Abstract
Protein intrinsic disorder is an important characteristic demonstrated by the absence of higher order structure, and is commonly detected in multifunctional proteins encoded by RNA viruses. Intrinsically disordered regions (IDRs) of proteins exhibit high flexibility and solvent accessibility, which permit several distinct protein functions, including but not limited to binding of multiple partners and accessibility for post-translational modifications. IDR-containing viral proteins can therefore execute various functional roles to enable productive viral replication. Respiratory syncytial virus (RSV) is a globally circulating, non-segmented, negative sense (NNS) RNA virus that causes severe lower respiratory infections. In this study, we performed a comprehensive evaluation of predicted intrinsic disorder of the RSV proteome to better understand the functional role of RSV protein IDRs. We included 27 RSV strains to sample major RSV subtypes and genotypes, as well as geographic and temporal isolate differences. Several types of disorder predictions were applied to the RSV proteome, including per-residue (PONDR®-FIT and PONDR® VL-XT), binary (CH, CDF, CH–CDF), and disorder-based interactions (ANCHOR and MoRFpred). We classified RSV IDRs by size, frequency and function. Finally, we determined the functional implications of RSV IDRs by mapping predicted IDRs to known functional domains of each protein. Identification of RSV IDRs within functional domains improves our understanding of RSV pathogenesis in addition to providing potential therapeutic targets. Furthermore, this approach can be applied to other NNS viruses that encode essential multifunctional proteins for the elucidation of viral protein regions that can be manipulated for attenuation of viral replication.
Introduction
The order Mononegavirales encompasses the spectrum of non-segmented, negative sense (NNS) RNA viruses. NNS viruses are characterized by a single-stranded RNA genome ranging from 15 to 19 kb in length, encoding a linear array of genes in antisense orientation to mRNA. The genomic organization of the Mononegavirales is similar and the gene products share sequence and functional homology. Among the NNS viruses are notable viruses such as measles, rabies, and Ebola viruses. There are several families within the Mononegavirales order. In particular, the Paramyxoviridae consists of a large array of human pathogens, including measles, mumps, Nipah, parainfluenza viruses 1–4, and respiratory syncytial virus (RSV).1 Human RSV is the prototype member of the Pneumovirus genus within the Pneumovirinae subfamily of paramyxoviruses.1,2
RSV is a global threat, causing severe lower respiratory tract infections in infants and young children. RSV is also a significant health problem in the elderly and immunocompromised patients. Unlike most paramyxoviruses, RSV commonly causes recurrent infection in healthy, immunocompetent individuals, resulting in mild, self-limiting upper respiratory tract disease. This is likely due to the induction of a short-lived anti-RSV adaptive immune response and poor memory to infection.3–5 However, RSV interaction with the host and the subsequent immune response to RSV are poorly understood.
RSV is divided into two subtypes, RSV A and RSV B, which are based on the antigenicity of the RSV attachment (G) and fusion (F) surface glycoproteins. These RSV subtypes co-circulate globally at relatively equal levels, although RSV A is slightly more prevalent and pathogenic.6 In addition, RSV subtypes will reappear consistently throughout the year, as opposed to other respiratory diseases such as influenza that circulate annually before being replaced by a variant strain.7–10 Until the late 2000’s there were few whole RSV genome sequences published and available for reference. Due to recent technological advances in whole genome sequencing, as well the use of the highly variable regions of the RSV surface glycoprotein G as a marker for diagnostics and genotyping, we have a much greater knowledge of the RSV phylogenetic tree.11–15 The RSV A subtype contains 9 known genotypes, with the two largest clades GA2 and GA5 encompassing most of the RSV A genotypes. The RSV B subtype consists of 10 known genotypes; interestingly, the RSV BA genotype first emerged in the 1990’s and has rapidly become the predominant B genotype globally.16–19
RSV is an enveloped virus containing a 15 kb NNS RNA genome that is replicated exclusively in the cytoplasm in the infected cell. The viral genome consists of a linear array of 10 genes, encoding 11 proteins, with cis-acting transcription initiation and transcription termination sequences flanking each gene. The 11 RSV proteins are categorized as structural when packaged within the virion, or non-structural if expressed solely within the infected cell. Most of the structural proteins of RSV have functional homologues among the paramyxoviruses. As with other paramyxoviruses, the RSV ribonucleoprotein (RNP) consists of the viral genome encapsidated by the nucleo-protein (N) to form the template for viral RNA synthesis while simultaneously protecting the RNA from cellular nucleases.20 The viral polymerase (L) protein encodes the enzymatic activities necessary for RNA synthesis and requires a cofactor, the phosphoprotein (P), which is essential for L interaction with the RNP and polymerase function.21–24 Unique to the Pneumovirinae, the viral polymerase complex is regulated by two small proteins encoded by the M2 gene, M2-1 and M2-2. M2-1 functions as an antitermination factor to allow for elongation of mRNA transcripts; M2-2 appears to act as a molecular switch for the polymerase to toggle between transcription and genome replication modes.24–28 The RSV matrix (M) protein is required for assembly of progeny virions, directing the RNP–polymerase complexes to the plasma membrane for budding from the cell.29–31 RSV encodes three surface glycoproteins, the small hydrophobic (SH), attachment (G), and fusion (F) proteins.32 RSV F is structurally and functionally homologous to other paramyxovirus F proteins, responsible for fusion of the viral and target cell membranes.33 G and SH are not structurally similar to their counterparts in other paramyxoviruses; however, all paramyxoviruses express an attachment protein while SH proteins are present in other subfamilies of Paramyxoviridae. The RSV nonstructural proteins, NS1 and NS2, are the first proteins expressed during infection and function primarily in blocking the innate immune response to allow for optimal viral transcription and replication.34–37 Their sequence does not share homology with any mammalian or viral proteins, and little is understood about their structure.
Small RNA viruses such as RSV encode relatively few viral proteins with which to carry out key life cycle events like RNA transcription and viral replication. In order to thrive in an intracellular environment, these viruses have evolved proteins that perform multiple functions. These multifunctional proteins often contain intrinsically disordered regions (IDRs) that are highly flexible to facilitate a diverse array of functions, such as transient protein–protein interactions, signal transduction, post-translational modifications, and other crucial biological roles.38,39 In general, IDRs are defined by unique physicochemical features such as low hydrophobicity, low sequence complexity, and high net charge.40–42 Using both computational and experimental techniques, several studies have shown that IDRs are prominent and play important roles in viral systems.43,44 Among other functions, IDRs allow for promiscuous binding between the various components of the host, including membranes, DNA, RNA, and protein. Additionally, it has been proposed that IDRs allow for higher tolerance of the rapid mutation that occurs in viral genomes, which is in the range of 10−5–10−3 mutations per position per generation for RNA viruses. Due to the numerous functional applications of intrinsic protein disorder, any particular functional advantage conferred by protein disorder can be determined by examining the degree of sequence conservation, or the location and frequency of polymorphisms within an IDR.45
For the Paramyxoviridae, the relevance of intrinsic disorder in viral function has already been described in the specific cases of the N and P proteins of Nipah (NiV), Hendra (HeV), and measles (MeV) viruses.46–55 The C-terminus of the N protein (NTAIL) consists of a region of disorder-to-order transition, also known as a molecular recognition feature (MoRF). This NTAIL MoRF binds to the C-terminal X domain of the P protein in a “fuzzy” fashion, a term which describes significant freedom of both bound and unbound states. In agreement with this paradigm, the NTAIL of MeV N protein also interacts with the folded RNA-binding domain of N (NCORE), providing a framework where IDRs regulate the assembly and positioning of the polymerase complex.
As intrinsic disorder is prominent in negative-strand RNA viral proteomes, and several IDRs have been identified which are important to viral function, understanding the level of intrinsic disorder in RSV may allow for the identification of viral targets for antiviral therapeutics and an avenue to better defining the interaction of RSV with the immune system. To determine key elements of RSV protein structure, we implemented several bioinformatics predictors of intrinsically disordered protein regions (IDRs) to evaluate the degree and location of intrinsic disorder among the 11 RSV proteins. We compared proteins from 27 fully sequenced RSV genomes across various genotypes within the two RSV subtypes to determine a potential relationship between intrinsic disorder status and protein function. We also evaluated the existence of polymorphisms within IDRs to expand on our assessment of functionally relevant, conserved RSV protein sequence. While traditional drug design targets structural regions required for protein function, our analysis instead reveals functional domains within intrinsically disordered regions of RSV proteins. In addition, mutation or deletion of these IDRs that have specific function may alter RSV infectivity and associated immune responses without affecting protein stability and expression, allowing for rational design of live-attenuated RSV vaccine candidates.
Materials and methods
Dataset
The 27 RSV clinical isolates used in this study are fully sequenced and reviewed.16 The two RSV subtypes, A and B, are represented by 17 RSV A and 10 RSV B genomes, including the classically referenced RSV A2 and B1 laboratory strains. Included are isolates from four RSV A genotypes – GA1, GA2, GA5 and ON1. RSV B isolates are from one of six genotypes – GB1, GB3, GB4, BA, BA2 and BA4. Accession IDs were collected from the National Center for Biotechnology Information (NCBI) GenBank and FASTA files were obtained for each RSV protein from Uniprot. GenBank accession IDs: M74568, KF826836, KF826846, KF826824, KF826847, KF826832, JQ901451, KC731482, KC731483, KF826848, KF826855, KF826821, KF826838, KF826840, KF826831, JX015499, JX015483, AY353550, AF013254, KF826853, JQ582844, KF826829, KF826845, KF530259, KF826851, KF826858, JX576761. Genotypes were chosen from several RSV phylogenetic clades based on predominance in current circulation patterns. Isolates were collected between 1961 and 2011 to include RSV global sequence divergence from the past 50 years in our analysis. We have also included an extensive sampling of locations throughout the world (Table 1).
Table 1.
RSV clinical isolates in this study. Clinical isolates were collected from the NCBI GenBank, with GenBank accession number shown
| GenBank Acc | Subtype | Genotype | Location | Collection date |
|---|---|---|---|---|
| M74568 | A | GA1 | Australia | 1961 |
| KF826836 | A | GA5 | Mexico | 2006 |
| KF826846 | A | GA5 | Argentina | 2008 |
| KF826824 | A | GA5 | USA | 1998 |
| KF826847 | A | GA5 | Australia | 2007 |
| KF826832 | A | GA5 | Italy | 2009 |
| JQ901451 | A | GA5 | Netherlands | 2001 |
| KC731482 | A | ON1 | India | 2011 |
| KC731483 | A | GA2 | India | 2011 |
| KF826848 | A | GA2 | Australia | 2007 |
| KF826855 | A | GA2 | Italy | 2009 |
| KF826821 | A | GA2 | USA | 2007 |
| KF826838 | A | GA2 | Argentina | 2006 |
| KF826840 | A | GA2 | Mexico | 2007 |
| KF826831 | A | GA2 | Germany | 2009 |
| JX015499 | A | GA2 | Belgium | 2008 |
| JX015483 | A | GA2 | Netherlands | 2008 |
| AY353550 | B | GB1 | USA | 1977 |
| AF013254 | B | GB4 | USA | 1985 |
| KF826853 | B | GB3 | Germany | 2008 |
| JQ582844 | B | BA2 | USA | 2002 |
| KF826829 | B | BA | Mexico | 2005 |
| KF826845 | B | BA | Argentina | 2008 |
| KF530259 | B | BA | South Africa | 2006 |
| KF826851 | B | BA | USA | 2007 |
| KF826858 | B | BA | Italy | 2009 |
| JX576761 | B | BA4 | Netherlands | 2002 |
RSV isolates range from 15 106 bp to 15 283 bp, depending on the genotype, with proteins ranging from 64 to 2166 amino acids (aa). Figures depicting averaged RSV A and RSV B data include the sequences from all 17 A and 10 B genomes. For the G protein, genotypes ON1 (one isolate) and BA (averaged five isolates) are represented separately from all other averaged RSV A and B genotypes. With 11 RSV proteins per isolate, there are a total of 297 proteins for classifications of IDRs by functional domain and post-translational modification.
Diversity analysis
The RSV proteomes of each isolate were concatenated to yield 27 strings, each containing the 11 RSV proteins. These strings were then aligned using a PAM matrix using the ClustalW function in the MEGA 7.0 software (default parameters).56 The A and B isolates were then separated into two separate alignment files, and the entropy (H(x)) plot function in BioEdit 7.0 was used to produce the plot values.
Per-residue analysis of RSV proteins
In order to predict level of disorder in single sequences, per-residue disorder predictors will be used. Different predictors take different sequence characteristics into account, but all consider scores above 0.5 to be disordered, whereas scores below 0.5 are considered ordered. Two predictors were primarily used. While PONDR® VL-XT57 sacrifices accuracy compared to more recent predictors, it is useful for detection of potential interaction regions because it is sensitive to local compositional biases.58,59 For example, IDRs are known to assume secondary structures known as molecular recognition features (MoRFs) when binding their partners, which are often represented in PONDR® VL-XT plots as sharp dips from disorder to order, and back to disorder.59,60 PONDR®-FIT61 is a highly accurate meta-predictor that utilizes PONDR® VL-XT as an input, as well as PONDR®-VSL2,62 PONDR®-VL3,63 FoldIndex,64 IUPred,65 and TopIDP.66 As it utilizes several different input features for prediction, it achieves ~85% accuracy, which is more accurate than the individual predictors. Throughout the study, PONDR® VL-XT is generally used to correlate IDRs to potential functional regions such as MoRFs, whereas PONDR®-FIT is generally used to understand overall level of intrinsic disorder.
Analysis of consensus intrinsic disorder and putative interaction regions of a representative RSV strain
The proteome of RSV strain A2 (UniProt IDs: P04544, P04543, P03418, P03421, P03419, P04852, P03423, P03420, P04545, P88812, P28887) was analyzed for consensus disorder using the MobiDB database, where consensus disorder is defined as incorporating data from multiple data sources including X-ray/NMR structures and intrinsic disorder predictors.67 Regions are classified as either ordered, disordered, or ‘ambiguous’ when different sources disagree regarding levels of intrinsic disorder. In order to quantitatively evaluate regions of potential interactions to support PONDR® VL-XT analysis, the proteome of RSV strain A2 was evaluated using ANCHOR, which determines regions unlikely to fold independently but potentially can in the presence of a partner,65,68 and MoRFpred, a predictor which can identify multiple MoRF types (α, β, coil, and complex).69
Charge-hydropathy (CH) plot
One established binary method of order–disorder classification is the CH plot, where ordered and disordered proteins plotted in charge-hydropathy space can be separated by a linear boundary.42 Absolute mean net charge for each protein was determined by calculating the total amount of charged amino acids (Lys, Arg, Asp, Glu), then dividing by the total number of residues to obtain the average charge per residue. Histidines were excluded as these residues are highly ionizable at physiological pH. Kyte-Doolittle hydropathy was calculated for each protein using a sliding window of 5 amino acids.70 The disorder/order boundary line is a modified, optimized version based on the original by Uversky and colleagues, represented by the equation ⟨charge⟩ = 2.743⟨hydropathy⟩ – 1.109.42,71 The boundary margins of the line were set to ±0.045, which reaches accuracy up to 95% for disordered proteins and 97% for ordered proteins.
Cumulative distribution function (CDF)
CDF is a binary analysis based on per-residue local sequence predictions,72 and in this case PONDR® VL-XT scores were used. Disorder scores were plotted against their cumulative frequency. The resulting distributions were classified based on the distance from a previously validated linear boundary, which is a measurement of the proportion of residues with high vs. low disorder scores. The x, y coordinates of the boundary line are 0.60, 0.6948; 0.65, 0.7323; 0.70, 0.7736; 0.75, 0.8141; 0.80, 0.8538; 0.85, 0.9051; 0.90, 0.9467. CDF curves above and below the boundary represent ordered and disordered, respectively, while curves that cross the boundary line are predicted to be a mixture of order and disorder.
Combined CH–CDF plot
While CH and CDF analyses are valuable separately, combination of these can yield even more information about the native state of a protein and roughly classify proteins as structured (Q2, lower right), mixture of order and disorder (Q3, lower left), disordered (Q4, upper left), and rare (Q1, upper right).73 Values for CH–CDF analysis were determined by calculating the distance between the selected point and the CH or CDF boundary line.
Graphics software
Clustal Omega (EMBL-EBI, ebi.ac.uk) was used for multiple sequence alignment for determination of amino acid polymorphisms. Illustrator for Biological Sequences (IBS) 1.0 and Adobe Illustrator CS6 were used for domain mapping. GraphPad Prism 6.0 was used for PONDR®-FIT and PONDR® VL-XT graphs. PyMOL was used for imaging of molecular structures (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Results and discussion
Intrinsic disorder of the respiratory syncytial virus proteome
We collected 27 fully sequenced RSV clinical isolates, 17 from the RSV subtype A and 10 from the RSV subtype B. Of the RSV A isolates, nine were from the GA2 genotype, six from the GA5 genotype, and one isolate from the ON1 genotype. In addition, we included the prototype A2 laboratory strain, which is of the GA1 genotype. The RSV B isolates include one GB3, one GB4, one BA2, one BA4, and five BA genotype isolates. Similarly, we included the RSV B prototype B1 laboratory strain, which is genotype GB1. All of the isolates have been reviewed and can be found in the NCBI GenBank database. Table 1 describes the 27 clinical isolates used in this study.
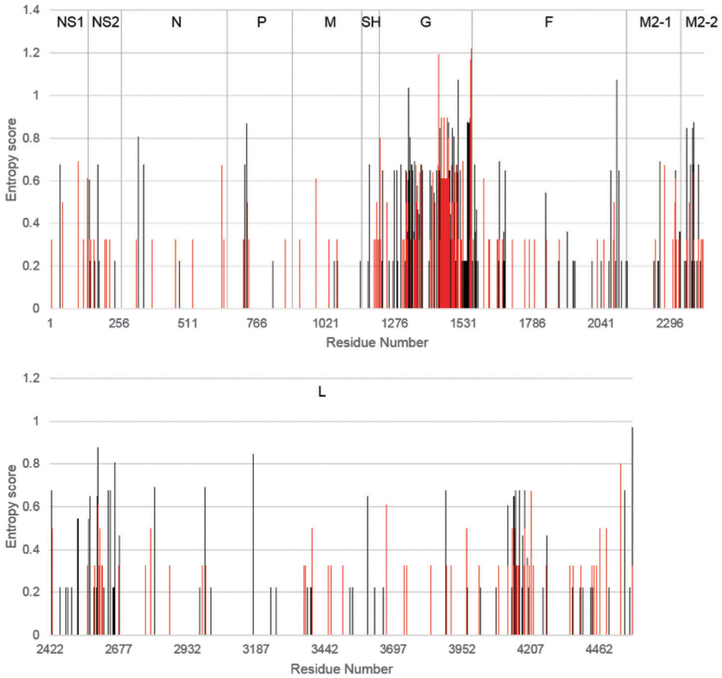
We strategically selected clinical isolates whose collection dates encompass the last fifty years to account for any sequence diversity over the past half-century. Furthermore, we chose isolates with an altogether worldwide distribution to include any global sequence diversity among samples. Despite these sampling methods, we performed diversity analysis on both RSV A and B isolates to validate a high degree of variation within selected samples, shown in Fig. 1. Our diversity analysis confirmed our sampling methods as effective, and was generally in agreement with previously published research, showing the G protein as expressing high levels of polymorphism relative to other RSV proteins.16
Fig. 1.
Protein entropy plots of 27 RSV proteomes. Entropy plots of concatenated and aligned proteomes of 17 RSVA isolates (red) and 10 RSVB isolates (black). Values were calculated using the entropy H(x) function in BioEdit 7.0. Entropy value (y-axis) is directly correlated to positional variation.
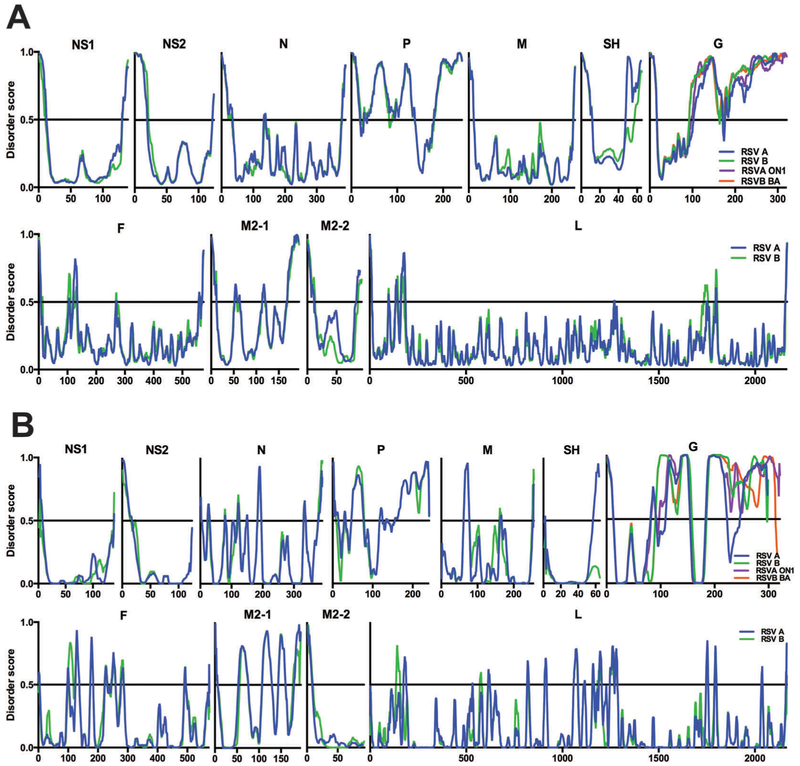
Disorder predictions for each RSV protein are shown in Fig. 2. PONDR®-FIT and PONDR® VL-XT disorder predictor tools consider an amino acid residue to be disordered if its disorder score is >0.5 and ordered if its score is <0.5, simplified by the horizontal line at the y-axis 0.5 traversing each graph. Intrinsic disorder regions (IDRs) are, by definition, four or more consecutive residues with disorder scores above 0.5. Peaks and valleys represent the degree of disorder and order, respectively. The proteins are organized in the order in which they are transcribed, and roughly scaled to represent the size of their amino acid sequence. The 17 RSV A genotypes (blue line) and 10 RSV B genotypes (green line) were averaged and compared for each RSV protein. Certain G protein genotypes contain a 24aa (RSV A) or 20aa (RSV B) duplication in their C-terminal sequence; these sequences are therefore shown separately to avoid distorting the graph in the C-terminal direction. In addition, we compared the IDR differences between the more recently isolated ON1 and BA genotypes and the remaining RSV A and RSV B genotypes, respectively. The RSV A ON1 genotype (one isolate) is shown in purple and the RSV B BA genotype (five averaged isolates) is shown in orange.
Fig. 2.
Intrinsic disorder of the RSV proteome. RSV proteome map of averaged PONDR®-FIT data (A) and PONDR® VL-XT data (B) for RSV A (blue) and B (green) subtypes. The G protein ON1 (purple) and BA (orange) genotypes are shown separately. Disorder score (disordered >0.5, ordered <0.5) on the y-axis and amino acid residue position on the x-axis.
PONDR®-FIT predictions for the RSV proteome are shown in Fig. 2A. For each protein, RSV A and B subtypes generally display a similar pattern of intrinsic disorder throughout the amino acid sequence, with a few minor differences. The SH protein RSV A isolates have highly disordered C-terminal peaks that remain disordered throughout the C-terminus. The SH RSV B isolates’ C-terminal peaks remain ordered, and the C-terminal IDRs are much shorter. Other minor variances involve the short IDR peaks in the RSV B proteins F and L near the N- and C-termini, respectively, that are present in their RSV A equivalents but as smaller, more ordered peaks.
Fig. 2B shows PONDR® VL-XT predictions for the RSV proteome. As we observed in the PONDR®-FIT predictions, there is an overall comparable trend between RSV A and RSV B. However, there are major differences between the two subtypes, namely in the SH and G graphs. For the SH protein, the RSV A isolates maintain the C-terminal IDRs displayed in the PONDR®-FIT data while the RSV B isolates remain highly ordered throughout the entirety of the protein sequence. The G graph for RSV A isolates contains two separate ordered regions near amino acid positions 100 and 220 whereas the parallel region of the RSV B G graph maintains its disordered status throughout those regions. The ON1 and BA isolate C-terminal disordered regions are not only different from one another, but they also display different C-terminal disorder patterns than their respective RSV A and B predecessor genotypes. There are noticeable differences in IDR prediction for each RSV protein between the two bioinformatics tools, many of which highlight the purpose of PONDR® VL-XT predictions. Many RSV proteins – N, M, SH, G, F, M2-1, and L – reveal an enhanced disorder profile, or higher peaks and lower valleys (Fig. 2A and B).
Overall disorder level for each RSV protein was calculated using the PONDR® VL-XT predictions (Table 2). Proteins were classified as either disordered (>15% disordered) or ordered (<15% disordered). Ordered proteins of the RSV proteome include NS1, NS2, N, M, SH, F, M2-2 and L. Proteins P, G and M2-1 are disordered, which is apparent in Fig. 2. All four G protein genotypes display a high degree of disorder, however ON1 and BA isolates containing the C-terminal duplication display an increased degree of disorder overall, compared to their RSV A and B counterparts.
Table 2.
Level of RSV protein intrinsic disorder. PONDR® VL-XT predictions for 17 RSV A and 10 RSV B isolates were averaged for each RSV protein, with ON1 and BA isolates shown separately for G. RSV proteins were classified as ordered (<15%) or disordered (>15%, bold) by percentage of total amino acids residues above the disorder score of 0.5
| RSV | NS1 (%) | NS2 (%) | N (%) | P (%) | M (%) | SH (%) | G (%) | F (%) | M2-1 (%) | M2-2 (%) | L (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 5.7 | 13.7 | 14.6 | 59.3 | 9.0 | 12.5 | 53.4 | 12.0 | 42.8 | 8.9 | 7.6 |
| B | 2.1 | 12.0 | 14.3 | 58.1 | 11 | 0.0 | 64.8 | 15.0 | 41.5 | 9.7 | 7.2 |
| ON1 | 76.0 | ||||||||||
| BA | 65.2 |
In order to gain the most conservative and accurate view of the level of intrinsic disorder in the RSV protein, we analyzed a representative proteome of RSV strain A2 using the ‘consensus’ prediction of the MobiDB database (Table 3). As expected, the consensus predicted values generally fluctuated from the PONDR® VL-XT predictions, likely because of its relatively low accuracy and propensity to predict MoRFs. The structural data generally agreed with the predictions, with the exception of the F protein, which had an X-ray structure with much higher intrinsic disorder than the predictions would suggest. Potential reasons for this discrepancy will be discussed further in subsequent sections.
Table 3.
Consensus disorder predictions of RSV strain A2. The MobiDB database was used to determine the consensus disorder prediction of RSV strain A2 (UniProt IDs: P04544, P04543, P03418, P03421, P03419, P04852, P03423, P03420, P04545, P88812, P28887), where regions of disagreement between two structure determination methods are defined as ‘ambiguous’ rather than structured or unstructured. Bold represents the overall consensus disorder prediction per protein
| RSV A2 | NS1 | NS2 | N | P | M | SH | G | F | M2-1 | M2-2 | L |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Predict | 6.47 | 11.29 | 5.88 | 66.39 | 3.91 | 17.19 | 60.07 | 2.61 | 14.43 | 4.4 | 2.4 |
| NMR | 0.0 | 10.82 | |||||||||
| X-ray | 3.07 | 0.78 | 19.34 | 10.31 |
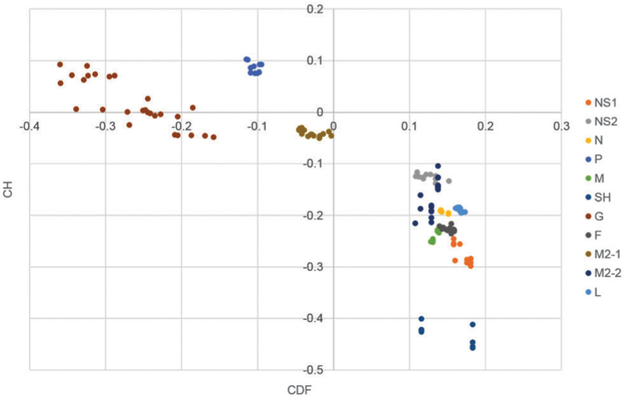
To expand the analysis of overall intrinsic disorder of the RSV proteome, binary disorder predictors of intrinsic disorder were used on the proteomes of the 27 isolates, thereby depicting all 297 proteins individually (Fig. 3). Charge-hydropathy analysis revealed that all proteins besides G, M2-1, and P clustered on the ordered side of the modified Uversky line. P was the only cluster to fall in the definitively disordered region, while G and M2-1 clustered within the line’s range of error, representing a mixture of order and disorder. Cumulative distribution function (CDF) analysis revealed similar results as CH analysis, with the notable exception of the M2-1 cluster, which was predicted as structured. Combined CH–CDF analysis revealed that M2-1 is clustered close to the center in the unusual quadrant, which is consistent with its high degree of variability in its disorder profile. P is clustered in the highly disordered, while G protein had very high degrees of variability, with several instances either surrounding or on the border of nonstructured and unusual. All other proteins were predicted as structured by CH–CDF analysis.
Fig. 3.
Combined CH–CDF analysis 27 RSV proteomes. Q1 (upper right) contains proteins predicted as structured by CDF, and unstructured by CH (unusual). Q2 (lower right) contains proteins predicted as structured by CDF, and structured by CH (ordered). Q3 (lower left) contains proteins predicted as unstructured by CDF, and structured by CH (mix). Q4 (upper left) contains proteins predicted as unstructured by CDF, and unstructured by CH (disordered). CH values were calculated by taking the vertical distance between the point and the modified Uversky line. CDF values were calculated by taking the average distance between the CDF line and the boundary line.
Classification of intrinsic disorder regions of the RSV proteome
To further understand the prevalence and function of identified IDRs in RSV proteins, we grouped the IDRs using the PONDR®-FIT predictions from each protein of all 27 isolates based on size. The size groupings, derived from a previous publication, are shown as colored bars ranging from 4–10aa to 91–300aa (Fig. 4A).74 The RSV proteins are arranged on the x-axis in genomic order. In addition to IDR size, the number of IDRs within that size group for each RSV protein is shown on the y-axis. The shorter IDRs in the 4–10aa and 11–20aa range are more abundant overall, and each appears in every protein except NS2.
Fig. 4.
Number and size of intrinsically disordered regions of RSV proteins. Number of IDRs shown on the y-axis. (A) On the x-axis, IDR size for each RSV protein in genomic order, with light blue as shortest IDRs and maroon as longest IDRs. (B) On the x-axis, each functional classification in order of most IDRs to least, with light blue for shortest IDRs and dark blue as the longest IDRs.
Every IDR was also classified by functional domain, including post-translational modifications with implications in protein function. In Fig. 4B, the number of IDRs are once more on the y-axis, and different functional classifications are arranged along the x-axis in order of high to low abundance within the RSV proteome. The colored bars depict IDR size groupings ranging from 5–10aa to 101–300aa. As above, phosphorylation and glycosylation events appear to frequently occur within longer IDRs. Unsurprisingly, protein-binding domains contained the most IDRs of all of the functional categories, as intrinsic disorder is known to facilitate multivalent, promiscuous interactions. This is also likely due to the large number of known RSV protein binding domains, including those with either cellular or viral binding partners. Notably, a common mechanism by which viruses exploit host machinery during infection is by expressing short linear motifs (SLiMs) of host proteins, which often reside within viral protein IDRs, to mimic cellular proteins and thereby interact with their functional counterparts.75 Using the eukaryotic linear motif (ELM) database (http://elm.eu.org), we found numerous cellular protein binding ELMs present within the NS2 protein N-terminal IDR (data not shown), indicative of ELM-expressing IDRs as potential sites of NS2 activity.76
One notable advantage of having a large number of complete RSV clinical isolate sequences available is the ease at which protein sequences can be aligned to determine the positions of amino acid polymorphisms across a diverse sampling of RSV strains. Table 4 reports each polymorphism identified for all RSV proteins within our 17 RSV A and 10 RSV B isolates examined in this study. IDRs containing polymorphisms are considered less functionally relevant due to a lower degree of conservation within that region of the protein. Importantly, targeting these polymorphism-containing IDRs for the purposes of therapeutic development would be less effective than targeting protein sequence exhibiting a high degree of conservation, therefore it is important to identify polymorphisms that fall within IDRs (italicized in Table 4). The majority of the polymorphic residues were outside of predicted IDRs; however, the polymorphisms in two proteins were largely, or exclusively, in IDRs. For the phosphoprotein (P), the polymorphisms are clustered in regions that have no known function, mostly in the N-terminal region (see below). Unsurprisingly, the attachment protein (G) is the most polymorphic protein in RSV, as it is one of the major antigens and is under constant immune selection. The G polymorphisms lie within the IDRs and thus would be considered poor candidates for therapeutic targeting.
Table 4.
RSV A and B protein polymorphisms. The Clustal Omega multiple sequence alignment tool was used for determination of amino acid polymorphisms within the 17 RSV A and 10 RSV B isolates sampled in this study. Polymorphisms within IDRs are italicized
| RSV A | RSV B | |
|---|---|---|
| NS1 | 5, 36, 105 | 6, 45, 105, 124, 139 |
| NS2 | 6, 7, 8, 10, 25, 38, 43, 100 | 6, 10, 24, 63, 68, 82 |
| N | 64, 84, 216 | 57, 115, 201, 264, 372, 380 |
| P | 66, 73, 75, 171 | 61–63, 75, 77, 80, 205 |
| M | 157, 168, 254 | 28, 136, 166 |
| SH | 27, 31 | 2, 49, 53, 57, 61, 65 |
| G | 4, 13, 15, 38, 52, 57, 67, 71, 81, 95, 99–102, 104, 106–108, 110–111, 113, 117, 120, 121–127, 131, 133, 136, 140–142, 146, 151, 153, 156–157, 160–161, 187, 191, 196, 205–206, 208, 215, 222–223, 225–226, 232–233, 236–238, 241, 244, 250–254, 256, 258, 262, 265, 269–271, 273–274, 279–280, 286, 289–290, 292–293, 295–297 | 4, 6, 32, 89, 95, 101, 103, 107, 109, 118, 131, 133, 135, 137–138, 140, 143, 150, 152, 158–159, 191, 200, 207–208, 219, 222–223, 227, 229, 233–235, 238–239, 268, 271–272, 276, 278–279, 282, 288, 290, 292–294, 301–302, 305, 307 |
| F | 2, 4, 8, 13, 15–17, 19–20, 25, 101–103, 105, 119, 122, 124–125, 127, 152, 276, 324, 356, 378–380, 384, 447, 482, 510, 518, 535, 540, 547, 555, 574 | 8, 45, 65, 67, 97, 100, 103, 117, 152, 197, 215, 234, 278, 326, 467, 490, 527, 529 |
| M2-1 | 4, 105, 118, 125, 180, 182, 185 | 107, 142, 172, 179–180, 185, 181–182 |
| M2-2 | 18–19, 23, 25, 39, 44, 48, 50, 52, 54, 64, 68–70, 77–80 | 28–29, 31, 38, 52, 56, 71, 82, 86, 88 |
| L | 6, 37, 59, 65, 81, 102–104, 144, 148, 162, 173–174, 177, 200, 216, 224, 232, 234, 236–237, 240, 257, 388, 445, 555, 575, 598, 754, 821, 839, 955, 967, 970, 1113–1114, 1124, 1180, 1206, 1238, 1471, 1489, 1551, 1599, 1657, 1700, 1715, 1718, 1721, 1723–1725, 1730–1731, 1745, 1754–1757, 1761, 1764, 1773, 1778, 1832, 1847, 1940, 1969, 1980, 2009, 2016, 2076, 2120, 2135, 2154, 2163 | 8, 138, 165–166, 177, 183–184, 191, 195, 254, 354, 374, 445, 564, 578, 943, 948, 973, 1032, 1043, 1087, 1250, 1314, 1325, 1413, 1471, 1489, 1546, 1593, 1667, 1700, 1716, 1718, 1723, 1726–1727, 1731, 1735, 1740, 1742, 1760, 1764, 1773, 1780, 1787, 1794, 1843, 1930, 1942–1943, 1972, 2014, 2021, 2030, 2042, 2065, 2119, 2164 |
Functional analysis of RSV protein intrinsic disorder predictions
Nonstructural proteins NS1 and NS2.
NS1 and NS2 accessory proteins are expressed solely within infected cells and while their repertoire of functions are not yet fully understood, it is clear both nonstructural proteins play a major role in antagonism of type I interferon (IFN-α/β) via several innate immune response pathways.77 NS1 contains a BC box consensus sequence for binding to the cellular protein Elongin C at aa22–29, an adaptor protein subunit of E3 ubiquitin ligases.78,79 NS1 also contains a putative binding domain for Cul2, a member of the cullin family of E3 ligase scaffolding proteins, identified by comparison of NS1 amino acid sequence with those of other Cul2-binding proteins. This putative binding sequence is likely required for NS1 association with Cul2.78 These two cellular proteins are subunits of an E3 ligase complex that polyubiquitinates signal transducer and activator of transcription 2 (STAT2), an essential transcription factor required in the type I and type III IFN signaling pathways, and targets STAT2 for proteasomal degradation during RSV infection. Expression of NS1 enhances proteasomal degradation of STAT2, an activity induced primarily by NS2.78,80 It is possible NS1 interacts with Elongin C and Cul2 E3 ligase components to aid in inhibition of IFN signaling via STAT2. However, mutation of NS1’s Elongin C-binding domain destabilized NS1, suggesting these residues may also play a structural role.79 Unlike NS1, NS2 does not contain direct binding domains for E3 ligase proteins, although its C-terminus is essential for STAT2 degradation, indicating NS1–NS2 form a multi-subunit STAT2-degradation complex.35,78,81
Upstream of STAT2 degradation, NS1 and NS2 alter the expression levels of two cellular signaling proteins, inhibitor of nuclear factor kappa-B kinase subunit epsilon (IKKε) and tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3), which are important for IFN-α/β production. While it is unclear the precise mechanisms by which NS1 and NS2 affect IKKε or TRAF3 activity, it is apparent that core of each NS1 and NS2 is essential. The 10 C-terminal and 20 N-terminal amino acids are disposable for NS1 and NS2 interaction with IKKε, respectively. This is consistent with the disorder predictions, where these disposable regions are predicted to be relatively flexible and are therefore not required for stable interactions. NS1 interaction with TRAF3 excludes the 20 N-terminal and C-terminal amino acids, whereas NS2 interaction with TRAF3 requires aa21–94.81,82 Residues aa64–65 are predicted as a potential MoRF (Table 5), and therefore may be an interesting target within this region for TRAF3 interaction.
Table 5.
Potential interaction sites within regions of intrinsic disorder. The ANCHOR and MoRFpred algorithms were applied to the proteome of RSV strain A2, and the list of potential interaction residues and the percentage of potential interaction sites are displayed. Bolded residues in the ANCHOR predictions represent regions of high confidence
| Protein | UniProt ID |
ANCHOR | % ANCHOR |
MoRFpred | % MoRFpred |
|---|---|---|---|---|---|
| NS1 | P04544 | 11–14 | 5.76 | ||
| 131–132 | |||||
| 135, 138 | |||||
| NS2 | P04543 | 12 | 8.87 | ||
| 64–65 | |||||
| 116–123 | |||||
| N | P03418 | 47–51 | 3.32 | 9–13 | 3.32 |
| 159–166 | 162–166 | ||||
| 245, 368, 391 | |||||
| P | P03421 | 1–8 | 40.66 | 12, 14, 18, 22 | 14.52 |
| 18–27 | 44–47 | ||||
| 39–51 | 60 | ||||
| 60–63 | 62–64 | ||||
| 79–87 | 81–85 | ||||
| 98–108 | 100–107 | ||||
| 118–121 | 220–222 | ||||
| 141–156 | 235–241 | ||||
| 170–175 | |||||
| 192–192 | |||||
| 220–228 | |||||
| 235–241 | |||||
| M | P03419 | ||||
| SH | P04852 | 11–15 | 7.81 | ||
| G | P03423 | 111–116 | 24.83 | 19 | 6.38 |
| 162–190 | 112–113 | ||||
| 239–255 | 164–171 | ||||
| 257–269 | 242 | ||||
| 278–286 | 279–284 | ||||
| 298 | |||||
| F | P03420 | 139–141 | 0.52 | 16, 18, 58 | 2.79 |
| 113–115 | |||||
| 138–143 | |||||
| 569–570 | |||||
| 572–573 | |||||
| M2-1 | P04545 | 127–134 | 12.37 | 11–12 | 4.64 |
| 148–156 | 128–134 | ||||
| 165–171 | |||||
| M2-2 | P88812 | 11–12 | 11.11 | ||
| 82–89 | |||||
| L | P28887 | 124–128 | 0.28 | 17–19 | 2.17 |
| 600–600 | 155–156 | ||||
| 208 | |||||
| 584–588 | |||||
| 668, 714 | |||||
| 731–732 | |||||
| 838–842 | |||||
| 847, 1041, 1059, | |||||
| 1174, 1247 | |||||
| 1294–1295 | |||||
| 1313–1314 | |||||
| 1331–1334 | |||||
| 1336, 1486, 1505, 1569 | |||||
| 2052–2057 | |||||
| 2157–2158 | |||||
| 2164–2165 |
In addition to interaction with innate immune response proteins, NS1 and NS2 both contain putative microtubule-associated protein 1B (MAP1B) binding domains at their C-terminal end, which both coincide with predicted MoRFs (Table 5).81 Interestingly, this binding site coincides with the DLNP sequence, the four extreme C-terminal amino acids of each nonstructural protein and the only sequence that is conserved amongst the two. The NS1 DLNP sequence is not required for any known function, however NS2 STAT2 and IKKε-related functions require its DLNP sequence. MAP1B expression appears to enhance NS2-induced STAT2 degradation,81 indicating a conserved region of the nonstructural proteins that is used for one of its synergistic functions.
The RSV nonstructural proteins are thought to exist as dimers or oligomers during infection.82 The functional overlap and synergistic behavior of NS1 and NS2 likely require dimerization or oligomerization for full activity; co-expression of the two proteins results in enhanced expression or stabilization. Similar to regions involved in cellular protein binding, NS1 and NS2 homo- and hetero-dimerization with one another require central aa21–119 and aa21–104, respectively, each excluding 20aa C-terminal regions.81
Small, viral proteins with multiple functions or binding partners often encode intrinsically disordered regions to allow different functions of the same amino acids, at specific points throughout infection. The IDRs of NS1 and NS2, which reside at the extreme N- and C-termini, are potentially sites of protein–protein binding, since both NS proteins bind several cellular interacting partners (Fig. 5). In the case of most putative functions of NS1 and NS2, the N- and C-terminal domains are not required and these activities require structured domains in the central region of the protein. However, several regions within the core of NS1 and NS2 are predicted to be relatively flexible, raising the possibility that these regions may not be directly involved in stable interactions.
Fig. 5.
Nonstructural proteins NS1 and NS2. PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Gray = unassigned protein sequence; dark blue domain = protein binding with labeled binding partner Elongin C (EC); line = putative domain with labeled binding partner or type of oligomerization. NS1 (A) NS2 (B).
Nucleoprotein.
The N protein protects the RSV genome from cellular RNAses and cytosolic antiviral sensors by encapsidating the RNA in a ring structure termed the nucleocapsid, with one N decamer per one ring of RNA.20 The crystal structure of the 391aa N in complex with RNA revealed several key residues in N-RNA binding, and more recent studies further characterized the specific amino acid residues of N required for oligomerization and interaction with viral RNA.20,83 Residues involved in RNA-binding include K170, D175, R184, R185, Y337 and R338, indicating two potential N domains critical for nucleocapsid formation. Mutation of residues K170 and R185 inhibits N oligomerization in addition to RNA binding, but do not affect other N interactions.83
The RNA-dependent RNA polymerase, consisting of the large subunit L polymerase and P phosphoprotein cofactor, must gain access to RNA within the nucleocapsid for transcription and RNA synthesis. The P protein interacts with N to unwind the nucleocapsid and allow entry and initiation of RdRp activity. The P-binding region of N has been mapped to its central aa36–253 core.23,84 Interaction with P while arranged as part of the nucleocapsid requires N residues K46, M50, I53, R132, Y135, R150 and H151, and the first two residues are predicted to be potential interaction residues (ANCHOR). N–P binding is also correlated with inclusion body formation as well as polymerase activity, for which both N residues I53 and R132 are essential.23
Alternatively, N–P binding occurs during recruitment of monomeric N to genomic RNA for nucleocapsid formation by P chaperone activity. Furthermore, P binds monomeric N to prevent N from self-oligomerization or interaction with cellular RNA. K170 and R185 are required for higher order N structure and RNA-binding, however they are dispensable for P interaction with monomeric N. It is possible P-binding monomeric and oligomeric N requires separate domains of N. Further studies must be done to distinguish P binding of monomeric N monomer versus N present in nucleocapsids.
Structural data depicts N (in complex with RNA) as a highly ordered protein with C- and N-terminal domains that are connected by a hinge region where the RNA groove exists. These domains interact with adjacent equivalents within the ring structure. Each domain is comprised of α-helices with a disordered region at the extreme C- or N-terminal end that projects from the decameric N-RNA ring.20 The N-terminal projection, aa1–35, functions to stabilize the nucleocapsid structure, while the disordered C-terminal aa361–391 reside in the space between helical nucleocapsid turns.20,55
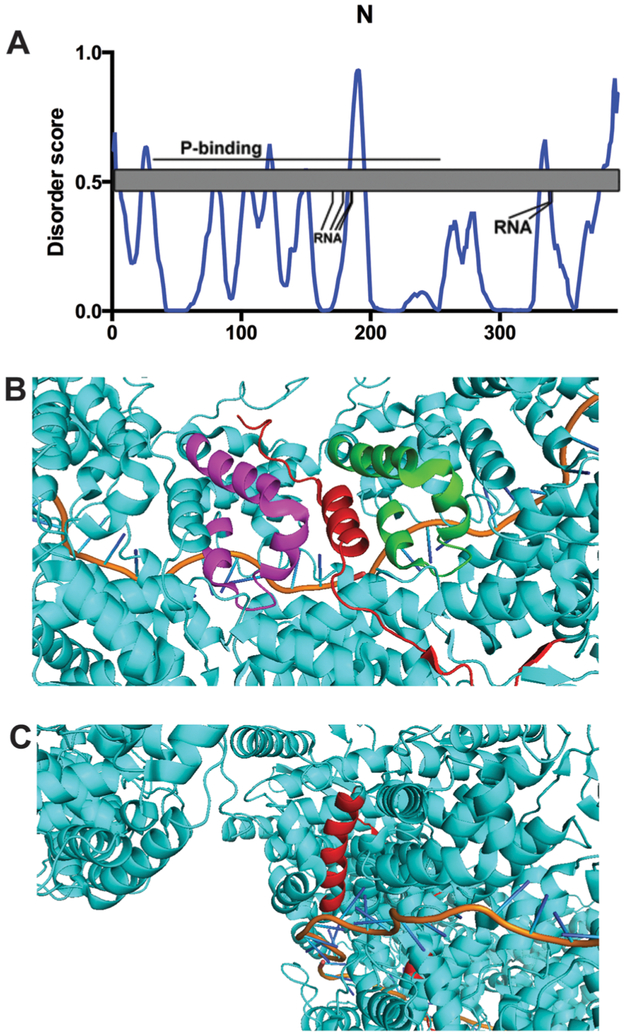
Overall the ring assumes a highly stable yet flexible structure, which is somewhat reflected in the PONDR® VL-XT data (Fig. 6A). The N-terminus is predicted to have a MoRF at aa9–13 (Table 5), and this region is within a helix in the crystal structure (Fig. 6B). Therefore, the intrinsic disorder of this region may have a stabilizing function on the nucleocapsid structure. The larger peak at aa184–195 could be the result of the RNA-binding residues at K170, D175, R184 and R185, since these sites would extend from the ring structure for interaction with the RSV genome. The salt bridge formed between N175 and R338 punctuates the IDR peak at residues aa332–338. The C-terminal 20 amino acids are disordered and the N-RNA ring structure supports the lack of order in this region.20
Fig. 6.
Nucleoprotein N. (A) PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Gray = unassigned protein sequence; line = putative domain with labeled binding partner; RNA = residues required for RNA-binding. (B) A predicted N-terminal MoRF makes intramolecular and intermolecular contacts with a region in the C-terminus. PDB structure 2WJ8 was used for this analysis. Red represents aa1–35, green represents aa282–312 on the same protein, while magenta represents aa282–312 on a different subunit. The orange line represents bound RNA. (C) A predicted MoRF in the N protein is surrounded by several helices. PDB structure 2WJ8 was imaged using Pymol analysis. Red indicates the region predicted to be a MoRF by ANCHOR and MoRFpred, aa159–166. The orange line represents bound RNA.
We observe IDR peaks at residues aa120–125 and aa184–195 that coincide with α-helices described by the nucleocapsid crystal structure. The first appears to be part of a larger α-helix, while the second is a kinked helix surrounded by long stretches of disordered residues. Both ANCHOR and MoRFpred predict a MoRF around aa159–166, which is helical in the crystal structure flanked by long stretches of disordered residues (Table 5). Interestingly, this region is surrounded by several α-helices in the tertiary structure, potentially providing several weak stabilizing interactions which promote helix formation (Fig. 6C). Of course, other external factors such as solvent conditions or the packing of the crystal lattice itself may also stabilize these flexible regions.85 We must assume variations in intrinsic disorder between monomeric and nucleocapsid-bound N based on the known experimental differences in residue usage for the different N assemblies. The residues required for protein- and RNA-binding to decameric N are disposable to monomeric N, therefore the IDR data can be interpreted alternatively for N–P assemblies (Fig. 6).
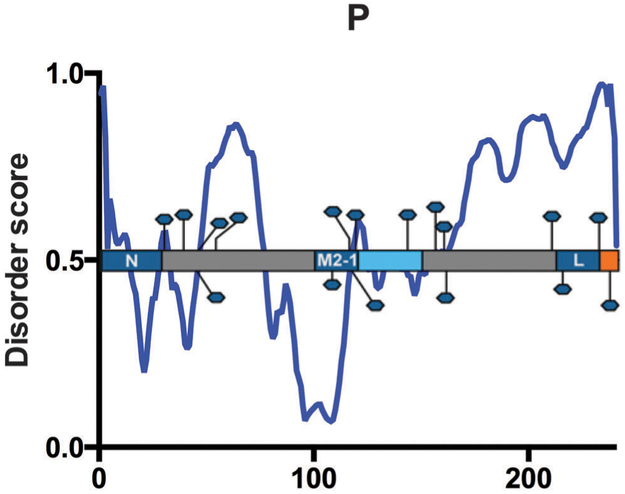
Phosphoprotein.
The 241 amino acid RSV P protein is the essential RdRp cofactor, vital to viral RNA synthesis. P interacts with each component of the ribonucleoprotein complex – L, N, viral genomic RNA as well as the M2-1 transcription anti-termination factor. As described for the N protein, P binds both monomeric N at the P N-terminus, and N in the nucleocapsid, at the P C-terminus, although the P C-terminus can also bind monomeric N.23,83,84 N-terminal P amino acids 1–29, which contains residues predicted for interaction (Table 5) are involved in P chaperone activity. Residues aa2–10 (ANCHOR) and aa20–26 (ANCHOR, MoRFpred) are directly involved in recruiting monomeric N to the ribonucleoprotein complex. P mutations in F4, F8, F20, L21 and I24 (ANCHOR) inhibit interaction with N (Table 5).83
Immediately upstream of the C-terminal RNA-binding domain, P contains an L-binding region, from aa212–239 (Table 5).86 The proximity of the RNA- and L-binding domains within the P protein sequence is suitable for P contribution to transcription and RNA synthesis. Concurrently, P binds M2-1, which is also required for RSV transcription. Mason et al. mapped an M2-1-binding region to aa100–120, just upstream of the P oligomerization domain.87 Additionally, mutation of residues L101, Y102, T108 or F109 results in inhibition of P interaction with M2-1, indicative of an M2-1 binding domain within this region of the P protein.88
P exists as a homotetramer when bound to the ribonucleo-protein complex. The oligomerization domain is at the core of the P protein, residues aa120–150 (ANCHOR), within what is predicted to be a coiled-coil domain (aa120–160).89 Fig. 7 maintains previously published predictions of intrinsic disorder regions flanking the P oligomerization domain, while the coiled coil domain itself is ordered.90 Circular dichroism studies indicated a high α-helical content in the central, ordered region of P, further supporting our PONDR® VL-XT predictions (Fig. 7).90
Fig. 7.
Phosphoprotein P. PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Gray = unassigned protein sequence; dark blue domain = protein binding with labeled binding partners N, M2-1 and L; light blue domain = oligomerization; orange domain = RNA-binding; blue hexagon = phosphorylation site.
P is phosphorylated at numerous sites: S30, S39, S45, T46, S54, T108, S116, S117, S119, S143, S156, T160, S161, T210, S215, S232, and S237.88,91–95 There is variation in transient and constitutive phosphorylation, and is unclear how these modifications are related to function, or whether they are sequential or co-dependent. Some studies have proven phosphorylation dispensable for oligomerization, while others have determined it is required.89,96 Phosphorylation is not necessary for replication, P–N or P–M2-1 interactions, suggesting L-binding, transcriptional activity or possibly budding as potential purposes for modification.89 However, phosphorylation at P residues S116, S117 and S119 is required for M2-2 regulation of the switch from viral transcription to replication, suggestive of a potential M2-2 binding site overlapping that of M2-1.97 In the PONDR® VL-XT plot, the phosphorylated residues are scattered throughout ordered and disordered regions of P without any noticeable pattern (Fig. 7). However, many of the phosphorylated residues fall within sharp dips likely to correspond with MoRFs, indicating that these residues may still be natively disordered (Table 5). Indeed, when phosphorylated residues are plotted against the more accurate PONDR®-FIT plot, there is a clear enrichment of phosphorylated residues within disordered region, a phenomenon that has been well-documented in other systems.98
P is a highly disordered protein, which is to be expected considering its various binding partners during infection. The high level of disorder in the region required for polymerase activity suggests structural flexibility necessary for binding multiple partners simultaneously. The PONDR® VL-XT plot demonstrates that the regions required for interaction with N, M2-1, and L fall within dips of the plot, which may indicate that the flexible P protein coordinates highly transient interactions during infection. Unlike previous reports, we find the majority of the sequence upstream of the oligomerization domain to be ordered, with disordered peaks at aa29–32 and aa47–77 (Fig. 7). The latter contains MoRFs predicted by ANCHOR and MoRFpred (Table 5).
Matrix protein.
The 256aa RSV M protein assembles the encapsidated RNA genome and associated structural proteins into the progeny virion in preparation for budding from the host plasma membrane. As part of the ribonucleoprotein complex, M2-1 binds M during virus assembly as a mediator of M interaction with genomic RNA. The M2-1 binding region has been assigned to the N-terminal 110aa of the M protein.29 The sequential process of virus assembly requires M interaction with RNA for facilitation of genomic and protein products into a virion.99 Sites of RNA-binding (K121, K130, K156, K157, R170) have delineated a putative RNA-binding domain from amino acids 120–170, which overlaps the zinc-finger and central oligomerization domains.99
The stable and biologically active form of M is a dimer. The oligomerization region at aa92–105 is responsible for M dimerization specifically, which is critical to virus-like particle (VLP) formation and budding.100 Subsequently, M forms higher-order oligomers to induce a switch from RNA synthesis to virus assembly and budding. M higher-order oligomerization is key to formation of a viral structure comprised of M oligomers, encompassing the viral nucleocapsid that will bud through the plasma membrane to form a virus particle. There are several well-defined oligomerization domains dispersed across the protein at aa63–68, aa129–134, aa144–163 and aa225–235, emphasizing the importance of M oligomerization.100 Interestingly, many of the oligomerization regions coincide with peaks on the disorder plots, potentially indicating that stable dimerization is coordinated by multiple, weak interactions by means of flexible regions in the protein. In addition, a putative oligomerization domain has been mapped to aa205–220, near the C-terminus.99,101
M nuclear trafficking during infection is one key feature of M distinct from any other RSV protein, although it is shared among other M proteins of the order Mononegavirales.102-104 Early in infection, M localizes to the nucleus, potentially for inhibition of host cell transcription.105 The M nuclear localization sequence is limited to aa110–183, encompassing the zinc-finger and central oligomerization domains, as well as the putative central RNA-binding domain.106 The nuclear export signal for trafficking back into the cytoplasm for virus assembly late in infection is located at aa194–206.107 M protein aa114–144 were initially identified as containing a putative zinc-finger domain via sequence alignment with closely related viruses.108 Indeed, M nuclear accumulation depends upon metal ion availability, indicating the presence of a metal-binding domain critical to nuclear trafficking.100,106
M undergoes phosphorylation at T205, the final residue of its nuclear export signal. T205 also marks the beginning of a putative oligomerization domain from aa205–220. This phosphorylation is essential for higher-order oligomerization of M during assembly, and mutation of T205 therefore attenuates RSV.101
The crystal structure of M was initially solved for the monomeric form of M, but has also recently been solved for the more biologically relevant dimeric form of M.100,109 The monomer structure describes M composition as β-sheets primarily, with some α-helices interspersed. Connecting the N-terminal domain (aa1–126) with the C-terminal domain (aa162–255) is an unstructured 36aa linker.109 Overall, M is a highly ordered protein with two predicted IDR peaks (Fig. 8A). While M is similar to P with its various domains and binding partners, it displays more order within and between those domains. The second, dimeric structure of M depicts the two IDR peaks as each coinciding with oligomerization domains.100 In particular, the disordered region from aa63–68 seems to facilitate inter-molecular contacts with other known oligomerization regions at aa129–134 and aa227–231 (Fig. 8B). Additionally, it should be noted that the M protein has no predicted MoRFs by neither ANCHOR nor MoRFpred (Table 5).
Fig. 8.
Matrix protein M. (A) PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Gray = unassigned protein sequence; line = putative domain with labeled function (NLS = nuclear localization sequence, Olig. = oligomerization); light blue domain = oligomerization; central red domain = zinc-finger domain (ZFD); C-terminal red domain = nuclear export signal; RNA = residues required for RNA-binding; blue hexagon = phosphorylation site. (B) Intrinsic disorder regulates inter-molecular contacts of dimerization regions. Analysis of PDB structure 4V23 using Pymol reveals that the intrinsically disordered loop (red, aa63–68) likely facilitates contacts with two other oligomerization regions (blue, aa127–131; magenta, aa227–231). Black spheres represent potassium ions.
Small hydrophobic protein.
The RSV SH protein is a type II integral membrane proteins and a member of the viroporin family of small, viral membrane proteins that oligomerize to enhance fusion and entry into the host. Little is understood regarding the function of SH, which at 64aa is the smallest of the three RSV viral surface proteins. Infection with SH-deleted RSV results in attenuation of RSV and decreased apoptosis of infected cells.110
The SH transmembrane domain from aa18–43 takes on an α-helical secondary structure, which is depicted as highly ordered in our IDR predictions in (Fig. 9). The N-terminal 17aa comprise the intracellular, cytoplasmic domain, while the C-terminal 21aa reside in the extracellular space.111 Our predictions suggest an IDR comprised of the C-terminal aa58–64 of the extracellular domain, with increases in intrinsic disorder beginning around residue 48. A flexible region on the extracellular surface may hint at its function, potentially as a target for transient protein–protein interactions. The only predicted MoRF (MoRFpred) is found at aa11–15, which may be an interesting target for future studies (Table 5).
Fig. 9.
Small hydrophobic protein SH. PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Bright green domain = cytoplasmic; yellow domain = transmembrane; purple domain = extracellular.
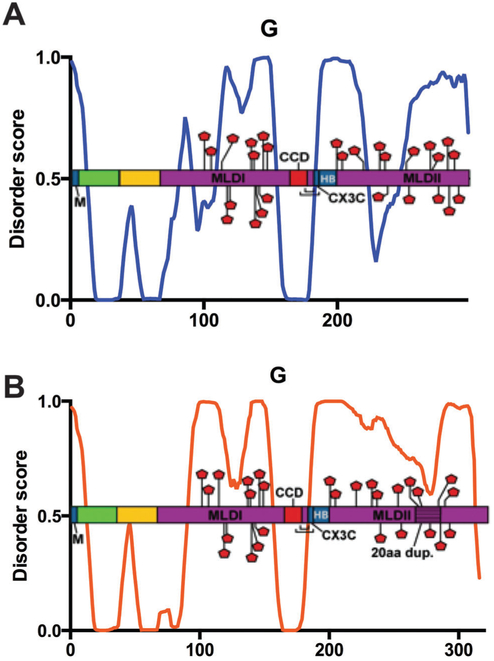
Glycoprotein G.
RSV G is the major surface glycoprotein for virus attachment with the host cell.112 Depending on the genotype, G is anywhere from 298 to 319 amino acids long. G is a type II integral membrane protein with the N-terminal 36aa residing in the cytoplasm and aa67–298 in the extracellular space. The helical transmembrane domain is found in aa37–66.113 Most of the cytoplasmic domain and the entire transmembrane domain are predicted to be ordered, as depicted in (Fig. 10A). A soluble form of G, sG, aa65–74 shorter at the N-terminus is secreted during infection, presumably to act as an antibody decoy.114–116 It is interesting to note that the majority of the ordered sequence of G is absent in the secreted form.
Fig. 10.
Glycoprotein G. PONDR® VL-XT plot of A2 (A) or averaged BA strains (B) overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Dark blue domain = protein binding with labeled binding partners M, CX3C motif and heparin (HB); bright green domain = cytoplasmic; yellow domain = transmembrane; purple = extracellular mucin-like domains (MLDI and MLDII) with textured 20aa duplication in (B); red domain = central conserved region; red pentagon = potential glycosylation sites; brackets = disulfide bond cysteine noose.
G is a heavily glycosylated protein, with 30–40 O-linked glycans and 4–5 N-linked glycans.117 The glycosylated sites are within one of the two mucin-like domains (MLDI and MLDII), which are highly variable in sequence. MLDI and MLDII distinguish G as the most variable RSV protein, which is useful for classification and diagnostic purposes.14,118 With few exceptions, the glycosylation sites reside within the highly disordered variable regions, which interact with many different antigenic sites (Fig. 10A). Appropriately, IDR regions are useful when interacting with a wide variety of binding partners, and are generally enriched in post-translational modifications.
The first six N-terminal residues of G, which are part of the cytoplasmic tail, interact with the M protein.30 This interaction is important during virus assembly, as M transports the viral nucleocapsids to sites on the plasma membrane from which budding will occur. Here, RSV surface proteins are embedded in the membrane, with exposed cytoplasmic tails for M-binding and subsequent nucleocapsid envelopment by the plasma membrane before budding from the infected cell.30,119 The M-binding site of G is the only region of the cytoplasmic domain that is predicted to be disordered (Fig. 10A). As described for other RSV protein–protein binding sites, disorder is likely required for the G–M interaction and subsequent budding.
The central conserved domain (CCD) sits between the two MLDs, from aa164–177 (Table 5).113 The C-terminal end of the CCD contains two cysteine residues, C173 and C176, which form disulfide bonds with C186 and C182, respectively. These four cysteines are linked in a 1–4 and 2–3 manner to create a cysteine noose.120 The C-terminal cysteine residues of the cysteine noose also function as part of the CX3C motif, which competes with the chemokine CX3CL1, also known as fractalkine, for its receptor CX3CR1.121 Immediately downstream of the CX3C motif is a heparin-binding domain at residues 184–198.122 This basic domain is the site of attachment to the cell surface receptor heparin sulfate on immortalized cells, although there are likely alternative cellular receptors for RSV attachment to the host.113,123,124 The highly conserved CCD displays the most consistent disorder predictions among the different G genotypes (Fig. 10A). Recent studies have shown that the CX3C domain is essential for RSV attachment to primary human airway epithelial cells, indicating that this region controls virus binding.125–127
Within the last two decades, new genotypes of G have evolved from both A and B subgroups. In 1999, the BA genotype from subgroup B was discovered, which contains an exact 20aa duplication inserted as aa260–279 (ANCHOR), within the MLDII domain.17,128–130 In 2009, isolation of a new A genotype, ON1, was first reported.131 Comparable to the BA genotype, the ON1 genotype contains a 24aa duplication of aa261–283, inserted as aa285–307, within its MLDII.131 The insertions provide up to seven additional glycosylation sites within the expanded variable region.131 The BA genotype has rapidly become the most prevalent circulating B genotype worldwide, suggesting that this duplication event confers a fitness advantage to RSV. It will be interesting to observe an potential ON1 circulation pattern resembling that of the current BA strain within the next decade.132 Increased glycosylation sites, combined with added sequence in the region responsible for RSV antigenic drift, may account for ON1 and BA rapid circulation throughout the world.
Shown in Fig. 10B, G is a highly disordered protein. When comparing ON1 and BA sequences with other A and B genotypes, respectively, G overall disorder status is increased in ON1 and BA strains with the MLDII duplication. We compared the averaged PONDR® VL-XT BA data to the A2 data both to showcase the changes resulting from the introduction of 20aa in the MLDII, and also to exhibit the RSV B subtype. Comparing G–A with G–BA, it is apparent that the increased overall disorder of BA is at least partially a result of the duplication near the C-terminus. While most of the extracellular domains of both graphs display disorder, the sequence from aa223–246 (Table 5), which drops into the ordered section of the graph in G–A, is no longer ordered in G–BA. Since the only difference between ON1 and BA genotypes and their older A and B counterparts is the duplication, we can theoretically correlate increased disorder with the insertion. Furthermore, this data suggests an indirect link between increased disorder of G and higher circulation of ON1 and BA genotypes.
Interestingly, there is another key difference between the G–A and G–BA graphs, upstream of the duplication. The dip in the disordered region of the N terminal end of G at aa91–109 disappears, and instead the G-BA sequence remains disordered from aa91–156 (Fig. 10B). Due to the location within the MLDI, it is probable that this inconsistency is a demonstration of the high level of variation in the two mucin-like domains of G, which may represent modified protein function.
Glycoprotein F.
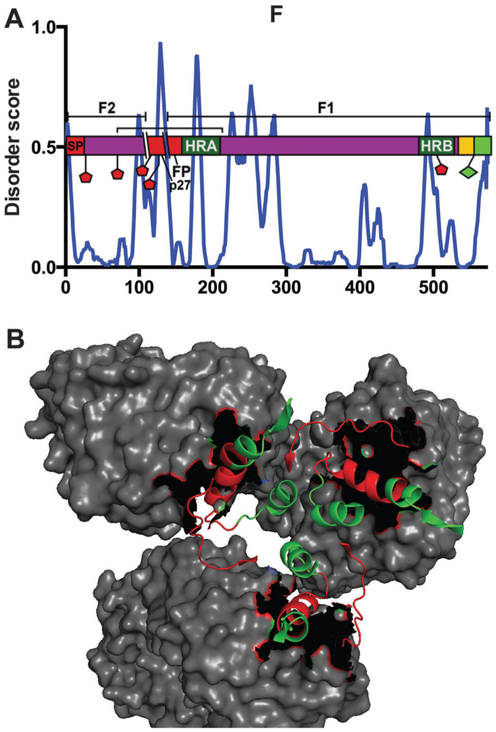
In contrast to SH and G, the 574aa F fusion protein is a type I integral membrane protein. F functions primarily to fuse the viral envelope with the plasma membrane to release the viral nucleocapsid into the cytoplasm of the host cell.2 The first 528 amino acids make up the extracellular domain, aa529–550 the transmembrane domain and aa551–574 reside in the cytoplasm.2 The RSV F protein is synthesized as the inactive precursor protein F0 and modified with a C-terminal palmitoylation at C550 and 5–6 N-linked glycans, depending on the strain of RSV.133,134 Here, we modeled domain maps around the A2 strain, which undergoes glycosylation at 5 residues – N27, N70, N116, N126 and N500.134 F0 is cleaved by a cellular furin-like protease at extracellular domain residues 109 and 136, yielding three distinct peptides – the F1 and F2 active subunits, and a 27aa peptide (p27) derived from the intervening sequence.135–138 The function of p27 is unknown and it dissociates shortly after cleavage.139,140 The PONDR® VL-XT data shows p27 to be highly disordered, although there is also a trend of disorder peaks at potential glycosylation sites, of which there are two within p27 (Fig. 11). The signal peptide, the N-terminal 25aa of F, is also cleaved at its C-terminal end in generation of the active F monomer.134 The resulting F1 and F2 chains remain attached via a disulfide bond linking C69 with C212.134,141 Fully mature F thus contains just three N-linked glycans.
Fig. 11.
Glycoprotein F. (A) PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Red domains from N- to C-termini = signal protein (SP), p27, fusion peptide (FP); dark green = heptad repeat domains (HRA and HRB) for intra-protein interactions; yellow domain = transmembrane; bright green domain = cytoplasmic; purple domain = extracellular; red pentagon = potential glycosylation sites; green diamond = palmitoylation site; bracket = disulfide bond; parallel lines depict proteolytic cleavage sites, resulting in F1 and F2 subunits designated by bracketed lines above domain map. (B) The prefusion state (PDB ID: 4JHW) bound to antibody D25 (gray), imaged using Pymol. The red residues (aa63–74, aa200–213) represent the antibody recognition epitope ASØ.
Upon triggering, the coiled-coil heptad repeat domain HRA near the N-terminus of the F1 subunit, aa157–209, lengthens and trimerizes with the adjacent F protein HRA domains.113,142 Specifically, the unstructured regions connecting the short α-helices that comprise HRA refold into α-helices themselves, generating an extended α-helix.113 This remodeling causes HRA trimerization and insertion of the hydrophobic stretch of amino acids at the N-terminus of the F1 subunit, termed the fusion peptide (FP, from aa136–157), into the plasma membrane of the host cell.134,142 FP incorporation into the host membrane allows for further intra-protein interactions, in which the HRB domains, from aa476–524 of each F protein, fold to interact with the HRA trimer to form a stable α-helical trimer consisting of HRA-HRB heterodimers. This drives the viral and host membranes together for fusion.143 Each heptad repeat domain induces a single IDR peak (Fig. 11). Although we know all HRA and HRB domains are α-helical, they are responsible for refolding F for its key fusion activity. In addition, the HRA IDR peak resides within the region of F that switches from unstructured to α-helical in the fusion process.
The C-terminal tail of F facilitates the release of M-ribonucleoprotein complexes from the inclusion bodies, which are the sites of RNA synthesis and translation.144,145 The phenylalanine residue F572 within the F cytoplasmic domain is critical for mediating assembly (Table 5). While our predictions display the transmembrane and cytoplasmic domains as ordered, there is a peak in the data corresponding to F572 (Fig. 11).
The crystal structure has been solved for both the pre- and post-fusion forms of F.33,146 To note, pre-fusion F consists an unstructured region from aa62–69 that connects the structured N- and C-terminal portions of F2. This region shifts drastically during the switch from pre- to post-fusion F, in comparison to the F2 peptide as a whole. Similarly, the F1 α-helix from aa196–210 alters its orientation during pre- to post-fusion transformation. These two regions are part of the antigenic site Ø (ASØ), located at the apex of the pre-fusion F trimer, and they account for most of the F variability in the otherwise highly conserved protein.33
Several crystal structures have been solved for a number of complexes in which an antibody is bound to F at an antigenic site.33,147,148 For our purposes, these complex structures are useful in determining correlations, if any, between our predicted IDRs and F-antibody binding sites. Interestingly, all identified antigenic sites are positioned within PONDR® VL-XT dips, potentially indicating a MoRF (Fig. 11A and B). This observation that the residues involved in ASØ fall within regions of predicted MoRFs is consistent with the high sequence variability and recognition of this region by multiple antibodies. Overall, F is a moderately ordered protein, which is reflected in its high global sequence conservation as well as its use of higher order oligomerization to carry out its fundamental F fusion activity.
M2-1 and M2-2 proteins.
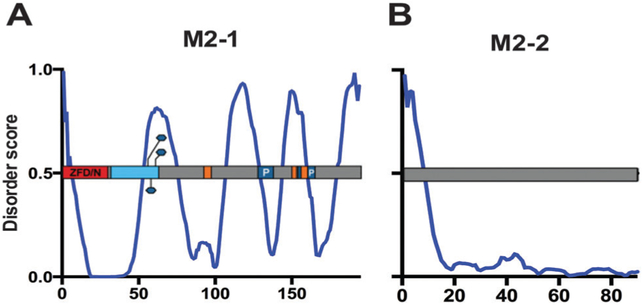
The M2 gene encodes two proteins with two distinct, overlapping ORFs – M2-1 and M2-2.149 M2-1 is an antitermination factor that is important for processive transcription by the RSV polymerase. M2-2 is approximately half the size of M2-1 and functions to switch RNP activity from viral transcription to genomic RNA synthesis.25 While it was previously noted that M2-2 inhibits viral transcription late in infection, which is dependent on P phosphorylation, the exact mechanism of this activity is unknown.97,150 Our intrinsic disorder predictions thus do not help to elucidate M2-2’s role in transcription and replication; therefore we will focus on M2-1 (Fig. 12B).
Fig. 12.
Transcriptional regulators M2-1 and M2-2. PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Gray = unassigned protein sequence; red = zinc-finger domain (ZFD)/N-binding domain; light blue domain = oligomerization; orange domains = RNA-binding; dark blue domains = protein binding with labeled binding partner P; blue hexagon = phosphorylation sites. M2-1 (A) M2-2 (B).
M2-1 regulates transcription through its anti-termination activity.25,151 The 194aa M2-1 protein is found within cytoplasmic inclusion bodies, where it associates with the RNP complex via P, the N-terminal domain of M and RNA.29,87,152,153 Core M2-1 residues aa53–177 (ANCHOR predicts 3 MoRFs, MoRFpred predicts 1, Table 5) bind P and RNA in a competitive manner that is independent of M2-1 phosphorylation.87,154 The more recently determined NMR structure of M2-1 revealed a partial overlap between RNA- and P-binding domains, supporting their competitive binding. In particular, residues K92-V97, L149-L152 and D155-K159 are sites of RNA-binding (ANCHOR). Interaction with P occurs at residues V127-S137 and L152-T164 (MoRFpred).155 M2-1 is recruited to viral inclusion bodies for RNA synthesis by association with P, while direct interaction with RNA is required for M2-1 transcription anti-termination and processivity activity.88,153,155 Based on our PONDR® VL-XT data, four out of the five RNA- and P-binding domains are of predicted structured sequence, within putative MoRFs. The disorder peak from aa144–160 contains one of the RNA-binding domains, as well as the N-terminal region of the P-binding domain from aa152–164 (ANCHOR) (Fig. 12A).
M2-1 forms a disc-like assembly as a tetramer.156 The oligomerization domain for M2-1 is located from aa32–63. M2-1 activity and optimal transcription will not occur without proper M2-1 tetramer formation.154 The majority of the oligomerization domain contains sequence predicted to have high disorder, although the phosphorylated residues within this domain appear to either reside within a disordered region, or they themselves prompt disorder within the M2-1 structure (Fig. 12A).
While interactions with P and RNA are independent of phosphorylation, M2-1’s transcription anti-termination function is dependent on its phosphorylation.153 Though the major M2-1 species is not phosphorylated, the functionally active form is the minor, phosphorylated species.154,157 Host kinases phosphorylate M2-1 at serines 58 and 61 of its oligomerization domain.156 An additional residue at position T56 is potentially phosphorylated.157 As previously mentioned, the three potential phosphorylated residues all lie within a peak of predicted high disorder, indicating that an unstructured region of M2-1 is likely required for phosphorylation (Fig. 12A).
M2-1 contains an N-terminal zinc-finger domain from aa1–32, which interacts with the viral nucleocapsid, although this interaction is not required for M2-1 transcriptional activity.22,158 The ZFD is required for phosphorylation of M2-1, as well as its ability to bind zinc. It is possible the M2-1 zinc-binding activity is necessary for its anti-termination function through the interaction with the RNP complex.158
Unlike the predictions of a highly structured sequence overall for M2-2, M2-1 is one of the three RSV proteins predicted to display high intrinsic disorder (Table 2). The crystal structure of M2-1 revealed four functionally significant regions, including the aforementioned zinc-finger and oligomerization domains. In addition, there is a core domain important for antitermination activity as it contains RNA- and P-binding domains. Finally, there is an unstructured C-terminal region, which is supported by our IDR predictions (Fig. 12A).154,156
Large RNA-dependent RNA polymerase subunit L.
The large subunit of the RdRp, L, is essential for RSV transcription and replication. Due to the transcription gradient from 3′ to 5′ end of the RSV genome, the 2,165aa L protein is itself transcribed last and expressed at low levels during infection.2 Although there are numerous peaks of IDR spanning the lengthy L amino acid sequence, L is predicted to be a highly ordered protein, shown in Table 2. Due to low copy number and low stability, few structural details are known, however several L functions critical to RSV infection have been described.
L contains two variable regions, VRI and VRII, at aa137–184 and aa1718–1764.159 Demonstrated by our PONDR® VL-XT predictions in Fig. 13, both variable regions are found within peaks of intrinsic disorder. There are six conserved regions throughout the sequence as well, labeled CRI-CRVI. The CRs were determined by sequence comparison of five NNS virus L proteins.160 While the variable regions each contain IDRs, there does not appear to be any correlation between the CRs and protein disorder and/or structure (Fig. 13). The CRs are often used as reference points across the vast L amino acid sequence for describing domain locations.
Fig. 13.
Large polymerase subunit L. PONDR® VL-XT A2 plot overlaid with protein domain map depicting amino acids assigned to specific function and post-translational modification, x-axis = amino acid position, y-axis = disorder score (ordered <0.5, disordered >0.5). Gray = unassigned protein sequence; red domains = variable regions (VR1 and VR2); orange domains = RNA-binding (from N- to C-termini contain RdRp, PRNTase and 2′-O-MTase catalytic activities); conserved regions CR1 to CRVI designated with bracketed lines above domain map.
The RNA-dependent RNA polymerase catalytic activity of L falls within CRII and CRIII, from aa693–877 (Table 5). This domain contains the signature GDNQ polymerase active motif, at aa810–813, which is responsible for phosphodiester bond formation during nucleotide incorporation.159,161 The GDNQ motif itself is predicted to be within an ordered structure; however immediately downstream of the active site is a predicted IDR within the RdRp catalytic domain (Fig. 13).
NNS virus mRNA cap formation requires a unique mechanism that differs from eukaryotic mRNA capping, which utilizes the L polyribonucleotidyl transferase (PRNTase) domain to create the 5′ mRNA cap independent of external transferase activity. Located at aa1152–1228 within CRIV, L mRNA capping activity is conserved amongst the NNS viruses.162,163 PONDR® VL-XT predictions show an IDR peak within the PRNTase domain, in CRIV (Fig. 13). After mRNA is transcribed and undergoes mRNA capping, the 5′-triphosphate is then methylated by a methyltransferase. Also conserved amongst all NNS viruses is the methyltransferase (2′-O-MTase) domain for cap 1 methylation of the mRNA 5′-cap, located after VRII in CRVI from aa1820–2008.164 This domain is found within a region of L predicted to be highly ordered. The inconsistencies between L conserved functional domains and predicted disorder regions support the poor correlation between L CRs and intrinsic protein disorder.
IDR predictions label L as one of the most ordered RSV proteins. However, there are several peaks throughout the disorder predictions, potentially indicating the presence of several small linkers or solvent-accessible sites for modification or interaction. Additionally, numerous sites uncharacterized by the literature are predicted to be MoRFs, potentially indicating sites of previously unknown function (Table 5). We also know that overall polymerase sequence and structure is well conserved amongst negative-strand RNA viruses, therefore we can infer high level of RSV L order from other MNV RdRp structures. This is demonstrated by the NNS vesicular stomatitis virus L RdRp structure, which contains two structural domains in addition to the three catalytic domains and six CRs illustrated by our RSV L domain map.165
Conclusions
The predictions presented in this study have established a comparable intrinsic disorder status between RSV subtypes A and B, which have persisted and co-circulated globally for over fifty years. SH was the only RSV protein exhibiting drastic divergence in intrinsic disorder between RSV subtypes, although the functional relevance of these findings is unknown since SH is not essential for RSV infectivity.110
With a better understanding the RSV IDRs that are well conserved and critical for viral activity, efforts can be made for IDR mutation and subsequent loss of protein function to limit essential viral activity. In terms of vaccine development, the objective is to achieve to a functionally incompetent protein while preserving structural stability and expression, to attenuate viral growth while maintaining virus viability. Therefore, targeting an unstructured yet functionally active sequence is favorable to that of a structured sequence. Due to consistency in degree and location of IDRs in the RSV proteome across all screened strains, targeting identified IDRs would be effective against all RSV strains.
Acknowledgements
J. N. W. acknowledges support from the University of South Florida Signature Research Doctoral Fellowship in Drug Design and Delivery. M. N. T. acknowledges support from NIAID R01 AI081977. We would like to acknowledge Insung Na for assistance with disorder prediction.
References
- 1.Noton SL and Fearns R, Virology, 2015, 479–480C, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins PL, Chanock RM and Murphy BR, Respiratory Syncytial Virus, Lippincott Williams and Wilkins, Philadelphia, 2001. [Google Scholar]
- 3.Welliver TP, Garofalo RP, Hosakote Y, Hintz KH, Avendano L, Sanchez K, Velozo L, Jafri H, Chavez-Bueno S, Ogra PL, McKinney L, Reed JL and Welliver RC Sr., J. Infect. Dis, 2007, 195, 1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glezen WP, Taber LH, Frank AL and Kasel JA, Am. J. Dis. Child, 1986, 140, 543–546. [DOI] [PubMed] [Google Scholar]
- 5.Henderson FW, Collier AM, Clyde WA Jr. and Denny FW, N. Engl. J. Med, 1979, 300, 530–534. [DOI] [PubMed] [Google Scholar]
- 6.Sande CJ, Mutunga MN, Medley GF, Cane PA and Nokes DJ, J. Infect. Dis, 2013, 207, 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waris M, J. Infect. Dis, 1991, 163, 464–469. [DOI] [PubMed] [Google Scholar]
- 8.Zlateva KT, Vijgen L, Dekeersmaeker N, Naranjo C and Van Ranst M, J. Clin. Microbiol, 2007, 45, 3022–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mlinaric-Galinovic G, Welliver RC, Vilibic-Cavlek T, Ljubin-Sternak S, Drazenovic V, Galinovic I and Tomic V, Virol. J, 2008, 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peret TC, Hall CB, Schnabel KC, Golub JA and Anderson LJ, J. Gen. Virol, 1998, 79(pt 9), 2221–2229. [DOI] [PubMed] [Google Scholar]
- 11.Cane PA, Matthews DA and Pringle CR, J. Gen. Virol, 1991, 72(pt 9), 2091–2096. [DOI] [PubMed] [Google Scholar]
- 12.Choi EH and Lee HJ, J. Infect. Dis, 2000, 181, 1547–1556. [DOI] [PubMed] [Google Scholar]
- 13.Tsukagoshi H, Yokoi H, Kobayashi M, Kushibuchi I, Okamoto-Nakagawa R, Yoshida A, Morita Y, Noda M, Yamamoto N, Sugai K, Oishi K, Kozawa K, Kuroda M, Shirabe K and Kimura H, Microbiol. Immunol, 2013, 57, 655–659. [DOI] [PubMed] [Google Scholar]
- 14.Tan L, Lemey P, Houspie L, Viveen MC, Jansen NJG, van Loon AM, Wiertz E, van Bleek GM, Martin DP and Coenjaerts FE, PLoS One, 2012, 7, e51439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebuffo-Scheer C, Bose M, He J, Khaja S, Ulatowski M, Beck ET, Fan J, Kumar S, Nelson MI and Henrickson KJ, PLoS One, 2011, 6, e25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose ME, He J, Shrivastava S, Nelson MI, Bera J, Halpin RA, Town CD, Lorenzi HA, Noyola DE, Falcone V, Gerna G, De Beenhouwer H, Videla C, Kok T, Venter M, Williams JV and Henrickson KJ, PLoS One, 2015, 10, e0120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Niekerk S and Venter M, J. Virol, 2011, 85, 8789–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnott A, Vong S, Mardy S, Chu S, Naughtin M, Sovann L, Buecher C, Beaute J, Rith S, Borand L, Asgari N, Frutos R, Guillard B, Touch S, Deubel V and Buchy P, J. Clin. Microbiol, 2011, 49, 3504–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khor CS, Sam IC, Hooi PS and Chan YF, Infect., Genet. Evol, 2013, 14, 357–360. [DOI] [PubMed] [Google Scholar]
- 20.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF and Rey FA, Science, 2009, 326, 1279–1283. [DOI] [PubMed] [Google Scholar]
- 21.Ruigrok RW, Crepin T and Kolakofsky D, Curr. Opin. Microbiol, 2011, 14, 504–510. [DOI] [PubMed] [Google Scholar]
- 22.Garcia J, Garcia-Barreno B, Vivo A and Melero JA, Virology, 1993, 195, 243–247. [DOI] [PubMed] [Google Scholar]
- 23.Galloux M, Tarus B, Blazevic I, Fix J, Duquerroy S and Eleouet JF, J. Virol, 2012, 86, 8375–8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosfeld H, Hill MG and Collins PL, J. Virol, 1995, 69, 5677–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins PL, Hill MG, Cristina J and Grosfeld H, Proc. Natl. Acad. Sci. U. S. A, 1996, 93, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy RW and Wertz GW, J. Virol, 1998, 72, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy RW, Harmon SB and Wertz GW, J. Virol, 1999, 73, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland KA, Collins PL and Peeples ME, Virology, 2001, 288, 295–307. [DOI] [PubMed] [Google Scholar]
- 29.Li D, Jans DA, Bardin PG, Meanger J, Mills J and Ghildyal R, J. Virol, 2008, 82, 8863–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghildyal R, Li DS, Peroulis I, Shields B, Bardin PG, Teng MN, Collins PL, Meanger J and Mills J, J. Gen. Virol, 2005, 86, 1879–1884. [DOI] [PubMed] [Google Scholar]
- 31.Ghildyal R, Mills J, Murray M, Vardaxis N and Meanger J, J. Gen. Virol, 2002, 83, 753–757. [DOI] [PubMed] [Google Scholar]
- 32.Collins PL, Olmsted RA and Johnson PR, J. Gen. Virol, 1990, 71, 1571–1576. [DOI] [PubMed] [Google Scholar]
- 33.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD and Graham BS, Science, 2013, 340, 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling Z, Tran KC and Teng MN, J. Virol, 2009, 83, 3734–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramaswamy M, Shi L, Varga SM, Barik S, Behlke MA and Look DC, Virology, 2006, 344, 328–339. [DOI] [PubMed] [Google Scholar]
- 36.Munir S, Hillyer P, Le Nouen C, Buchholz UJ, Rabin RL, Collins PL and Bukreyev A, PLoS Pathog, 2011, 7, e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling Z, Tran KC, Arnold JJ and Teng MN, Protein Expression Purif, 2008, 57, 261–270. [DOI] [PubMed] [Google Scholar]
- 38.Uversky VN, Protein Sci, 2002, 11, 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM and Obradovic Z, Biochemistry, 2002, 41, 6573–6582. [DOI] [PubMed] [Google Scholar]
- 40.Bourhis JM, Canard B and Longhi S, Curr. Protein Pept. Sci, 2007, 8, 135–149. [DOI] [PubMed] [Google Scholar]
- 41.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ and Dunker AK, Proteins, 2003, 53(suppl 6), 566–572. [DOI] [PubMed] [Google Scholar]
- 42.Uversky VN, Gillespie JR and Fink AL, Proteins, 2000, 41, 415–427. [DOI] [PubMed] [Google Scholar]
- 43.Xue B, Blocquel D, Habchi J, Uversky AV, Kurgan L, Uversky VN and Longhi S, Chem. Rev, 2014, 114, 6880–6911. [DOI] [PubMed] [Google Scholar]
- 44.Xue B, Dunker AK and Uversky VN, J. Biomol. Struct. Dyn, 2012, 30, 137–149. [DOI] [PubMed] [Google Scholar]
- 45.Bellay J, Han S, Michaut M, Kim T, Costanzo M, Andrews BJ, Boone C, Bader GD, Myers CL and Kim PM, Genome Biol, 2011, 12, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erales J, Blocquel D, Habchi J, Beltrandi M, Gruet A, Dosnon M, Bignon C and Longhi S, Adv. Exp. Med. Biol, 2015, 870, 351–381. [DOI] [PubMed] [Google Scholar]
- 47.Habchi J and Longhi S, Int. J. Mol. Sci, 2015, 16, 15688–15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dosnon M, Bonetti D, Morrone A, Erales J, di Silvio E, Longhi S and Gianni S, ACS Chem. Biol, 2015, 10, 795–802. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Chu X, Longhi S, Roche P, Han W, Wang E and Wang J, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, E3743–E3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longhi S, Adv. Exp. Med. Biol, 2012, 725, 126–141. [DOI] [PubMed] [Google Scholar]
- 51.Habchi J and Longhi S, Mol. BioSyst, 2012, 8, 69–81. [DOI] [PubMed] [Google Scholar]
- 52.Longhi S and Oglesbee M, Protein Pept. Lett, 2010, 17, 961–978. [DOI] [PubMed] [Google Scholar]
- 53.Bourhis JM, Canard B and Longhi S, Virology, 2006, 344, 94–110. [DOI] [PubMed] [Google Scholar]
- 54.Bourhis JM, Receveur-Brechot V, Oglesbee M, Zhang X, Buccellato M, Darbon H, Canard B, Finet S and Longhi S, Protein Sci, 2005, 14, 1975–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourhis JM, Johansson K, Receveur-Brechot V, Oldfield CJ, Dunker KA, Canard B and Longhi S, Virus Res, 2004, 99, 157–167. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Nei M, Dudley J and Tamura K, Briefings Bioinf, 2008, 9, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ and Dunker AK, Proteins, 2001, 42, 38–48. [DOI] [PubMed] [Google Scholar]
- 58.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN and Dunker AK, Biochemistry, 2007, 46, 13468–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN and Dunker AK, Biochemistry, 2005, 44, 12454–12470. [DOI] [PubMed] [Google Scholar]
- 60.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK and Uversky VN, J. Mol. Biol, 2006, 362, 1043–1059. [DOI] [PubMed] [Google Scholar]
- 61.Xue B, Dunbrack RL, Williams RW, Dunker AK and Uversky VN, Biochim. Biophys. Acta, 2010, 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng K, Radivojac P, Vucetic S, Dunker AK and Obradovic Z, BMC Bioinf, 2006, 7, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK and Obradovic Z, J. Bioinf. Comput. Biol, 2005, 3, 35–60. [DOI] [PubMed] [Google Scholar]
- 64.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I and Sussman JL, Bioinformatics, 2005, 21, 3435–3438. [DOI] [PubMed] [Google Scholar]
- 65.Dosztanyi Z, Csizmok V, Tompa P and Simon I, Bioinformatics, 2005, 21, 3433–3434. [DOI] [PubMed] [Google Scholar]
- 66.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN and Dunker AK, Protein Pept. Lett, 2008, 15, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potenza E, Di Domenico T, Walsh I and Tosatto SC, Nucleic Acids Res, 2015, 43, D315–D320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dosztanyi Z, Csizmok V, Tompa P and Simon I, J. Mol. Biol, 2005, 347, 827–839. [DOI] [PubMed] [Google Scholar]
- 69.Disfani FM, Hsu WL, Mizianty MJ, Oldfield CJ, Xue B, Dunker AK, Uversky VN and Kurgan L, Bioinformatics, 2012, 28, i75–i83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kyte J and Doolittle RF, J. Mol. Biol, 1982, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 71.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN and Dunker AK, Biochemistry, 2005, 44, 1989–2000. [DOI] [PubMed] [Google Scholar]
- 72.Xue B, Oldfield CJ, Dunker AK and Uversky VN, FEBS Lett, 2009, 583, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang F, Oldfield C, Meng J, Hsu WL, Xue B, Uversky VN, Romero P and Dunker AK, Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, 2012, 128–139. [PubMed] [Google Scholar]
- 74.Fan X, Xue B, Dolan PT, LaCount DJ, Kurgan L and Uversky VN, Mol. BioSyst, 2014, 10, 1345–1363. [DOI] [PubMed] [Google Scholar]
- 75.Davey NE, Trave G and Gibson TJ, Trends Biochem. Sci, 2011, 36, 159–169. [DOI] [PubMed] [Google Scholar]
- 76.Dinkel H, Van Roey K, Michael S, Kumar M, Uyar B, Altenberg V Schneider Milchevskaya, M., Kuhn H, Behrendt A, Dahl SL, Damerell V, Diebel S, Kalman S, Klein S, Knudsen AC, Mader C, Merrill S, Staudt A, Thiel V, Welti L, Davey NE, Diella F and Gibson TJ, Nucleic Acids Res, 2016, 44, D294–D300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramaswamy M, Shi L, Monick MM, Hunninghake GW and Look DC, Am. J. Respir. Cell Mol. Biol, 2004, 30, 893–900. [DOI] [PubMed] [Google Scholar]
- 78.Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, Buick R, Stevenson NJ, Touzelet O, Gadina M, Power UF and Johnston JA, J. Virol, 2007, 81, 3428–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Straub CP, Lau WH, Preston FM, Headlam MJ, Gorman JJ, Collins PL and Spann KM, Virol. J, 2011, 8, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lo MS, Brazas RM and Holtzman MJ, J. Virol, 2005, 79, 9315–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swedan S, Andrews J, Majumdar T, Musiyenko A and Barik S, J. Virol, 2011, 85, 10090–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swedan S, Musiyenko A and Barik S, J. Virol, 2009, 83, 9682–9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galloux M, Gabiane G, Sourimant J, Richard CA, England P, Moudjou M, Aumont-Nicaise M, Fix J, Rameix-Welti MA and Eleouet JF, J. Virol, 2015, 89, 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garcia-Barreno B, Delgado T and Melero JA, J. Virol, 1996, 70, 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uversky VN, Int. J. Biochem. Cell Biol, 2011, 43, 1090–1103. [DOI] [PubMed] [Google Scholar]
- 86.Sourimant J, Rameix-Welti MA, Gaillard AL, Chevret D, Galloux M, Gault E and Eleouet JF, J. Virol, 2015, 89, 4421–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mason SW, Aberg E, Lawetz C, DeLong R, Whitehead P and Liuzzi M, J. Virol, 2003, 77, 10670–10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Asenjo A, Calvo E and Villanueva N, J. Gen. Virol, 2006, 87, 3637–3642. [DOI] [PubMed] [Google Scholar]
- 89.Castagne N, Barbier A, Bernard J, Rezaei H, Huet JC, Henry B, Da Costa B and Eleouet JF, J. Gen. Virol, 2004, 85, 1643–1653. [DOI] [PubMed] [Google Scholar]
- 90.Llorente MT, Garcia-Barreno B, Calero M, Camafeita E, Lopez JA, Longhi S, Ferron F, Varela PF and Melero JA, J. Gen. Virol, 2006, 87, 159–169. [DOI] [PubMed] [Google Scholar]
- 91.Asenjo A, Rodriguez L and Villanueva N, J. Gen. Virol, 2005, 86, 1109–1120. [DOI] [PubMed] [Google Scholar]
- 92.Navarro J, Lopezotin C and Villanueva N, J. Gen. Virol, 1991, 72, 1455–1459. [DOI] [PubMed] [Google Scholar]
- 93.Dupuy LC, Dobson S, Bitko V and Barik S, J. Virol, 1999, 73, 8384–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mazumder B and Barik S, Virology, 1994, 205, 104–111. [DOI] [PubMed] [Google Scholar]
- 95.Asenjo A, Mendieta J, Gomez-Puertas P and Villanueva N, Virus Res, 2008, 132, 160–173. [DOI] [PubMed] [Google Scholar]
- 96.Asenjo A and Villanueva N, FEBS Lett, 2000, 467, 279–284. [DOI] [PubMed] [Google Scholar]
- 97.Asenjo A and Villanueva N, Virus Res, 2015, 211, 117–125. [DOI] [PubMed] [Google Scholar]
- 98.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z and Dunker AK, Nucleic Acids Res, 2004, 32, 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rodriguez L, Cuesta I, Asenjo A and Villanueva N, J. Gen. Virol, 2004, 85, 709–719. [DOI] [PubMed] [Google Scholar]
- 100.Forster A, Maertens GN, Farrell PJ and Bajorek M, J. Virol, 2015, 89, 4624–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bajorek M, Caly L, Tran KC, Maertens GN, Tripp RA, Bacharach E, Teng MN, Ghildyal R and Jans DA, J. Virol, 2014, 88, 6380–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lyles DS, Puddington L and Mccreedy BJ, J. Virol, 1988, 62, 4387–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peeples ME, Can W, Gupta KC and Coleman N, J. Virol, 1992, 66, 3263–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoshida T, Nagai Y, Yoshii S, Maeno K, Matsumoto T and Hoshino M, Virology, 1976, 71, 143–161. [DOI] [PubMed] [Google Scholar]
- 105.Ghildyal R, Baulch-Brown C, Mills J and Meanger J, Arch. Virol, 2003, 148, 1419–1429. [DOI] [PubMed] [Google Scholar]
- 106.Ghildyal R, Ho A, Wagstaff KM, Dias MM, Barton CL, Jans P, Bardin P and Jans DA, Biochemistry, 2005, 44, 12887–12895. [DOI] [PubMed] [Google Scholar]
- 107.Ghildyal R, Ho A, Dias M, Soegiyono L, Bardin PG, Tran KC, Teng MN and Jans DA, J. Virol, 2009, 83, 5353–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Latiff K, Meanger J, Mills J and Ghildyal R, Clin. Microbiol. Infect, 2004, 10, 945–948. [DOI] [PubMed] [Google Scholar]
- 109.Money VA, McPhee HK, Mosely JA, Sanderson JM and Yeo RP, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bukreyev A, Whitehead SS, Murphy BR and Collins PL, J. Virol, 1997, 71, 8973–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gan SW, Tan E, Lin X, Yu DJ, Wang JJ, Tan GMY, Vararattanavech A, Yeo CY, Soon CH, Soong TW, Pervushin K and Torres J, J. Biol. Chem, 2012, 287, 24671–24689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Levine S, Klaiberfranco R and Paradiso PR, J. Gen. Virol, 1987, 68, 2521–2524. [DOI] [PubMed] [Google Scholar]
- 113.McLellan JS, Ray WC and Peeples ME, Curr. Top. Microbiol. Immunol., 2013, 372, 83–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hendricks DA, Baradaran K, Mcintosh K and Patterson JL, J. Gen. Virol, 1987, 68, 1705–1714. [DOI] [PubMed] [Google Scholar]
- 115.Bukreyev A, Yang L and Collins PL, J. Virol, 2012, 86, 10880–10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roberts SR, Lichtenstein D, Ball LA and Wertz GW, J. Virol, 1994, 68, 4538–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wertz GW, Collins PL, Yung HA, Gruber C, Levine S and Ball LA, Proc. Natl. Acad. Sci. U. S. A, 1985, 82, 4075–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luchsinger V, Noy AE and Avendano LF, J. Clin. Virol, 2008, 42, 260–263. [DOI] [PubMed] [Google Scholar]
- 119.Henderson G, Murray J and Yeo RP, Virology, 2002, 300, 244–254. [DOI] [PubMed] [Google Scholar]
- 120.Gorman JJ, Ferguson BL, Speelman D and Mills J, Protein Sci, 1997, 6, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tripp RA, Jones LP, Haynes LM, Zheng H, Murphy PM and Anderson LJ, Nat. Immunol, 2001, 2, 732–738. [DOI] [PubMed] [Google Scholar]
- 122.Feldman SA, Hendry RM and Beeler JA, J. Virol, 1999, 73, 6610–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Teng MN, Whitehead SS and Collins PL, Virology, 2001, 289, 283–296. [DOI] [PubMed] [Google Scholar]
- 124.Zhang LQ, Peeples ME, Boucher RC, Collins PL and Pickles RJ, J. Virol, 2002, 76, 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson SM, McNally BA, Ioannidis I, Flano E, Teng MN, Oomens AG, Walsh EE and Peeples ME, PLoS Pathog, 2015, 11, e1005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chirkova T, Lin S, Oomens AG, Gaston KA, Boyoglu-Barnum S, Meng J, Stobart CC, Cotton CU, Hartert TV, Moore ML, Ziady AG and Anderson LJ, J. Gen. Virol, 2015, 96, 2543–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jeong KI, Piepenhagen PA, Kishko M, DiNapoli JM, Groppo RP, Zhang L, Almond J, Kleanthous H, Delagrave S and Parrington M, PLoS One, 2015, 10, e0130517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Trento A, Galiano M, Videla C, Carballal G, Garcia-Barreno B, Melero JA and Palomo C, J. Gen. Virol, 2003, 84, 3115–3120. [DOI] [PubMed] [Google Scholar]
- 129.Trento A, Viegas M, Galiano M, Videla C, Carballal G, Mistchenko AS and Melero JA, J. Virol, 2006, 80, 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trento A, Casas I, Calderon A, Garcia-Garcia ML, Calvo B, Perez-Brena P and Melero JA, J. Virol, 2010, 84, 7500–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li AM, Patel SN, Low DE and Gubbay JB, PLoS One, 2012, 7, e32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cui G, Zhu R, Deng J, Zhao L, Sun Y, Wang F and Qian Y, Infect., Genet. Evol, 2015, 33, 163–168. [DOI] [PubMed] [Google Scholar]
- 133.Arumugham RG, Seid RC, Doyle S, Hildreth SW and Paradiso PR, J. Biol. Chem, 1989, 264, 10339–10342. [PubMed] [Google Scholar]
- 134.Collins PL, Huang YT and Wertz GW, P. Natl. Acad. Sci.-Biol, 1984, 81, 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Collins PL and Mottet G, J. Gen. Virol, 1991, 72(pt 12), 3095–3101. [DOI] [PubMed] [Google Scholar]
- 136.Bolt G, Pedersen LO and Birkeslund HH, Virus Res, 2000, 68, 25–33. [DOI] [PubMed] [Google Scholar]
- 137.Gonzalez-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, Skehel JJ, Wiley DC and Melero JA, Proc. Natl. Acad. Sci. U. S. A, 2001, 98, 9859–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zimmer G, Budz L and Herrler G, J. Biol. Chem, 2001, 276, 31642–31650. [DOI] [PubMed] [Google Scholar]
- 139.Ruiz-Arguello MB, Gonzalez-Reyes L, Calder LJ, Palomo C, Martin D, Saiz MJ, Garcia-Barreno B, Skehel JJ and Melero JA, Virology, 2002, 298, 317–326. [DOI] [PubMed] [Google Scholar]
- 140.Zimmer G, Conzelmann KK and Herrler G, J. Virol, 2002, 76, 9218–9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lopez JA, Bustos R, Orvell C, Berois M, Arbiza J, Garcia-Barreno B and Melero JA, J. Virol, 1998, 72, 6922–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM and Peeples ME, J. Virol, 2011, 85, 3968–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao X, Singh M, Malashkevich VN and Kim PS, Proc. Natl. Acad. Sci. U. S. A, 2000, 97, 14172–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baviskar PS, Hotard AL, Moore ML and Oomens AG, J. Virol, 2013, 87, 10730–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shaikh FY, Cox RG, Lifland AW, Hotard AL, Williams JV, Moore ML, Santangelo PJ and Crowe JE, mBio, 2012, 3, e00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McLellan JS, Yang Y, Graham BS and Kwong PD, J. Virol, 2011, 85, 7788–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McLellan JS, Chen M, Kim A, Yang YP, Graham BS and Kwong PD, Nat. Struct. Mol. Biol, 2010, 17, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McLellan JS, Chen M, Chang JS, Yang YP, Kim A, Graham BS and Kwong PD, J. Virol, 2010, 84,12236–12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Collins PL and Wertz GW, J. Virol, 1985, 54, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cheng X, Park H, Zhou H and Jin H, J. Virol, 2005, 79, 13943–13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fearns R and Collins PL, J. Virol, 1999, 73, 5852–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kiss G, Holl JM, Williams GM, Alonas E, Vanover D, Lifland AW, Gudheti M, Guerrero-Ferreira RC, Nair V, Yi H, Graham BS, Santangelo PJ and Wright ER, J. Virol, 2014, 88, 7602–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cartee TL and Wertz GW, J. Virol, 2001, 75, 12188–12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tran TL, Castagne N, Dubosclard V, Noinville S, Koch E, Moudjou M, Henry C, Bernard J, Yeo RP and Eleouet JF, J. Virol, 2009, 83, 6363–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Blondot ML, Dubosclard V, Fix J, Lassoued S, Aumont-Nicaise M, Bontems F, Eleouet JF and Sizun C, PLoS Pathog, 2012, 8, e1002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tanner SJ, Ariza A, Richard CA, Kyle HF, Dods RL, Blondot ML, Wu W, Trincao J, Trinh CH, Hiscox JA, Carroll MW, Silman NJ, Eleouet JF, Edwards TA and Barr JN, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 1580–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cuesta I, Geng XH, Asenjo A and Villanueva N, J. Virol, 2000, 74, 9858–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hardy RW and Wertz GW, J. Virol, 2000, 74, 5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fix J, Galloux M, Blondot ML and Eleouet JF, Open Virol. J, 2011, 5, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Poch O, Blumberg BM, Bougueleret L and Tordo N, J. Gen. Virol, 1990, 71(pt 5), 1153–1162. [DOI] [PubMed] [Google Scholar]
- 161.Stec DS, Hill MG and Collins PL, Virology, 1991, 183, 273–287. [DOI] [PubMed] [Google Scholar]
- 162.Lij J, Rahmeh A, Morelli M and Whelan SPJ, J. Virol, 2008, 82, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Barik S, J. Gen. Virol, 1993, 74, 485–490. [DOI] [PubMed] [Google Scholar]
- 164.Bujnicki JM and Rychlewski L, Protein Eng, 2002, 15, 101–108. [DOI] [PubMed] [Google Scholar]
- 165.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC and Whelan SP, Cell, 2015, 162, 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]