coli isolates from vendor A encoded the pks island, 100% of isolates from vendor B were pks+. Institutional pks+ E. coli prevalence in rats was seemingly low risk, consistent with reported rat vendor usage and origin, with vendor A institutions having lower E. coli genotoxin prevalence in rats compared with rats housed in vendor B institutions. In addition, API code and serotype patterns tended to correlate with vendor origin rather than institution (Table 1). This pattern underscores that genotoxin-positive E. coli efficiently colonize and likely persist in the bowel throughout life;65 these strains likely colonize rats at the vendors and inhabit the alimentary tract of the animals for the duration of studies performed at destination institutions.
The clb genes encoded in the pks island constitute an ‘assembly line’ of enzymes that produce precolibactin and colibactin metabolites by complex and incompletely defined biosynthetic pathways. Furthermore, these metabolites can be formed or modified by the clb enzymes through alterative pathways, leading to a large structural diversity of molecules that has not been entirely cataloged. In particular, recent reports have indicated that the pks module in clbK can be biochemically bypassed to yield an alternative precolibactin metabolite with unknown cytopathogenic properties.74,83 This alternative pathway still requires the nonribosomal peptide synthetase modules and oxidase activity from clbJ and clbK. The putative clbJK-hybrid gene detected in isolate S15 is predicted to contain 2 nonribosomal peptide synthetase modules and an oxidase domain but lacks the pks module from clbK. As a result, it may be possible for the putative clbJK-hybrid gene to synthesize precolibactin metabolites in analogous fashion to the alternate scheme mentioned earlier.
All 3 cdt genes in the cdt island were intact and conserved among isolates (Figure 4 B). Although the cnf island itself was intact, the hemolysin hlyA gene was disrupted by an approximately 500-bp insertion consisting of the insertion element IS1 protein InsB, which is the most common transposase in the E. coli genome (Figure 4 C). Transposable IS1 elements have been reported to disrupt other portions of the hemolysin operon.9
In addition to pks, cdt, and cnf, other virulence factor genes that are known to enhance colonization and survival and promote disease in the host were identified in the rat E. coli isolates. Glutamate decarboxylase (gad) and increased serum survival/bor protein precursor (iss) promote survival in the host by neutralizing stomach acid during oral transmission4,31,66 and by promoting resistance against host complement protein,39,48,54 respectively, whereas enterobactin siderophore receptor protein (iroN) allows uptake of the essential nutrient iron into the pathogen.12,26,41 Long polar fimbriae (lpfA) and S-fimbriae minor subunit (sfaS) are both adhesion factors for colonization of host epithelial cells.40,50,64,71 Colicin B (cba), colicin E (celb), colicin M (cma), and microcin H47 (mchB, mchC, mchF, mcmA) are bacteriocins produced by pathogenic E. coli strains that target and kill susceptible bacteria.8,11,55,56 As a result, bacteriocin producers may have competitive advantages in niches with scare essential nutrients, like iron. Enteroaggregative E. coli heat-stable enterotoxin 1 (astA) is a cytotoxin that actives guanylyl cyclase in the gastrointestinal epithelium, resulting in ion secretion that contributes to watery diarrheal disease.16,41,75 Protease involved in intestinal colonization (pic) and vacuolating autotransporter toxin (vat) are both serine protease autotransporters of Enterobacteriaceae (SPATE) that degrade the mucous barrier to facilitate invasion18 and cause intracellular vacuolation,18,32 respectively. Of particular interest, cdt genes were identified in the genomic sequences of 3 isolates (S11, S14, S15).
The presence or absence of cyclomodulin genotoxins in laboratory rats may have unintended effects on experimental results and repeatability across institutions. Because E. coli is not included on vendor surveillance reports, rats from various institutions may have vastly different gastrointestinal microbiota, producing inherent variability in results and conclusions. Genotoxic-E. coli–colonizing rats arriving from vendors may interfere with studies of experimental E. coli infection. This situation is especially relevant, because neonatal rats are an extremely popular model of E. coli K1 infection and sequelae,79 in which the K1 capsule protects the bacteria from the host's immune response. This strain is another early colonizer of the neonatal gastrointestinal tract that can translocate from lumen to blood. These rats are used to characterize changes in oxidative responses after E. coli inoculation throughout life,29 track vertical transmission of pks+ E. coli from mothers to offspring,59 and evaluate the efficacy of a variety of antimicrobial agents against genotoxic E. coli infection.33,80,82
If genotoxic E. coli species are present at the initiation of these and other relevant studies, comparisons between sham and experimental groups may be erroneous. In addition, the possibility of zoonotic transfer from rats to humans should not be overlooked, especially given that O4:H5 E. coli isolated from rats in this study are associated with urosepsis in humans.19,37 This possibility emphasizes the importance of proper hygiene and personal protective equipment, even in seemingly low-risk areas. Together, the identification of virulence factor genes from genotoxin-encoding rat E. coli isolates with significantly variable prevalence across multiple vendors suggests that these pathobionts have the potential to cause clinical or subclinical disease in rats and significantly confound rat research models.
Acknowledgments
We thank Alyssa Pappa for her excellent assistance in preparing the manuscript.
Ethics approval and consent to participate in work involving animals was approved by the Massachusetts Institute of Technology's Committee on Animal Care and Use Office.
Research reported in this publication was supported by NIH under awards T32OD010978 and R01OD01141 and the National Institute of Environmental Health Sciences under award P30ES0022109. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergholz TM, Tarr CL, Christensen LM, Betting DJ, Whittam TS. 2007. Recent gene conversions between duplicated glutamate decarboxylase genes (gadA and gadB) in pathogenic Escherichia coli. Mol Biol Evol 24:2323–2333. 10.1093/molbev/msm163. [DOI] [PubMed] [Google Scholar]
- 5.Bidet P, Metais A, Mahjoub-Messai F, Durand L, Dehem M, Aujard Y, Bingen E, Nassif X, Bonacorsi S. 2007. Detection and identification by PCR of a highly virulent phylogenetic subgroup among extraintestinal pathogenic Escherichia coli B2 strains. Appl Environ Microbiol 73:2373–2377. 10.1128/AEM.02341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, 3rd, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. 2013. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8:1–10. 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budič M, Rijavec M, Petkovšek Ž, Žgur-Bertok D. 2011. Escherichia coli bacteriocins: antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS One 6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgos Y, Beutin L. 2010. Common origin of plasmid encoded α-hemolysin genes in Escherichia coli. BMC Microbiol 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MIZ, Gomes TAT, Amaral LA, Ottoboni LMM. 2010. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol 10:1–10. 10.1186/1471-2180-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caza M, Kronstad JW. 2013. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol 3:1–23. 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chumduri C, Gurumurthy RK, Zietlow R, Meyer TF. 2016. Subversion of host genome integrity by bacterial pathogens. Nat Rev Mol Cell Biol 17:659–673. doi: 10.1038/nrm.2016.100. [DOI] [PubMed] [Google Scholar]
- 14.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, Sauvanet P, Darcha C, Déchelotte P, Bonnet M, Pezet D, Wodrich H, Darfeuille-Michaud A, Bonnet R. 2014. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63:1932–1942. 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 16.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. 10.1038/nrmicro2265. Erratum in: Nat Rev Microbiol 2013.11:141. [DOI] [PubMed] [Google Scholar]
- 17.Dalgakiran F, Witcomb LA, McCarthy AJ, Birchenough GMH, Taylor PW. 2014. Noninvasive model of neuropathogenic Escherichia coli infection in the neonatal rat. J Vis Exp 92 e52018:1–10. doi:10.3791/52018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dautin N. 2010. Serine protease autotransporters of Enterobacteriaceae (SPATEs): biogenesis and function. Toxins (Basel) 2:1179–1206. 10.3390/toxins2061179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Paula Coelho C, Motta PD, Petrillo M, de Oliveira Iovine R, Dalboni LC, Santana FR, Correia MSF, Casarin RCV, Carvalho VM, Bonamin LV. 2017. Homeopathic medicine Cantharis modulates uropathogenic E. coli (UPEC)-induced cystitis in susceptible mice. Cytokine 92:103–109. 10.1016/j.cyto.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 20.De Rycke J, González EA, Blanco J, Oswald E, Blanco M, Boivin R. 1990. Evidence for 2 types of cytotoxic necrotizing factor in human and animal clinical isolates of Escherichia coli. J Clin Microbiol 28:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubois D, Delmas J, Cady A, Robin F, Sivignon A, Oswald E, Bonnet R. 2010. Cyclomodulins in urosepsis strains of Escherichia coli. J Clin Microbiol 48:2122–2129. 10.1128/JCM.02365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escobar-Páramo P, Le Menac'h A, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ Microbiol 8:1975–1984. 10.1111/j.1462-2920.2006.01077.x. [DOI] [PubMed] [Google Scholar]
- 23.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. 1993. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun 61:4909–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, Mannion A, Madden CM, Swennes AG, Townes C, Byrd C, Marini RP, Fox JG. 2017. Cytotoxic Escherichia coli strains encoding colibactin and cytotoxic necrotizing factor (CNF) colonize laboratory macaques. Gut Pathog 9:1–15. 10.1186/s13099-017-0220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Féria C, Machado J, Correia JD, Gonçalves J, Gaastra W. 2001. Virulence genes and P fimbriae PapA subunit diversity in canine and feline uropathogenic Escherichia coli. Vet Microbiol 82:81–89. 10.1016/S0378-1135(01)00375-3. [DOI] [PubMed] [Google Scholar]
- 26.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. 2017. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol 15:109–128. 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 28.García A, Mannion A, Feng Y, Madden CM, Bakthavatchalu V, Shen Z, Ge Z, Fox JG. 2016. Cytotoxic Escherichia coli strains encoding colibactin colonize laboratory mice. Microbes Infect 18:777–786. 10.1016/j.micinf.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giridharan VV, Simões LR, Dagostin VS, Generoso JS, Rezin GT, Florentino D, Muniz JP, Collodel A, Petronilho F, Quevedo J, Barichello T. 2017. Temporal changes of oxidative stress markers in Escherichia coli K1-induced experimental meningitis in a neonatal rat model. Neurosci Lett 653:288–295. 10.1016/j.neulet.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Gómez-Moreno R, Robledo IE, Baerga-Ortiz A. 2014. Direct detection and quantification of bacterial genes associated with inflammation in DNA isolated from stool. Adv Microbiol 4:1065–1075. 10.4236/aim.2014.415117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant MA, Weagant SD, Feng P. 2001. Glutamate decarboxylase genes as a prescreening marker for detection of pathogenic Escherichia coli groups. Appl Environ Microbiol 67:3110–3114. 10.1128/AEM.67.7.3110-3114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guignot J, Segura A, Tran Van Nhieu G. 2015. The serine protease EspC from enteropathogenic Escherichia coli regulates pore formation and cytotoxicity mediated by the type III secretion system. PLoS Pathog 11:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He X, Zeng Q, Puthiyakunnon S, Zeng Z, Yang W, Qiu J, Du L, Boddu S, Wu T, Cai D, Huang SH, Cao H. 2017. Lactobacillus rhamnosus GG supernatant enhance neonatal resistance to systemic Escherichia coli K1 infection by accelerating development of intestinal defense. Sci Rep 7:1–14 10.1038/srep43305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinenoya A, Shima K, Asakura M, Nishimura K, Tsukamoto T, Ooka T, Hayashi T, Ramamurthy T, Faruque SM, Yamasaki S. 2014. Molecular characterization of cytolethal distending toxin gene+ Escherichia coli from healthy cattle and swine in Nara, Japan. BMC Microbiol 14:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang YY, Wintner A, Seed PC, Brauns T, Gelfand JA, Hamblin MR. 2018. Antimicrobial photodynamic therapy mediated by methylene blue and potassium iodide to treat urinary tract infection in a female rat model. Sci Rep 8:1–9. 10.1038/s41598-018-25365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JR, Kaster N, Kuskowski MA, Ling GV. 2003. Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. J Clin Microbiol 41:337–345. 10.1128/JCM.41.1.337-345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JR, Stell AL, Delavari P, Murray AC, Kuskowski M, Gaastra W. 2001. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J Infect Dis 183:897–906. https://doi.org/10.1086/319263 [DOI] [PubMed] [Google Scholar]
- 39.Johnson TJ, Wannemuehler YM, Nolan LK. 2008. Evolution of the iss gene in Escherichia coli. Appl Environ Microbiol 74:2360–2369. 10.1128/AEM.02634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordan DM, Cornick N, Torres AG, Dean-Nystrom EA, Kaper JB, Moon HW. 2004. Long polar fimbriae contribute to colonization by Escherichia coli O157:H7 in vivo. Infect Immun 72:6168–6171. 10.1128/IAI.72.10.6168-6171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 42.Knöbl T, Micke Moreno A, Paixo R, Gomes TAT, Vieira MAM, Da Silva Leite D, Blanco JE, Ferreira AJP. 2012. Prevalence of avian pathogenic Escherichia coli (APEC) clone harboring sfa gene in Brazil. ScientificWorldJournal 2012:1–7. 10.1100/2012/437342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolmogorov M, Raney B, Paten B, Pham S. 2014. Ragout—a reference-assisted assembly tool for bacterial genomes. Bioinformatics 30:i302–i309. 10.1093/bioinformatics/btu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM. 2016. Diet–microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 64:982–992. 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landraud L, Gauthier M, Fosse T, Boquet P. 2000. Frequency of Escherichia coli strains producing the cytotoxic necrotizing factor (CNF1) in nosocomial urinary tract infections. Lett Appl Microbiol 30:213–216. 10.1046/j.1472-765x.2000.00698.x. [DOI] [PubMed] [Google Scholar]
- 46.Leser TD, Molbak L. 2009. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol 11:2194–2206. 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- 47.López-Banda DA, Carrillo-Casas EM, Leyva-Leyva M, Orozco-Hoyuela G, Manjarrez-Hernández ÁH, Arroyo-Escalante S, Moncada-Barrón D, Villanueva-Recillas S, Xicohtencatl-Cortes J, Hernández-Castro R. 2014. Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Biomed Res Int 2014:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynne AM, Skyberg JA, Logue CM, Nolan LK. 2007. Detection of Iss and Bor on the surface of Escherichia coli. J Appl Microbiol 102:660–666. 10.1111/j.1365-2672.2006.03133.x. [DOI] [PubMed] [Google Scholar]
- 49.Marini RP, Taylor NS, Liang AY, Knox KA, Peña JA, Schauer DB, Fox JG. 2004. Characterization of hemolytic Escherichia coli strains in ferrets: recognition of candidate virulence factor CNF1. J Clin Microbiol 42:5904–5908. 10.1128/JCM.42.12.5904-5908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marre R, Kreft B, Hacker J. 1990. Genetically engineered S and F1C fimbriae differ in their contribution to adherence of Escherichia coli to cultured renal tubular cells. Infect Immun 58:3434–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin HR, Taylor NS, Buckley EM, Marini RP, Patterson MM, Fox JG. 2009. Characterization of cytotoxic necrotizing factor 1-producing Escherichia coli strains from faeces of healthy macaques. J Med Microbiol 58:1354–1358. 10.1099/jmm.0.012088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massot M, Daubié AS, Clermont O, Jauréguy F, Couffignal C, Dahbi G, Mora A, Blanco J, Branger C, Mentré F, Eddi A, Picard B, Denamur E, The Coliville Group 2016. Phylogenetic, virulence and antibiotic resistance characteristics of commensal strain populations of Escherichia coli from community subjects in the Paris area in 2010 and evolution over 30 y. Microbiology 162:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCarthy AJ, Martin P, Cloup E, Stabler RA, Oswald E, Taylor PW. 2015. The genotoxin colibactin is a determinant of virulence in Escherichia coli K1 experimental neonatal systemic infection. Infect Immun 83:3704–3711. 10.1128/IAI.00716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miajlovic H, Smith SG. 2014. Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol Lett 354:1–9. 10.1111/1574-6968.12419. [DOI] [PubMed] [Google Scholar]
- 55.Micenková L, Bosák J, Štaudová B, Kohoutová D, Čejková D, Woznicová V, Vrba M, Ševčíková A, Bureš J, Šmajs D. 2016. Microcin determinants are associated with B2 phylogroup of human fecal Escherichia coli isolates. MicrobiologyOpen 5:490–498. 10.1002/mbo3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Micenková L, Štaudová B, Bosák J, Mikalová L, Littnerová S, Vrba M, Ševčíková A, Woznicová V, Šmajs D. 2014. Bacteriocin-encoding genes and ExPEC virulence determinants are associated in human fecal Escherichia coli strains. BMC Microbiol 14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 58.Pandey M, Khan A, Das SC, Sarkar B, Kahali S, Chakraborty S, Chattopadhyay S, Yamasaki S, Takeda Y, Nair GB, Ramamurthy T. 2003. Association of cytolethal distending toxin locus cdtB with enteropathogenic Escherichia coli isolated from patients with acute diarrhea in Calcutta, India. J Clin Microbiol 41:5277–5281. 10.1128/JCM.41.11.5277-5281.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Payros D, Secher T, Boury M, Brehin C, Ménard S, Salvador-Cartier C, Cuevas-Ramos G, Watrin C, Marcq I, Nougayrède JP, Dubois D, Bedu A, Garnier F, Clermont O, Denamur E, Plaisancié P, Theodorou V, Fioramonti J, Olier M, Oswald E. 2014. Maternally acquired genotoxic Escherichia coli alters offspring's intestinal homeostasis. Gut Microbes 5:313–325. 10.4161/gmic.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection? Infect Immun 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raisch J, Buc E, Bonnet M, Sauvanet P, Vazeille E, de Vallée A, Déchelotte P, Darcha C, Pezet D, Bonnet R, Bringer MA, Darfeuille-Michaud A. 2014. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol 20:6560–6572. 10.3748/wjg.v20.i21.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosadi F, Fiorentini C, Fabbri A. 2015. Bacterial protein toxins in human cancers. Pathog Dis 74:1–11. 10.1093/femspd/ftv105. [DOI] [PubMed] [Google Scholar]
- 63.Rosenberg S, Horowitz R, Coppenhagen-Glazer S, Pizov G, Elia A, Gofrit ON, Ginsburg I, Pode D. 2014. Intravesical administration of green tea extract attenuates the inflammatory response of bacterial cystitis—a rat model. BJU Int 114:601–607. 10.1111/bju.12544. [DOI] [PubMed] [Google Scholar]
- 64.Ross BN, Rojas-Lopez M, Cieza RJ, McWilliams BD, Torres AG. 2015. The role of long polar fimbriae in Escherichia coli O104:H4 adhesion and colonization. PLoS One 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Secher T, Brehin C, Oswald E. 2016. Early settlers: which E. coli strains do you not want at birth? Am J Physiol Gastrointest Liver Physiol 311:G123–G129. 10.1152/ajpgi.00091.2016. [DOI] [PubMed] [Google Scholar]
- 66.Shin S, Castanie-Cornet MP, Foster JW, Crawford JA, Brinkley C, Kaper JB. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator per. Mol Microbiol 41:1133–1150. 10.1046/j.1365-2958.2001.02570.x. [DOI] [PubMed] [Google Scholar]
- 67.Siqueira AK, Ribeiro MG, Leite D da S, Tiba MR, de Moura C, Lopes MD, Prestes NC, Salerno T, da Silva AV. 2009. Virulence factors in Escherichia coli strains isolated from urinary tract infection and pyometra cases and from feces of healthy dogs. Res Vet Sci 86:206–210. 10.1016/j.rvsc.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 68.Skowron B, Baranowska A, Kaszuba-Zwoińska J, Więcek G, Malska-Woźniak A, Heczko P, Strus M. 2017. Experimental model for acute kidney injury caused by uropathogenic Escherichia coli. Postepy Hig Med Dosw 71:520–529. 10.5604/01.3001.0010.3833. [DOI] [PubMed] [Google Scholar]
- 69.Taieb F, Petit C, Nougayrède J, Oswald E. 2016. The enterobacterial genotoxins: cytolethal distending toxin and colibactin. Ecosal Plus 7: 10.1128/ecosalplus.ESP-0008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 71.Torres AG, Blanco M, Valenzuela P, Slater TM, Patel SD, Dahbi G, López C, Barriga XF, Blanco JE, Gomes TAT, Vidal R, Blanco J. 2009. Genes related to long polar fimbriae of pathogenic Escherichia coli strains as reliable markers to identify virulent isolates. J Clin Microbiol 47:2442–2451. 10.1128/JCM.00566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tóth I, Hérault F, Beutin L, Oswald E. 2003. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J Clin Microbiol 41:4285–4291. 10.1128/JCM.41.9.4285-4291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tóth I, Oswald E, Mainil JG, Awad-Masalmeh M, Nagy B. 2000. Characterization of intestinal cnf1+ Escherichia coli from weaned pigs. Int J Med Microbiol 290:539–542. [DOI] [PubMed] [Google Scholar]
- 74.Trautman EP, Healy AR, Shine EE, Herzon SB, Crawford JM. 2017. Domain-targeted metabolomics delineates the heterocycle assembly steps of colibactin biosynthesis. J Am Chem Soc 139:4195–4201. 10.1021/jacs.7b00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner SM, Scott-Tucker A, Cooper LM, Henderson IR. 2006. Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol Lett 263:10–20. 10.1111/j.1574-6968.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 76.Veltri D, Wight MM, Crouch JA. 2016. SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res 44 W1:W41–W45. 10.1093/nar/gkw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vysakh A, Suma D, Jayesh K, Jyothis M, Latha MS. 2017. The influence of tissue antioxidant enzymes and inflammatory cascade in pathology of cystitis: an experimental rat model. Microb Pathog 113:102–106. 10.1016/j.micpath.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 78.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45 D1:D535–D542. 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Witcomb LA, Collins JW, McCarthy AJ, Frankel G, Taylor PW. 2015. Bioluminescent imaging reveals novel patterns of colonization and invasion in systemic Escherichia coli K1 experimental infection in the neonatal rat. Infect Immun 83:4528–4540. 10.1128/IAI.00953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon J, Cho HCs, Park C, Park BY, Kim YH, Min J. 2017. Efficacy of yeast vacuoles as antimicrobial agents to Escherichia coli bacteremia in rat. Curr Microbiol 74:22–27.doi:10.1007/s00284-016-1146-1 [DOI] [PubMed] [Google Scholar]
- 81.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng Q, He X, Puthiyakunnon S, Xiao H, Gong Z, Boddu S, Chen L, Tian H, Huang SH, Cao H. 2017. Probiotic mixture Golden Bifido prevents neonatal Escherichia coli K1 translocation via enhancing intestinal defense. Front Microbiol 8:1–13. 10.3389/fmicb.2017.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zha L, Wilson MR, Brotherton CA, Balskus EP. 2016. Characterization of polyketide synthase machinery from the pks island facilitates isolation of a candidate precolibactin. ACS Chem Biol 11:1287–1295. 10.1021/acschembio.6b00014. [DOI] [PubMed] [Google Scholar]