Abstract
Exposure to particulate matter < 2.5 μm (PM2.5) is associated with a variety of airway diseases. Although studies have demonstrated that high doses of PM2.5 cause cytotoxicity and changes to gene expression in bronchial epithelial cells, the effect of lower doses and repeated exposure to PM2.5 are less well studied. Here, we treated BEAS-2B cells with varying doses of PM2.5 for 1–7 days and examined the expression of a variety of genes implicated in airway disorders. At high doses, PM2.5 increased the expression of IL6, TNF, TSLP, CSF2, PTGS2, IL4R, and SPINK5. Other genes such as ADAM33, ORMDL3, DPP10 and CYP1A1, however, were increased by PM2.5 at much lower doses (≤1 μg/cm2). Repeated exposure to PM2.5 at 1 or 5 μg/cm2 every day for 7 days increased the sensitivity and magnitude of change for all of the aforementioned genes. Genes such as IL13 and TGFB1, increased only when cells were repeatedly exposed to PM2.5. Treatment with an antioxidant, or inhibitors to aryl hydrocarbon receptor or NF-κB attenuated the effect of PM2.5. These data demonstrate that PM2.5 exerts pleiotropic actions that differ by dose and duration that affect a variety of genes important to the development of airway disease.
Keywords: PM2.5, BEAS-2B, Aryl hydrocarbon receptor (AhR), Reactive oxygen species (ROS), Nuclear factor κB (NF-κB)
1. Introduction
Ambient air pollution leads to 3.3 million premature deaths worldwide and the burden of air pollution continues to rise due to increased global industrialization (Lelieveld et al., 2015). Particulate matter (PM), one of the most toxic forms of air pollution, consists of a mixture of volatile organic compounds, polycyclic aromatic hydrocarbons (PAH), and inorganic chemicals such as heavy metals that, both individually and together, contribute to adverse health effects (Chen and Lippmann, 2009; Schwarze et al., 2006). These effects not only depend on the source and composition of PM, but also on the dose and duration of exposure (Graff et al., 2007; Tolbert, 2007). PM less than or equal to 2.5 μm in diameter (PM2.5) are of particular concern to public health because of its ability to travel down the lower respiratory tract and pass into systemic circulation (Karakatsani et al., 2012; Strak et al., 2012; Xing et al., 2016).
Bronchial epithelial cells line the respiratory airways and are the first cells of the lung exposed to PM2.5. Studies of bronchial epithelial cells directly exposed to PM2.5 have shown that PM2.5 impairs bronchial epithelial cell function (Zhang et al., 2017), causes mitochondrial dysfunction (Lavrich et al., 2018), and at high doses, induces cell toxicity and death (Dergham et al., 2015; Wu et al., 2017). PM2.5 has also furthermore been shown to induce bronchial epithelial cells to synthesize proinflammatory cytokines, including interleukin (IL)-6, IL-8, and tumor necrosis factor (TNF)-α (Ovrevik et al., 2009; Wang et al., 2017). Transcriptomic analyses of bronchial epithelial cells have shown that PM2.5 induces a variety of proinflammatory chemokines and genes involved in proliferation, inflammation, and immune response (Ding et al., 2014; Li et al., 2017; Longhin et al., 2016a; Zhou et al., 2015). The majority of these in vitro studies utilize brief exposures of high doses of PM2.5 (Huang, 2013) and the effects of lower doses and repeated exposure, which may be more relevant to that of an individual exposed to pollution on a daily basis, is less well-studied.
Here, we examined how varying doses and duration of exposure to PM2.5 affect the expression of key genes relevant to airway disease (Table 1) in BEAS-2B cells, a primary bronchial epithelial cell line. We utilized PM2.5 obtained from air filters collected on a January day in Beijing, China, a populous city commonly challenged with high pollution levels over the past several years (Chen et al., 2013). In addition to examining the effects of PM2.5 on cytokines and genes traditionally associated with inflammation, we also examined whether PM2.5 altered the expression of ADAM metallopeptidase domain 33 (ADAM33), serine peptidase inhibitor Kazal type 5 (SPINK5), ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3), and dipeptidyl peptidase-like 10 (DPP10) – all genes identified through genome-wide association studies or positional cloning approaches as important to the development of asthma and other airway disorders (Allen et al., 2003; Birben et al., 2012; Holgate et al., 2006; Kim et al., 2015; Song et al., 2017; Tripathi et al., 2014). We observed that different doses of PM2.5 increases distinct groups of genes, and that different signaling pathways are responsible for the effects of PM2.5. The current study shows that even low-level exposure of PM2.5 is sufficient to exert changes in gene expression that may play important roles in the development of airway diseases.
Table 1.
Classification of genes examined based on function and relation to airway disorders.
| HGNC IP (chromosomal location) | Name (symbol) | Function |
|---|---|---|
| Genes involved in triggering the immune response and directing CD4+ T helper (Th)-cell differentiation→ | ||
| 1176 (19q13.2) | Transforming growth factor beta 1 (TGFβ-1) | Multifunctional cytokine that regulates proliferation, differentiation and immunity. Expression known to be associated with a variety of airway and other related lung diseases |
| Regulate Th2-cell differentiation and Th2-cell effector functions→ | ||
| 5973 (5q31.1) | Interleukin 13 (IL13) | Mediates production of IgE and airway remodeling |
| 6015 (16p12.1) | Interleukin 4 receptor (IL4R) | Receptor for IL4; mediates allergic inflammation by promoting IgE production and Th2 cell development |
| 30743 (5q22.1) | Thymic stromal lymphopoietin (TSLP) | Promotes T helper type 2 (TH2) cell responses |
| Expressed in epithelial cells, factors involved in maintaining the integrity of the epithelial-cell barriers→ | ||
| 15464 (5q32) | Serine peptidase inhibitor, Kazal type 5 (SPINK5) | Polymorphisms in this gene are associated with atopy and asthma |
| The group of asthma susceptibility genes discovered through positional-cloning approaches or genome wide association studies→ | ||
| 15478 (20p13) | ADAM metallopeptidase domain 33 (ADAM33) | Type I transmembrane protein implicated in asthma and bronchial hyperresponsiveness |
| 20823 (2q14.1) | dipeptidyl peptidase like 10 (DPP10) | Members of the S9B family of DPP serine proteases, which includes DPP4, a widely expressed enzyme that plays a central role in chemokine processing as part of the innate immune system. |
| Polymorphisms strongly associated with asthma | ||
| 16038 (17q21.1) | ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) | ORMDL3-dysregulated sphingolipid synthesis on bronchial hyperreactivity. Polymorphisms associated with asthma and other bronchial airway diseases |
| Top Particulate Matter Interacting Genes (cytochrome P450,inflammatory genes and cytokines)→ | ||
| 2595 (15q24.1) | cytochrome P450 family 1 subfamily A member 1 (CYP1A1) | Gene known to be induced by pollution through aryl hydrocarbon receptor signaling |
| 11892 (6p21.33) | tumor necrosis factor (TNF) | Proinflammatory cytokine-increase inflammation. Important in the pathogenesis of airway remodeling |
| 2434 (5q31.1) | colony stimulating factor 2 (granulocyte- macrophage) (CSF2) | Promotes the survival and activation of macrophages, neutrophils, and eosinophils, as well as dendritic cell maturation. Association of GM-CSF and Th2 immunity is reported in allergic airway inflammation |
| 6018 (7p15.3) | interleukin 6 (IL6) | Pro-inflammatory cytokine during inflammation |
| 9605 (1q31.1) | prostaglandin-endoperoxide synthase 2 (PTGS2) | Responsible for the prostanoid biosynthesis involved in inflammation and mitogenesis |
2. Materials and methods
2.1. Cell culture
Human bronchial epithelial cells BEAS-2B were cultured in serum-free Bronchial Epithelial Growth Medium (BEGM; CC-3170, Lonza, Walkersville, MD) consisting of basal medium supplemented with standardized growth factors provided by the manufacturer (BEGM BulletKit CC-3171 & CC-4175; Lonza) and maintained in a 37° C incubator with 5% CO2. All cells were cultured on collagen-coated plates. Tissue culture plates were coated with pre-made bovine collagen solution (PureCol-Type I Bovine Collagen Solution, Advanced BioMatrix, San Diego, CA) diluted to a concentration of 3 mg/ml with 0.1 N HCl. Plates were incubated in collagen solution at 4° C overnight. The liquid was later aspirated and plates were UV-irradiated for 30 min before washed three times with sterile water.
2.2. PM2.5 collection and preparation
PM2.5 was collected on 90 mm Emfab filters, made of borosilicate fibers reinforced with woven cloth and bonded with polytetrafluoroethylene (TX40HI20WW, part #7234, Pall Company, Beijing Office, Beijing, China). A manual sampler located on the rooftop of the School of Public Health Building of Peking University in Beijing, China was used to collect PM2.5 from January 19–21, 2015. PM2.5 was collected over 24 h and filters were replaced each day. Filters were folded in half, shipped to the United States in sterile, secure packaging and stored at −20° C until extraction. Before extraction, each filter was equilibrated for 24 h in sterile amber jars located in a sterile biosafety containment hood at constant humidity and room temperature. Each filter was weighed on a microbalance (AC 100, Mettler-Toledo, Columbus, OH) before extraction. To extract PM2.5, each filter was placed face down in amber jars, wetted with 20 ml of double distilled water, and sonicated (VWR, model no. 97043–968, VWR International, Radnor, Pennsylvania, USA) on ice at 15 min intervals for a total of 3 h. In some experiments, clean filters not exposed to any ambient air pollution were also sonicated for 3 h and used as a control. After sonication, filters were air-dried in amber jars located in the same biosafety containment hood at constant humidity and room temperature for 3 days before being weighed on a microbalance. The difference in weight (averaged from 3 to 5 measurements) before and after extraction was used to calculate the concentration (mg/ml). Samples were aliquoted and stored for future use at −80° C.
2.3. Endotoxin levels of PM2.5
Levels of endotoxin in PM2.5 were measured using the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (Cat # L00350C, GenScript, Piscataway, NJ), per manufacturer’s protocol.
2.4. Treatment of cells with PM2.5
For “24-hour” experiments, 5 × 105 BEAS-2B cells were allowed to adhere on collagen-coated 6-well plates (35 mm) overnight in BEGM before being treated with varying concentrations of PM2.5 (0.1–30 μg/cm2), diluted in BEGM for 24 h. In some experiments, cells were treated with liquid derived from sonication of clean, naïve filters at volumes equal to that used for PM2.5. Cells were then collected for RNA or protein analysis. For experiments in which cells were treated repeatedly over several days, 2.5 × 105 cells were cultured on collagen-coated 6-well plates and treated on a daily basis for up to 7 days with either medium or PM2.5 at a dose of 1 or 5 μg/cm2. To prevent accumulation of PM2.5 during this treatment period, medium was removed and cells were washed each day with PBS before the next treatment. Control cells had medium replaced each day with BEGM alone. In some experiments, the cells were pre-incubated with the antioxidant N-acetylcysteine (NAC) (100 μM, Sigma-Aldrich, St Louis, MO), the aryl hydrocarbon receptor (AhR) antagonist CH223191 (10 μM, Tocris Bioscience, Ellisville, MO) or the NF-κB inhibitor Bay11–7082 (10 μM, Sc-200,615 Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h prior to addition of PM2.5 to the medium. The specificity of these inhibitors and chosen dosage were based on literature (Lee et al., 2016).
2.5. Cytotoxicity
Cell cytotoxicity, as measured by levels of lactate dehydrogenase (LDH) in the supernatant, was assayed using the LDH Cytotoxicity Assay Kit (Catalog number 88953, Thermo Fisher Scientific, Pierce Biotechnology, Rockford, IL, USA) according to the manufacturer’s instructions. Apoptosis was assayed by immunoblot of cell lysates for presence of cleaved poly-ADP ribose polymerase (PARP).
2.6. RNA extraction and quantitative real-time PCR
RNA was isolated from cells using Trizol (Catalog Number 15596018, Invitrogen, Carlsbad, CA, USA). RNA concentration was quantified with a Nanodrop Spectrophotometer (NanoDrop 2000, Nanodrop Technologies LLC, Thermo Fisher Scientific, Wilmington, DE, USA). Isolated RNA was stored at −80° C until processing. RNA was reverse-transcribed to cDNA by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s recommendations; quantitative real-time PCR was performed on cDNA using SYBR green PCR Master Mix (Applied Biosystems) on a StepOne Real-time PCR System (Applied Biosystems). Primer specificity was verified by observing a single peak on the melting curve. The fold change in expression of the target genes was calculated by the ΔΔCt method relative to β-actin as the endogenous control. GAPDH was used as an alternative endogenous control to verify the findings. Table 2 lists the primers used.
Table 2.
Primers for the genes examined.
| TSLP | F: TATCTGGTGCCCAGGCTATT; R:ACGCCACAATCCTTGTAATTG |
| TNF | F: CTGCTGCACTTTGGAGTGAT; R:GGTTTGCTACAACATGGGCTA |
| IL4 | F: CCGTATCCCCCTGACAATTA; R: ATCCCAGACTTCAGGGTGCT |
| CSF2 | F: CAGCCACTACAAGCAGCACT; R: AGCAGTCAAAGGGGATGACA |
| IL6 | F: AGTGAGGAACAAGCCAGAGC; R: GCATTTGTGGTTGGGTCAG |
| SPINK5 | F: GCAATGTGTGCTGAGCTGTT; R: ACTGGATCACTCTCCCGTGT |
| PTGS2 | F: CAGCACTTCACGCATCAGTT; R: ATCCTTGAAAAGGCGCAGT |
| CYP1A1 | F: GATTGAGCACTGTCAGGAGAAGC; R: ATGAGGCTCCAGGAGATAGCAG |
| ADAM33 | F: CGTTGCTGCTGCTGCTACTA; R: CCAGGGTTGTCCATCCAG |
| DPP10 | F: CATCAAGACATTCAGTTTCACCA; R: GCGTACTGCAAGACGGAGTC |
| ORMDL3 | F: TTGTGAGTGTCCCTGTCGTC; R: TGGACCCCATAATCCATCTG |
| IL13 | F: CATCACCCAGAACCAGAAGG; R: CTGCAGCCTGACACGTTGAT |
| TGF-β1 | F: GAGCCCTGGACACCAACTAT; R: GCAGAAGTTGGCATGGTAGC |
| β-actin | F: GCCACGGCTGCTTCCA; R: GAACCGCTCATTGCCATTG |
| GAPDH | F: CAGCCTCAAGATCATCAGCA; R: ACAGTCTTCTGGGTGGCAGT |
2.7. Immunoblot analysis
Cells were lysed in lysis buffer (PBS containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN) and Phosphatase Inhibitor I and II Cocktails (EMD Millipore, Billerica, MA). Protein concentration was determined by the DC Protein Assay (5000111, Biorad, Hercules, CA) and equal protein was loaded onto 4–20% tris-glycine gels, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were blocked with 5% bovine serum albumin before being probed with the following antibodies overnight at 4° C: thymic stromal lymphoprotein (TSLP, 1:1000, ab47943, Abcam, Cambridge, United Kingdom), prostaglandin-endoperoxide synthase 2 (PTGS2, 1:1000, Cell Signaling, Danvers, MA), cytochrome P450 1A1 (CYP1A1, 1:1000, sc-393979, Santa Cruz Biotechnology, Dallas, TX), ADAM33 (1:1000, ab137772, Abcam, Cambridge, United Kingdom), PARP (1:1000, Cell Signaling), or α-tubulin (1:5000, Sigma-Aldrich). Membranes were then incubated with appropriate secondary antibody conjugated to horseradish peroxidase (Cell Signaling) for 1 h at room temperature before developing with enhanced chemiluminescent reagent (GE Healthcare, Pittsburgh, PA). For all protein bands, densitometry was analyzed by Image J Software (NIH, Bethesda, MD) and normalized to α-tubulin.
2.8. Cell proliferation
Cells were plated at 2.5 × 104 cells per well in collagen-coated 96-well plates (Thermo Fisher Scientific) overnight in BEGM and then treated with PM2.5 (0–30 μg/cm2) for the indicated times. Cells were then washed and incubated for 60 min with 100 μl of CyQUANT NF dye (CyQUANT Proliferation Assay Kit, Thermo Fisher Scientific). Fluorescence was detected at 530 nm on the SpectraMax plate reader (Molecular Devices, Sunnyvale, CA). A separate plate of cells treated with PM2.5 (0–30 μg/cm2) for 30 min was also assayed by the CyQuant assay to obtain baseline values of fluorescence prior to cell proliferation.
2.9. ELISA
Supernatants from cells were frozen and collected for protein analysis by ELISA for IL-6, granulocyte-macrophage colony stimulating factor (GM-CSF), and IL-13. ELISA was performed at the University of Michigan Cancer Center Immunology Core.
2.10. Statistical analysis
Expression of each gene was expressed as the mean ± SEM. The effect of treatment on the expression of each gene of interest was tested by one-way ANOVA followed by Dunnett’s method to elucidate the pattern of significant effects (GraphPad Prism Software v7, La Jolla, CA). Differences with P < 0.05 were considered statistically significant.
3. Results
3.1. Treatment with PM2.5 did not increase cytotoxicity
The PM collected for our experiments was on the same day or similar days as several studies that comprehensively characterized the chemical composition of atmospheric PM2.5 in Beijing, China in late January-early February 2015. Those studies identified vehicle emissions as a major source of PM2.5 (Ji et al., 2018) and described the PM2.5 as consisting of high levels of SO42− and NO3−, elemental metals including Ag, As, Cd, Cu, Hg, Pb, Se, and Zn, and polyaromatic hydrocarbons including Benz(a)anthracene, Chrysene, and 1,8-Naphthalic anhydride (Ji et al., 2018; Niu et al., 2017). Levels of endotoxin in our particulate sample were 0.433 EU/ml in our highest treatment condition.
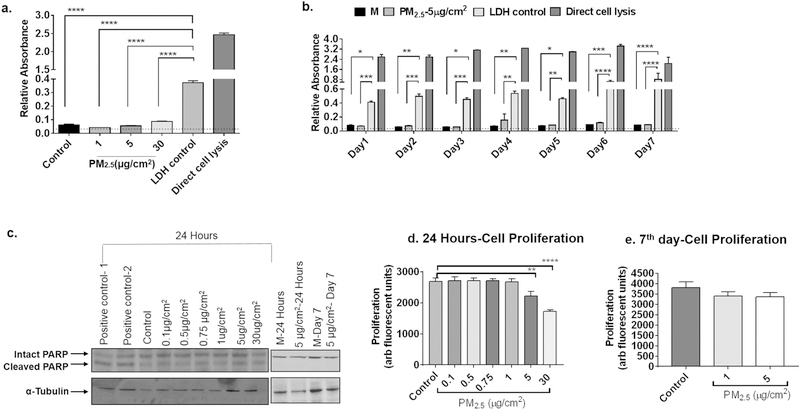
We first examined whether PM2.5 from Beijing, China caused cytotoxicity by treating BEAS-2B cells with 0, 1, 5 and 30 μg/cm2 of PM2.5 for 24 h and assaying levels of LDH in the supernatant. There was no significant increase in LDH levels from cells treated at any of the doses compared to both the LDH control provided by the assay manufacturer and the positive control from direct lysis of untreated cells (Fig. 1a). We also assessed for cytotoxicity in cells treated sequentially for 7 days with 5 μg/cm2 of PM2.5 and also did not observe an increase in LDH levels in the supernatants (Fig. 1b). To assess for apoptosis, cell lysates were immunoblotted for total and cleaved PARP, a marker of apoptosis. Levels of total PARP were similar for all of the treatments, with no increase in levels of cleaved PARP product at any of the treatment conditions (Fig. 1c and d). The morphology of cells treated for 24 h and 7 days were examined by brightfield microscopy and are shown in Supplemental Fig. 1. Microscopic images show relatively equal cell density and confluence among all of the conditions, and illustrate the distribution of different doses of PM2.5 in each well. Finally, we assayed cell proliferation using the CyQuant assay and noted that PM2.5 inhibited proliferation to a mild degree at a dose of 5 μg/cm2 and to a more moderate degree at a dose of 30 μg/cm2 when treated for 24 h (Fig. 1e), but not when treated at lower doses for 7 days (Fig. 1f). Baseline levels of fluorescence were also measured, and were not altered by the presence of PM2.5. These data demonstrate that at the doses and treatment durations assayed, PM2.5 did not appreciably affect apoptosis or cytotoxicity. These doses were chosen as the doses for all subsequent experiments examining gene expression.
Fig. 1.
Measures of cell cytotoxicity, apoptosis, and proliferation after PM2.5 exposure. a) BEAS-2B cells were treated for 24 h with medium (BEGM) alone or medium with 1, 5, or 30 μg/cm2 of PM2.5 and levels of lactate dehydrogenase (LDH) were assayed in cell supernatants (n = 3 independent experiments). Absorbance was measured relative to an LDH control provided by the assay manufacturer and direct lysis of cells by detergent. b) BEAS-2B cells were treated every day for the indicated number of days with BEGM (M) alone or BEGM with 5 μg/cm2 of PM2.5 and supernatants were assayed for levels of LDH (n = 3 independent experiments). c) Cell lysates were assayed for apoptosis by immunoblotting for intact and cleaved PARP in cells after 24 h of exposure and in cell exposed to PM2.5 daily for 7 days. Representative immunoblots from three independent experiments are shown. Positive controls were derived from fibroblast lysates treated with temozolamide, a known inducer of apoptosis. d and e) Cells were treated at the indicated concentrations of PM2.5 for 24 h (d) and daily for 7 days (e) and cell proliferation was assayed by Cy-Quant assay. Results are representative of two independent experiments. Statistical analysis was determined by ANOVA (*p < 0.05, **p < 0.01, ***p < 0.01, ****p < 0.001).
3.2. Gene expression changes after 24 h of PM2.5 exposure
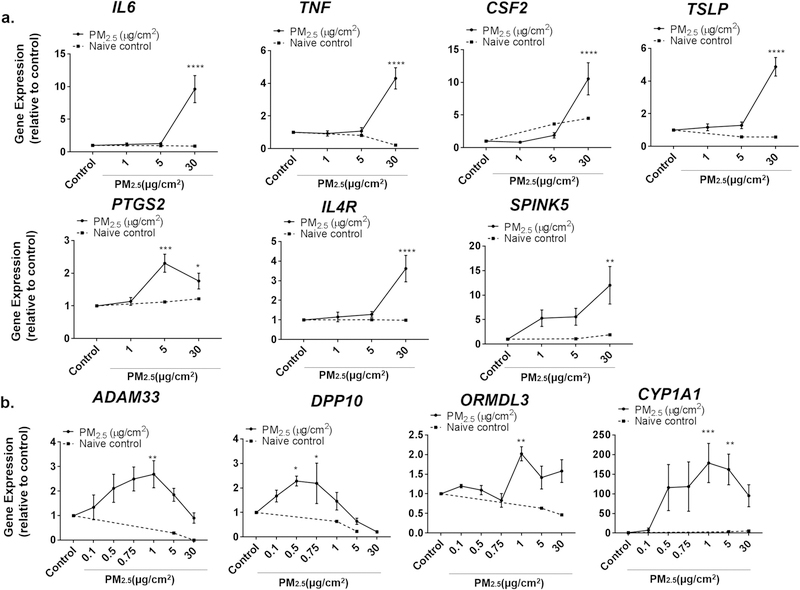
To examine the effects of short-term PM2.5 exposure on the gene expression of bronchial epithelial cells, we treated BEAS-2B cells with 0, 1, 5 and 30 μg/cm2 of PM2.5 for 24 h (Fig. 2). We first sought to examine the expression of specific genes expressed by epithelial cells that are associated with airway disorders, are often increased with inflammation, and which have been shown in other studies to be increased with exposure to high doses of PM2.5. This included examining the expression of the pro-inflammatory cytokines TNF-α (TNF) and IL-6 (IL6), and GM-CSF (CSF2) and TSLP, cytokines that are highly expressed by lung epithelium and that are associated with T-helper (Th) 2 responses and airway disorders. We also examined expression of PTGS2, an enzyme involved in prostaglandin synthesis that is often elevated in inflammation. Consistent with other studies (Boland et al., 2000; Longhin et al., 2016a; Ovrevik et al., 2009; Wang et al., 2017; Zhao et al., 2009), a high dose of PM2.5 (30 μg/cm2) increased the expression of all of these genes (Fig. 2a). This was consistent whether RNA levels were expressed relative to β-actin, or to GAPDH, another endogenous control (Supplemental Fig. 2). Use of samples from sonicated filters not previously exposed to PM2.5 did not significantly alter the expression of these genes. Given that PM2.5 has been associated with the development of allergic airway disorders such as asthma, we also examined the expression of IL-4 receptor (IL4R), a receptor that mediates Th2 responses and is highly associated by linkage analysis with asthma and allergic inflammation (Howard et al., 2002), and SPINK5, a gene whose product plays a critical role in mucosal barrier function and which has been shown by genome-wide association studies to be highly associated with asthma (Birben et al., 2012). Expression of IL4R and SPINK5 also increased at high doses of PM2.5 exposure (Fig. 2a).
Fig. 2.
Effect of various doses of PM2.5 on expression of different genes. a) The expression of IL6, TNF, CSF2, TSLP, PTGS2, IL4R and SPINK5 were assayed by RT-PCR from BEAS-2B cells after treatment with 0 (control), 1, 5, or 30 μg/cm2 of PM2.5 for 24 h. b) Lower doses of PM2.5 (≤1 μg/cm2) were used to examine the expression of ADAM33, DPP10, ORMDL3, and CYP1A1 in BEAS-2B cells by RT-PCR. Dotted lines represent relative expression of genes when cells were treated with liquid from sonication of naïve, unexposed filters at equal volumes as that used to dose PM2.5. Statistical significance was determined by ANOVA (*p < 0.05; **p < 0.01, ***p < 0.001, ****p < 0.0001, n ≥7 independent experiments for all genes examined).
Genome-wide association and positional cloning studies for asthma and COPD have recently identified several novel genes, including ADAM33, DPP10, and ORMDL3 as being important in disease pathogenesis. Variant polymorphisms in these genes result in their increased expression and susceptibility of individuals to asthma and COPD (Balantic et al., 2013; Holgate et al., 2006; Kim et al., 2015; Ono et al., 2014). As the effect of PM2.5 on the expression of these genes has not previously been reported, we sought to determine whether PM2.5 alters the expression of these genes in BEAS-2B cells. Interestingly, the expression of ADAM33, DPP10, and ORMDL3 all increased in a dose-dependent manner at a much lower dose range (Fig. 2b), but not at high doses of PM2.5. In fact, the maximal effect of PM2.5 occurred at a dose of 1 μg/cm2. We next examined the dose-response to PM2.5 of CYP1A1, a gene reported by other studies to be highly induced by PM2.5. The peak increase in CYP1A1 also occurred at a concentration of 1μg/cm2, with higher doses having less of an effect (Fig. 2b).
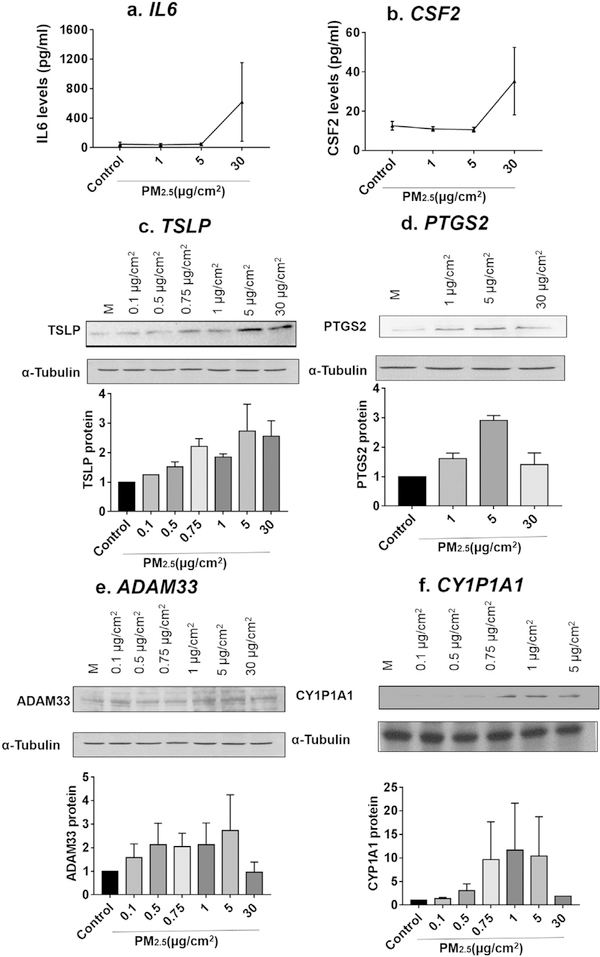
To ensure the changes in mRNA levels were also reflected by changes at the protein level, we performed ELISA for IL-6 and GM-CSF and observed an increase in their expression at doses that parallel increases in mRNA (Fig. 3a–b). Similarly, levels of TSLP, PTGS2, ADAM33, and CYP1A1 increased, as assayed by immunoblot, in a dose-dependent manner and these increases parallel the increase observed in mRNA (Fig. 3c–f). PM2.5 thus increases the expression of genes at both the mRNA and protein level.
Fig. 3.
Effect of PM2.5 on the protein expression of different genes. Supernatants from cells treated for 24 h at the indicated doses of PM2.5 were collected and assayed by ELISA for IL-6 (a, n= 3) and CSF2 (b, n = 3 independent experiments). Lysates from cells treated with PM2.5 were assayed by immunoblot for TSLP (c), PTGS2 (d), ADAM33 (e), and CYP1A1 (f). Representative immunoblots of three independent experiments are shown for each protein, with densitometric analysis shown beneath each blot.
3.3. Gene expression changes after repeat exposure of PM2.5 for seven days
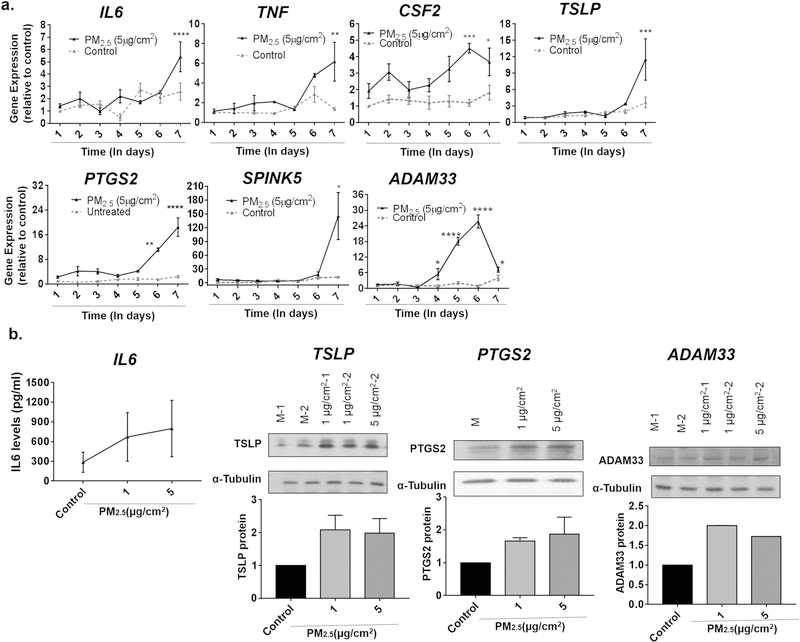
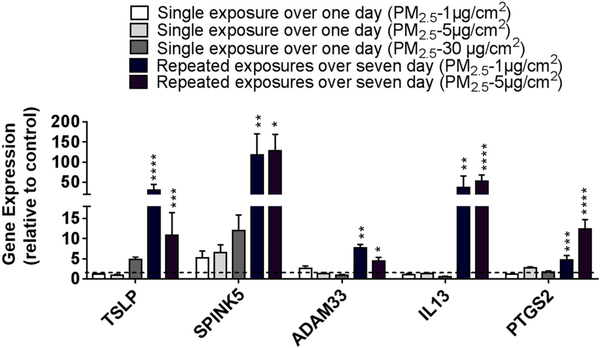
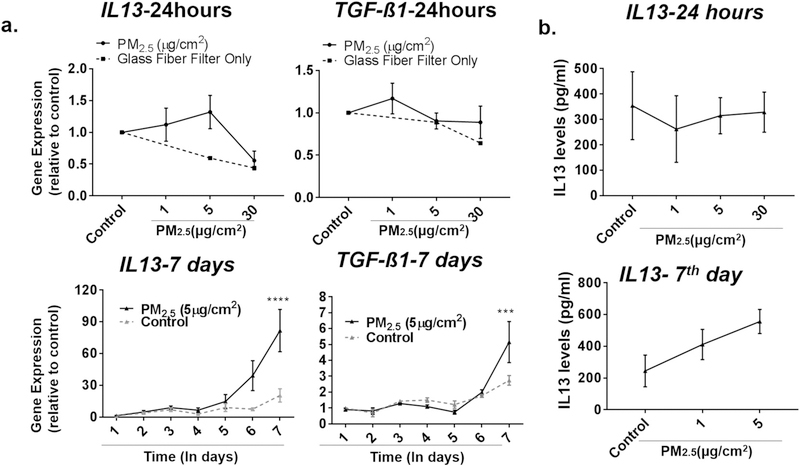
Although a single dose of PM2.5, often at a high dose, for 24 h was sufficient to increase the expression of a number of genes, repeated exposure to PM2.5, especially at lower doses, may better model pollution exposure of individuals in the general population. We thus treated BEAS-2B cells with repeated doses of 1 or 5 μg/cm2 of PM2.5 for seven consecutive days, washing the cells between each day to prevent accumulation, and examined the expression of the same genes we had previously examined after 24 h exposure. Inflammatory cytokines, such as IL6, TNF, TSLP, and CSF2, which previously were observed to only increase with a dose of 30 μg/cm2 of PM2.5, were increased when cells were exposed to lower doses of PM2.5 (5 μg/cm2) repeatedly for seven days (Fig. 4). Other genes such as PTGS2, SPINK5, and ADAM33 were also upregulated when exposed to PM2.5 at lower doses for consecutive days (Fig. 4). Although repeated doses of 5 μg/cm2 for seven days may lead one to consider this to be comparable to a concentration of ~35 μg/cm2, the magnitude of increase in gene expression was still higher in cells treated at lower doses over seven days compared to higher doses given in a single day. In fact, treatment with even just 1 μg/cm2 of PM2.5 repeated over seven days resulted in an increased of expression greater than any dose of PM2.5 given over a single day (Fig. 5). This was observed even when cells were washed with PBS between daily doses of PM2.5. Of note, the level of baseline expression of these genes did not change significantly over the seven days in culture. Finally, two genes, IL13 and transforming growth factor-β1 (TGFB1), which have also been shown to contribute to the development of allergic airway disease but were not increased after a single daily exposure to PM2.5, were noted to be increased after repeated exposures of PM2.5 over seven days (Fig. 6). These data suggest that chronic daily exposure to PM2.5, even at low doses, may have profound effects on gene expression that may not be captured solely by experiments of brief exposure. A summary of the gene expression changes after both one day and seven days of repeated exposure is included in Table 3.
Fig. 4.
Gene expression changes after repeated exposure to PM2.5 for seven days. a) BEAS-2B cells were treated with medium alone (control) or 5 μg/cm2 of PM2.5 on a daily basis for seven days and IL6, TNF, CSF2, TSLP, PTGS2, SPINK5 and ADAM33 were examined by RT-PCR. Statistical significance was determined by ANOVA (*p < 0.05; **p < 0.01, ***p < 0.001, ****p < 0.0001, n ≥5 for all genes examined). b) Supernatants from cells treated with PM2.5 on a daily basis for seven days were assayed for IL-6 by ELISA (n = 3 independent experiments). Cell lysates from cells treated daily for seven days with PM2.5 were immunoblotted for TLSP, PTGS2, and ADAM33. Representative blots from three independent experiments are shown.
Fig. 5.
Comparison of gene expression changes in cells treated with PM2.5 at different doses and durations. BEAS-2B cells were treated for one day or daily for seven days with PM2.5 at the indicated concentrations. Gene expression was assayed by RT-PCR and expressed relative to control cells with no PM2.5 exposure. Statistical significance was determined by ANOVA (**p < 0.01, ***p < 0.001, ***p < 0.001, n ≥5 independent experiments for all genes examined).
Fig. 6.
Expression of IL13 and TGFB1 increased after repeated, but not single, day of PM2.5 exposure. a) Top graphs, BEAS-2B cells were treated with 0 (control), 1, 5, or 30 μg/cm2 of PM2.5 for 24 h and IL13 and TGFB1 expression were assayed by RT-PCR. Bottom graphs, BEAS-2B cells were treated with control or 5 μg/cm2 of PM2.5 for consecutive days and IL13 and TGFB1 expression at each day were assayed by RT-PCR. Statistical significance was determined by ANOVA (***p < 0.001, ****p < 0.0001, n≥ 5 for all genes examined) b) IL-13 was assayed by ELISA in cells treated with PM2.5 for 24 h (top graph) and daily for seven days (bottom graph) (n = 3 independent experiments).
Table 3.
Summary of the response for various genes to different doses and duration of PM2.5 exposure.
| Genes | Single exposure over one day |
Repeated exposure over 7 days |
||||
|---|---|---|---|---|---|---|
| ≤ 1 μg/cm2 | 1 μg/cm2 | 5 μg/cm2 | 30 μg/cm2 | 1 μg/cm2 | 5 μg/cm2 | |
| TSLP | – | – | – | ↑ | ↑↑↑ | ↑↑ |
| TNF | – | – | – | ↑ | – | ↑↑ |
| IL4R | – | – | – | ↑ | – | – |
| CSF2 | – | – | – | ↑↑ | – | ↑ |
| IL6 | – | – | – | ↑↑ | – | ↑ |
| SPINK5 | – | – | – | ↑↑ | ↑↑↑↑ | ↑↑↑↑ |
| PTGS2 | – | – | ↑ | ↑ | ↑ | ↑↑ |
| CYP1A1 | – | ↑↑↑↑ | ↑↑↑↑ | – | – | – |
| ADAM33 | – | ↑ | – | – | ↑↑ | ↑↑ |
| DPP10 | ↑ | – | – | – | – | – |
| ORMDL3 | – | ↑ | – | – | – | – |
| IL13 | – | – | – | – | ↑↑↑ | ↑↑↑↑ |
| TGFβ | – | – | – | – | – | ↑ |
Gene Expression (Relative to Control): ↑ 1.7–6 fold; ↑↑ 7–20 fold; ↑↑↑ 20–50 fold; ↑↑↑↑ > 50 fold.
3.4. PM2.5 induced the expression of many genes via diverse signaling pathways
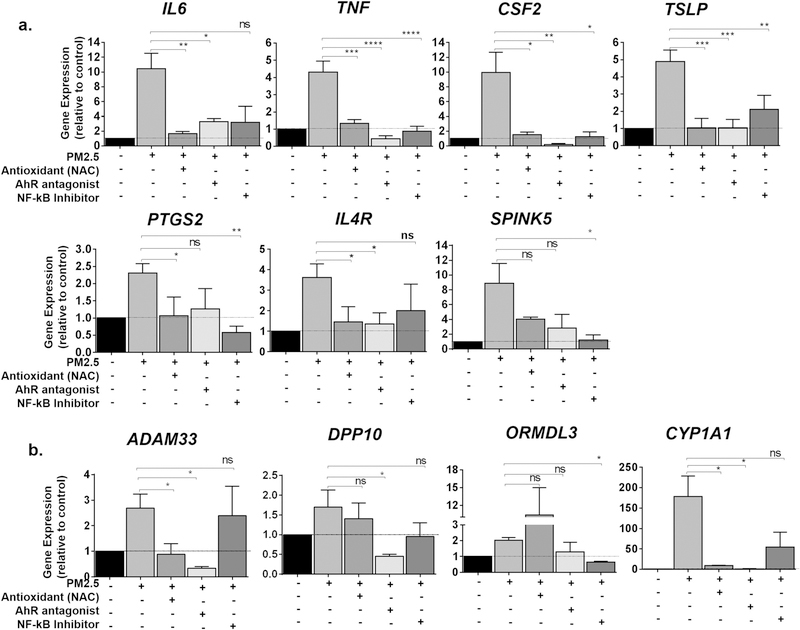
PM2.5 consists of free radicals, metal ions, and organic compounds that generate reactive oxygen species (ROS). These in turn can activate several transcription factors, including NF-κB (Quay et al., 1998; Silbajoris et al., 2011). Polyaromatic hydrocarbons (PAH) from PM2.5 also activate AhR, which itself is a transcription factor that activates many genes (Andrysik et al., 2011; Ferecatu et al., 2010). Given that we observed different genes to be upregulated by different doses and kinetics of PM2.5 exposure, we sought to examine which of these signaling pathways may be responsible for the increase in each particular gene. Cells were pre-treated with either NAC to diminish ROS production, the NF-κB inhibitor Bay11–7082, or the AhR inhibitor CH223191, before subsequent treatment with 24 h of PM2.5 and expression of genes were examined by RT-PCR.
For many of the inflammatory cytokines that were upregulated by a single high dose of PM2.5, use of either NAC, Bay11–7082, or CH223191 was effective in decreasing the expression of each gene (Fig. 7). That use of each inhibitor alone was sufficient to block the increase in the expression of these genes suggests that the increase in expression of these genes may depend on the integrated action of ROS, NF-κB, and AhR signaling, rather than the actions of each of these pathways alone.
Fig. 7.
Effect of the antioxidant N-acetylcysteine (NAC) and inhibitors to aryl hydrocarbon receptor (AhR) and nuclear factor (NF)-κB in the expression of genes after PM2.5. BEAS-2B cells were pretreated for 1 h with either the NF-κB inhibitor Bay11–7082 (BAY) (10 μM), the AhR antagonist CH223191 (10 μM), or NAC (100 μM) and exposed to 30 μg/cm2 (a) or 1 μg/cm2 (b) of PM2.5 for 24 h. a) Expression of IL6, TNF, CSF2, TSLP, PTGS2, IL4R and SPINK5 were assayed by RT-PCR. b) Expression of ADAM33, DPP10, ORMDL3, and CYP1A1 were assayed by RT-PCR. Statistical significance was determined by ANOVA (*p < 0.05; **p < 0.01, ***p < 0.001, n ≥ 5 independent experiments for all genes examined).
As noted earlier, ADAM33, SPINK5, and ORMDL3 were genes identified by positional cloning and genome-wide association studies as having genetic variants important to asthma and COPD, and whose expression increased with much lower doses of PM2.5. Although NAC and the AhR antagonist CH223191 were sufficient in blocking the increase in ADAM33 by PM2.5, the NF-κB antagonist Bay11–7082 had no effect. Conversely, ORMDL3 was effectively inhibited by Bay11–7082, but its expression, if anything, was increased by NAC. Although all of the pre-treatments inhibited the expression of SPINK5, the effects among the different compounds were variable. Finally, CYP1A1, a known target of AhR, was effectively inhibited by the AhR antagonist, with only intermediate response to Bay11–7082. These data suggest that different signaling pathways, especially during low doses of exposure, are responsible for the expression of different genes in bronchial epithelial cells (Table 4).
Table 4.
Ability of an antioxidant or various inhibitors to attenuate the effects of PM2.5 on different genes.
| Gene | PM2.5 | NAC | AhR antagonist | NFk-B Inhibitor |
|---|---|---|---|---|
| TSLP | 30 μg/cm2 | ✓ | ✓ | ✓ |
| TNF | 30 μg/cm2 | ✓ | ✓ | ✓ |
| CSF2 | 30 μg/cm2 | ✓ | ✓ | ✓ |
| IL4R | 30 μg/cm2 | ✓ | ✓ | @ |
| I16 | 30 μg/cm2 | ✓ | ✓ | @ |
| CYP1A1 | 1 μg/cm2 | ✓ | ✓ | @ |
| PTGS2 | 5 μg/cm2 | ✓ | @ | ✓ |
| ADAM33 | 1 μg/cm2 | ✓ | ✓ | - |
| SPINK5 | 30 μg/cm2 | @ | @ | ✓ |
| ORMDL3 | 1 μg/cm2 | - | @ | ✓ |
✓ = complete inhibition; @ = partial inhibition; - no effect.
4. Discussion
Air pollution contributes an estimated 3.3 million premature deaths per year worldwide and the number of deaths from pollution will double by 2050 if the issue remains unattended (Lelieveld et al., 2015). PM2.5 is one of the most important components of pollution based on its size, composition, and toxicity. Although the effects of PM2.5 on human health are well described (Karakatsani et al., 2012; Strak et al., 2012; Xing et al., 2016), the mechanisms of its deleterious actions are not fully understood. Here, we examine how different doses and duration of PM2.5 exposure affect the expression of select genes, chosen because of their pathobiological relevance to airway disorders including asthma and COPD. Consistent with that observed in other studies (Boggaram et al., 2016; Boland et al., 2000; Boublil et al., 2013; Longhin et al., 2016a; Ovrevik et al., 2009; Quay et al., 1998; Wang et al., 2017), the inflammatory cytokines IL6, TNF, CSF2, and TSLP all increased with 24 h exposure to 30 μg/cm2 of PM2.5. However, other genes including ADAM33, DPP10, ORMDL3, and SPINK5 that are relevant to asthma and COPD based on genetic studies (Balantic et al., 2013; Birben et al., 2012; Holgate et al., 2006; Kim et al., 2015; Ono et al., 2014), but have not been studied in the context of pollution, were also increased by PM2.5 exposure. Interestingly, the responsiveness of these genes occurred at a much lower dose (≤1 μg/cm2) of exposure. Sequential treatment of cells with PM2.5 over seven days enhanced the magnitude of gene expression changes for all of the genes. These data demonstrate that PM2.5 exerts pleiotropic actions that differ by dose and duration and that these variables are important in considering the implications and mechanisms PM2.5 has in disease.
Lung diseases such as COPD and asthma are characterized by chronic inflammation and the generation of pro-inflammatory cytokines by PM2.5 may be one explanation for how PM2.5 contributes to the development of these disorders. PM2.5 increased not only IL6 and TNF, but also TSLP and CSF2, genes linked with asthma that are associated with a Th2 phenotype. We show for the first time that PM2.5 also increased expression of IL4R, which also mediates Th2 responses. However, other genes, such as ADAM33, DPP10, SPINK5, and ORMDL3, code for proteins involved in epithelial barrier function and mucosal integrity and genetic studies have shown that polymorphisms in these genes also contribute to the development of asthma and COPD (Balantic et al., 2013; Birben et al., 2012; Holgate et al., 2006; Kim et al., 2015; Ono et al., 2014). To our knowledge, the effect of PM2.5 on the expression of these genes was previously unknown. ADAM33 codes for a member of the disintegrin and metalloprotease family, DPP10 is a member of a family of serine proteases, and SPINK5 is a serine protease inhibitor. Collectively, increased expression of these genes may lead to epithelial barrier disruption, which could allow PM2.5 to affect other cells in the submucosa or interstitial layer. That these genes were also upregulated by PM2.5, especially at low doses (1μg/cm2), suggest that even low levels of PM2.5 exposure may contribute to asthma and COPD development through diverse mechanisms. Finally, the presence of certain single nucleotide polymorphisms may further amplify (or negate) the effect of PM2.5 on these genes. Pollution has been shown to increase the risk susceptibility of ADAM33 polymorphism rs597980 for asthma (Tripathi et al., 2014). Smoking increases the relative risks for asthma in individuals with genetic variants rs12603332 and rs4065275 for ORMDL3. These studies demonstrate how pollution, combined with genetics, can affect susceptibility to disease (Song et al., 2017).
As individuals are often exposed to ambient air pollution over long periods on a daily basis, we exposed bronchial epithelial cells to lower doses of PM2.5 repeatedly over seven days and compared the effects to cells exposed over 24 h. Although the data suggest that long-term exposure increases the sensitivity of cells to lower doses, we recognize that PM2.5 may accumulate within cells over time even despite washing cells between treatment days, and that daily exposure to 5 μg/cm2 over seven days compared to ~35 μg/cm2 in a single day may be a more appropriate comparison. Nonetheless, the magnitude of increase in cells treated over seven days with 5 μg/cm2, or even 1 μg/cm2, was still higher than single doses of 30 μg/cm2 for most all of the genes studied, including TSLP, TNF, CSF2, IL6, SPINK5, PTGS2, and ADAM33. Additionally, two genes, IL13 and TGFB1, which were unchanged after 24 h of PM2.5 exposure at any of the doses studied, were upregulated only when cells were treated for 7 days. Both IL13 and TGFB1 are implicated in airway disorders such as asthma and COPD as well (Aschner and Downey, 2016; Brightling et al., 2010; Howard et al., 2002). Other investigators have conducted similar experiments of treating cells with lower doses (2.5 μg/cm2) over two weeks and also observed an increase in gene expression of IL-6, IL-8, CYP1A1, and COX-2 after longer exposure periods (Longhin et al., 2016b). Repeated exposures of 1, 5, and 10 μg/cm2 for 4 h each for up to 5 weeks also demonstrated a persistent and sustained increase in GM-CSF and IL-6 expression (Boublil et al., 2013).
We did not observe significant cytotoxicity or cellular apoptosis at any of the treatment doses, but did note a decrease in cell proliferation, especially at higher doses (e.g. 30 μg/cm2). Some investigators dose PM in in vitro experiments based on concentration, and for the volume of medium we used in our experiments, the doses of 1 μg/cm2, 5 μg/cm2, and 30 μg/cm2 were equivalent to 4.5 μg/ml, 22.5 μg/ml, and 135 μg/ml, respectively. Microscopic photographs of the distribution of different concentrations of PM2.5 in the treatment well are shown in Supplemental Fig. 1. Higher doses (e.g. > 30 μg/cm2, or 250–500 μg/ml) have been utilized to demonstrate increased cell toxicity, mitochondrial injury (Dergham et al., 2015; Lavrich et al., 2018; Niu et al., 2017; Sayes et al., 2007; Seriani et al., 2016) and DNA damage (Yang et al., 2016), with doses of 7.5 μg/cm2 contributing to cell-cycle arrest (Longhin et al., 2013). We specifically chose lower doses for our experiments to examine the effects of PM2.5 at sub-lethal conditions. The lower doses we used approximate those employed in more recent studies that focus on transcriptomic profiling (Longhin et al., 2016a) and other epithelial cell functions (Boublil et al., 2013; Longhin et al., 2016b; Longhin et al., 2013), including miRNA transcripts (Borgie et al., 2015; Longhin et al., 2016a). Indeed, the increase in inflammatory cytokines at the doses we observed are congruent with these other studies; however, we also made a point of emphasis to examine the effects the effects of PM2.5 at doses < 1 μg/cm2, which haven’t been routinely examined by other investigators and which upregulated expression of other important genes. The importance of experimentally examining lower doses is further supported by a recent study that demonstrated how levels of PM2.5 exposure even below National Ambient Air Quality Standards in the United States continue to exert a dose-dependent risk for mortality (Di et al., 2017).
A limitation in our study is that we performed experiments on cultured cells submerged in medium, but increasing data suggest that bronchial epithelial cells grown in an air-liquid interface (ALI) may better approximate physiologic conditions (Upadhyay and Palmberg, 2018). Doses of PM2.5 for in vitro experiments utilizing culture medium are often chosen based on similar in vitro to in vivo toxicity profiles (Sayes et al., 2007) and/or mathematical estimates of in vivo to in vitro exposure (Teeguarden et al., 2007). However, for experiments utilizing ALI, pollutants can be aerosolized, which may allow for dosing that more accurately approximates in vivo exposure, while also limiting the risk of mechanical or chemical transformation of PM2.5 during solubilization in water or alcohol (Upadhyay and Palmberg, 2018). Aerosolized PM2.5 may also mitigate clumping or conglomeration of PM2.5 that may occur when PM2.5 is resuspended in a liquid medium. ALI also allows primary bronchial epithelial cells to undergo transformation to columnar epithelium, which may also serve as a more representative cell in the airway. We utilized BEAS-2B cells, a normal bronchial epithelial cell line that is transformed by virus, and whether our observations can extend to primary patient derived cells, including those with pre-existing asthma or COPD, is unknown. Finally, several studies have shown how growth of bronchial epithelial cells in ALI alters the response to PM2.5 exposure (Boublil et al., 2013; Ghio et al., 2013).
The toxicity of urban PM2.5 often derives from both inorganic metals and organic pollutants, including volatile organic compounds and PAH, which together, often promote the formation of ROS that induce oxidant damage. The composition of PM2.5 from Beijing in the winter of 2015, which is when our samples were obtained, has been well described and are notable for particularly high levels of SO42− and NO3−, elemental metals including Ag, As, Cd, Cu, Hg, Pb, Se, and Zn, and polyaromatic hydrocarbons including Benz(a)anthracene, Chrysene, and 1,8-Naphthalic anhydride (Ji et al., 2018; Niu et al., 2017). ROS are capable of activating several signaling pathways, both dependent and independent of the transcription factor NF-κB (Quay et al., 1998; Silbajoris et al., 2011). PAH can also directly activate the AhR, itself a transcription factor (Baeza-Squiban et al., 1999; Lee et al., 2016; Tao et al., 2003). Given that we observed different time and dose-dependent responses to PM2.5 for various genes, we sought to determine whether the response of different genes is dependent on NF-κB, AhR, or oxidant signaling in general. CYP1A1 is a well-described target of AhR transcriptional activity, and served as a useful control whose expression was upregulated by PM2.5 and inhibited by the AhR antagonist, CH-223191. Genes such as IL6, TNF, TSLP, CSF2, PTGS2, and IL4R appear to be broadly inhibited by the presence of either NAC, the NF-κB inhibitor, or the AhR inhibitor alone. These data suggest that these pathways overlap, and that inhibition of either oxidants in general (via NAC) or of individual transcription factors NF-κB and AhR was sufficient to inhibit the effects of PM2.5 on the transcription of these genes. On the other hand, it was interesting to note that the NF-κB inhibitor had no effect on ADAM33 expression while it was most sensitive in inhibiting SPINK5. Although the NF-κB and AhR inhibitor both inhibited ORMDL3 expression, ORMDL3 actually increased in the presence of NAC. The sensitivity of response among different genes also appeared to vary with different inhibitors. Finally, the increased expression of some genes may be a result of endotoxin that we detected in our PM2.5 sample. A more comprehensive approach for each gene is needed to understand how PM2.5 affects its expression, but these data suggest that the expression of different genes, especially those increased by low-dose PM2.5, depend on the activation of unique signaling pathways.
In conclusion, we show that varying dose and duration of PM2.5 exposure result in increased gene expression of different genes relevant to airway remodeling and respiratory disorders. PM2.5 induced the expression of ADAM33, DPP10, and ORMDL3 at considerably lower concentrations than that required for inflammatory cytokines, including IL6, TNF, CSF2, TSLP, PTGS2, and IL4R. The expression of some of these genes, including IL4R, SPINK5, ADAM33, DPP10, and ORMDL3 have not previously been shown to be upregulated by PM2.5 in bronchial epithelial cells, but their role in the genetics of asthma and COPD helps to identify a means by which gene-environment interactions may contribute to disease development. The effects of PM2.5 are dependent on the oxidative tone and the activation of AhR and NF-κB, which may overlap, but also may have independent functions. These data emphasizes the need to study a range of doses to determine how even low levels of pollution may adversely affect health.
Supplementary Material
Acknowledgments
This work was supported by the University of Michigan-Peking University Joint Institute and from the National Institutes of Health (grant number HL127203).
Funding
This work was supported by the University of Michigan Health System-Peking University Health Science Center Joint Institute for Translational and Clinical Research (BMU 20140481) and from the National Institutes of Health (grant number HL127203).
Footnotes
All authors declare no conflicts of interest.
Supplementary data to this article can be found online at http://doi.org/10.1016/j.tiv.2018.05.004.
References
- Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, Tinsley J, Zhang Y, Holt R, Jones EY, Lench N, Carey A, Jones H, Dickens NJ, Dimon C, Nicholls R, Baker C, Xue L, Townsend E, Kabesch M, Weiland SK, Carr D, von Mutius E, Adcock IM, Barnes PJ, Lathrop GM, Edwards M, Moffatt MF, Cookson WO, 2003. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat. Genet 35, 258–263. [DOI] [PubMed] [Google Scholar]
- Andrysik Z, Vondracek J, Marvanova S, Ciganek M, Neca J, Pencikova K, Mahadevan B, Topinka J, Baird WM, Kozubik A, Machala M, 2011. Activation of the aryl hydrocarbon receptor is the major toxic mode of action of an organic extract of a reference urban dust particulate matter mixture: the role of polycyclic aromatic hydrocarbons. Mutat. Res 714, 53–62. [DOI] [PubMed] [Google Scholar]
- Aschner Y, Downey GP, 2016. Transforming growth factor-beta: master regulator of the respiratory system in health and disease. Am. J. Respir. Cell Mol. Biol 54, 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza-Squiban A, Bonvallot V, Boland S, Marano F, 1999. Airborne particles evoke an inflammatory response in human airway epithelium. Activation of transcription factors. Cell Biol. Toxicol 15, 375–380. [DOI] [PubMed] [Google Scholar]
- Balantic M, Rijavec M, Flezar M, Camlek T, Hudoklin I, Kosnik M, Korosec P, Suskovic S, 2013. A polymorphism in ORMDL3 is associated not only with asthma without rhinitis but also with chronic obstructive pulmonary disease. J Investig Allergol Clin Immunol 23, 256–261. [PubMed] [Google Scholar]
- Birben E, Sackesen C, Turgutoglu N, Kalayci O, 2012. The role of SPINK5 in asthma related physiological events in the airway epithelium. Respir. Med 106, 349–355. [DOI] [PubMed] [Google Scholar]
- Boggaram V, Loose DS, Gottipati KR, Natarajan K, Mitchell CT, 2016. Gene expression profiling of the effects of organic dust in lung epithelial and THP-1 cells reveals inductive effects on inflammatory and immune response genes. Physiol. Genomics 48, 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland S, Bonvallot V, Fournier T, Baeza-Squiban A, Aubier M, Marano F, 2000. Mechanisms of GM-CSF increase by diesel exhaust particles in human airway epithelial cells. Am. J. Phys. Lung Cell. Mol. Phys 278, L25–L32. [DOI] [PubMed] [Google Scholar]
- Borgie M, Ledoux F, Verdin A, Cazier F, Greige H, Shirali P, Courcot D, Dagher Z, 2015. Genotoxic and epigenotoxic effects of fine particulate matter from rural and urban sites in Lebanon on human bronchial epithelial cells. Environ. Res 136, 352–362. [DOI] [PubMed] [Google Scholar]
- Boublil L, Assemat E, Borot MC, Boland S, Martinon L, Sciare J, Baeza-Squiban A, 2013. Development of a repeated exposure protocol of human bronchial epithelium in vitro to study the long-term effects of atmospheric particles. Toxicol. in Vitro 27, 533–542. [DOI] [PubMed] [Google Scholar]
- Brightling CE, Saha S, Hollins F, 2010. Interleukin-13: prospects for new treatments. Clin. Exp. Allergy 40, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LC, Lippmann M, 2009. Effects of metals within ambient air particulate matter (PM) on human health. Inhal. Toxicol 21, 1–31. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wang JN, Ma GX, Zhang YS, 2013. China tackles the health effects of air pollution. Lancet 382, 1959–1960. [DOI] [PubMed] [Google Scholar]
- Dergham M, Lepers C, Verdin A, Cazier F, Billet S, Courcot D, Shirali P, Garcon G, 2015. Temporal-spatial variations of the physicochemical characteristics of air pollution Particulate Matter (PM2.5–0.3) and toxicological effects in human bronchial epithelial cells (BEAS-2B). Environ. Res 137, 256–267. [DOI] [PubMed] [Google Scholar]
- Di Q, Wang Y, Zanobetti A, Wang Y, Koutrakis P, Choirat C, Dominici F, Schwartz JD, 2017. Air pollution and mortality in the medicare population. N. Engl. J. Med 376, 2513–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Wang M, Chu H, Chu M, Na T, Wen Y, Wu D, Han B, Bai Z, Chen W, Yuan J, Wu T, Hu Z, Zhang Z, Shen H, 2014. Global gene expression profiling of human bronchial epithelial cells exposed to airborne fine particulate matter collected from Wuhan, China. Toxicol. Lett 228, 25–33. [DOI] [PubMed] [Google Scholar]
- Ferecatu I, Borot MC, Bossard C, Leroux M, Boggetto N, Marano F, Baeza-Squiban A, Andreau K, 2010. Polycyclic aromatic hydrocarbon components contribute to the mitochondria-antiapoptotic effect of fine particulate matter on human bronchial epithelial cells via the aryl hydrocarbon receptor. Part Fibre Toxicol. 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghio AJ, Dailey LA, Soukup JM, Stonehuerner J, Richards JH, Devlin RB, 2013. Growth of human bronchial epithelial cells at an air-liquid interface alters the response to particle exposure. Part Fibre Toxicol. 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff DW, Schmitt MT, Dailey LA, Duvall RM, Karoly ED, Devlin RB, 2007. Assessing the role of particulate matter size and composition on gene expression in pulmonary cells. Inhal. Toxicol 19 (Suppl. 1), 23–28. [DOI] [PubMed] [Google Scholar]
- Holgate ST, Davies DE, Powell RM, Holloway JW, 2006. ADAM33: a newly identified protease involved in airway remodelling. Pulm. Pharmacol. Ther 19, 3–11. [DOI] [PubMed] [Google Scholar]
- Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER, 2002. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am. J. Hum. Genet 70, 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, 2013. The role of in vitro gene expression profiling in particulate matter health research. J. Toxicol. Environ. Health B Crit. Rev 16, 381–394. [DOI] [PubMed] [Google Scholar]
- Ji D, Cui Y, Li L, He J, Wang L, Zhang H, Wang W, Zhou L, Maenhaut W, Wen T, Wang Y, 2018. Characterization and source identification of fine particulate matter in urban Beijing during the 2015 spring festival. Sci. Total Environ 628–629, 430–440. [DOI] [PubMed] [Google Scholar]
- Karakatsani A, Analitis A, Perifanou D, Ayres JG, Harrison RM, Kotronarou A, Kavouras IG, Pekkanen J, Hameri K, Kos GP, de Hartog JJ, Hoek G, Katsouyanni K, 2012. Particulate matter air pollution and respiratory symptoms in individuals having either asthma or chronic obstructive pulmonary disease: a European multicentre panel study. Environ. Health 11, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Choi H, Yoon MG, Ye YM, Park HS, 2015. Dipeptidyl-peptidase 10 as a genetic biomarker for the aspirin-exacerbated respiratory disease phenotype. Ann Allergy Asthma Immunol 114, 208–213. [DOI] [PubMed] [Google Scholar]
- Lavrich KS, Corteselli EM, Wages PA, Bromberg PA, Simmons SO, Gibbs-Flournoy EA, Samet JM, 2018. Investigating mitochondrial dysfunction in human lung cells exposed to redox-active PM components. Toxicol. Appl. Pharmacol 342, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Lin ZC, Hu SC, Chiang YC, Hsu LF, Lin YC, Lee IT, Tsai MH, Fang JY, 2016. Urban particulate matter down-regulates filaggrin via COX2 expression/PGE2 production leading to skin barrier dysfunction. Sci. Rep 6, 27995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelieveld J, Evans JS, Fnais M, Giannadaki D, Pozzer A, 2015. The contribution of outdoor air pollution sources to premature mortality on a global scale. Nature 525, 367–371. [DOI] [PubMed] [Google Scholar]
- Li Y, Duan J, Yang M, Li Y, Jing L, Yu Y, Wang J, Sun Z, 2017. Transcriptomic analyses of human bronchial epithelial cells BEAS-2B exposed to atmospheric fine particulate matter PM2.5. Toxicol. in Vitro 42, 171–181. [DOI] [PubMed] [Google Scholar]
- Longhin E, Holme JA, Gutzkow KB, Arlt VM, Kucab JE, Camatini M, Gualtieri M, 2013. Cell cycle alterations induced by urban PM2.5 in bronchial epithelial cells: characterization of the process and possible mechanisms involved. Part Fibre Toxicol 10, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhin E, Capasso L, Battaglia C, Proverbio MC, Cosentino C, Cifola I, Mangano E, Camatini M, Gualtieri M, 2016a. Integrative transcriptomic and protein analysis of human bronchial BEAS-2B exposed to seasonal urban particulate matter. Environ. Pollut 209, 87–98. [DOI] [PubMed] [Google Scholar]
- Longhin E, Gualtieri M, Capasso L, Bengalli R, Mollerup S, Holme JA, Ovrevik J, Casadei S, Di Benedetto C, Parenti P, Camatini M, 2016b. Physico-chemical properties and biological effects of diesel and biomass particles. Environ. Pollut 215, 366–375. [DOI] [PubMed] [Google Scholar]
- Niu X, Ho SSH, Ho KF, Huang Y, Sun J, Wang Q, Zhou Y, Zhao Z, Cao J, 2017. Atmospheric levels and cytotoxicity of polycyclic aromatic hydrocarbons and oxygenated-PAHs in PM2.5 in the Beijing-Tianjin-Hebei region. Environ. Pollut 231, 1075–1084. [DOI] [PubMed] [Google Scholar]
- Ono JG, Worgall TS, Worgall S, 2014. 17q21 locus and ORMDL3: an increased risk for childhood asthma. Pediatr. Res 75, 165–170. [DOI] [PubMed] [Google Scholar]
- Ovrevik J, Lag M, Holme JA, Schwarze PE, Refsnes M, 2009. Cytokine and chemokine expression patterns in lung epithelial cells exposed to components characteristic of particulate air pollution. Toxicology 259, 46–53. [DOI] [PubMed] [Google Scholar]
- Quay JL, Reed W, Samet J, Devlin RB, 1998. Air pollution particles induce IL-6 gene expression in human airway epithelial cells via NF-kappaB activation. Am. J. Respir. Cell Mol. Biol 19, 98–106. [DOI] [PubMed] [Google Scholar]
- Sayes CM, Reed KL, Warheit DB, 2007. Assessing toxicity of fine and nanoparticles: comparing in vitro measurements to in vivo pulmonary toxicity profiles. Toxicol. Sci 97, 163–180. [DOI] [PubMed] [Google Scholar]
- Schwarze PE, Ovrevik J, Lag M, Refsnes M, Nafstad P, Hetland RB, Dybing E, 2006. Particulate matter properties and health effects: consistency of epidemiological and toxicological studies. Hum. Exp. Toxicol 25, 559–579. [DOI] [PubMed] [Google Scholar]
- Seriani R, de Souza CE, Krempel PG, Frias DP, Matsuda M, Correia AT, Ferreira MZ, Alencar AM, Negri EM, Saldiva PH, Mauad T, Macchione M, 2016. Human bronchial epithelial cells exposed in vitro to diesel exhaust particles exhibit alterations in cell rheology and cytotoxicity associated with decrease in antioxidant defenses and imbalance in pro- and anti-apoptotic gene expression. Environ. Sci. Pollut. Res. Int 23, 9862–9870. [DOI] [PubMed] [Google Scholar]
- Silbajoris R, Osornio-Vargas AR, Simmons SO, Reed W, Bromberg PA, Dailey LA, Samet JM, 2011. Ambient particulate matter induces interleukin-8 expression through an alternative NF-kappaB (nuclear factor-kappa B) mechanism in human airway epithelial cells. Environ. Health Perspect 119, 1379–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Schwager MJ, Backer V, Guo J, Porsbjerg C, Khoo SK, Laing IA, Moses EK, LeSouef P, Zhang GB, 2017. Environment changes genetic effects on respiratory conditions and allergic phenotypes. Sci. Rep 7, 6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strak M, Janssen NA, Godri KJ, Gosens I, Mudway IS, Cassee FR, Lebret E, Kelly FJ, Harrison RM, Brunekreef B, Steenhof M, Hoek G, 2012. Respiratory health effects of airborne particulate matter: the role of particle size, composition, and oxidative potential-the RAPTES project. Environ. Health Perspect 120, 1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao F, Gonzalez-Flecha B, Kobzik L, 2003. Reactive oxygen species in pulmonary inflammation by ambient particulates. Free Radic. Biol. Med 35, 327–340. [DOI] [PubMed] [Google Scholar]
- Teeguarden JG, Hinderliter PM, Orr G, Thrall BD, Pounds JG, 2007. Particokinetics in vitro: dosimetry considerations for in vitro nanoparticle toxicity assessments. Toxicol. Sci 95, 300–312. [DOI] [PubMed] [Google Scholar]
- Tolbert PE, 2007. Invited commentary: heterogeneity of particulate matter health risks. Am. J. Epidemiol 166, 889–891 (discussion 892–883). [DOI] [PubMed] [Google Scholar]
- Tripathi P, Awasthi S, Gao P, 2014. ADAM metallopeptidase domain 33 (ADAM33): a promising target for asthma. Mediat. Inflamm 2014, 572025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S, Palmberg L, 2018. Air-liquid interface: relevant in vitro models for investigating air pollutant-induced pulmonary toxicity. Toxicol. Sci 10.1093/toxsci/kfy053. (2018 Mar 9, Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- Wang J, Huang J, Wang L, Chen C, Yang D, Jin M, Bai C, Song Y, 2017. Urban particulate matter triggers lung inflammation via the ROS-MAPK-NF-kappaB signaling pathway. J. Thorac. Dis 9, 4398–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Shi Y, Asweto CO, Feng L, Yang X, Zhang Y, Hu H, Duan J, Sun Z, 2017. Fine particle matters induce DNA damage and G2/M cell cycle arrest in human bronchial epithelial BEAS-2B cells. Environ. Sci. Pollut. Res. Int 24, 25071–25081. [DOI] [PubMed] [Google Scholar]
- Xing YF, Xu YH, Shi MH, Lian YX, 2016. The impact of PM2.5 on the human respiratory system. J. Thorac. Dis 8, E69–E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu G, Lin Z, Wang Y, He H, Liu T, Kamp DW, 2016. Pro-inflammatory response and oxidative stress induced by specific components in ambient particulate matter in human bronchial epithelial cells. Environ. Toxicol 31, 923–936. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Feng H, Hu G, Wang L, Liu J, Gao X, Shang J, Zhu T, Tang S, Jia G, 2017. Effects of 1,4-naphthoquinone aged carbon black particles on the cell membrane of human bronchial epithelium. Environ. Toxicol. Pharmacol 54, 21–27. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Usatyuk PV, Gorshkova IA, He D, Wang T, Moreno-Vinasco L, Geyh AS, Breysse PN, Samet JM, Spannhake EW, Garcia JG, Natarajan V, 2009. Regulation of COX-2 expression and IL-6 release by particulate matter in airway epithelial cells. Am. J. Respir. Cell Mol. Biol 40, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Liu Y, Duan F, Qin M, Wu F, Sheng W, Yang L, Liu J, He K, 2015. Transcriptomic analyses of the biological effects of airborne PM2.5 exposure on human bronchial epithelial cells. PLoS One 10, e0138267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.