Abstract
The restriction factor TRIM5α recognizes incoming retroviral capsids and blocks virus replication. In this issue of Cell Host and Microbe, Fletcher et al. (2018) show how the TRIM5α RING domain mediates formation of various ubiquitin conjugates that differentially regulate TRIM5α turnover, inhibition of viral reverse transcription, and innate immune activation.
Retroviruses exert strong selective pressure, and hosts have therefore evolved protective mechanisms to inhibit their propagation and interspecies transfer. TRIM5α is a mammalian restriction factor that has garnered considerable attention because alleles from some species can potently block HIV-1 replication. The protein is a multi-domain ubiquitin (Ub) E3 ligase of the TRIM family, comprising RING, B-box 2, coiled-coil, and SPRY domains. Restriction occurs when TRIM5α binds and inactivates the incoming retroviral core. Considerable progress has been made in understanding how TRIM5α can recognize the capsid shell that surrounds and protects the viral core RNA and replicative enzymes. Recognition requires direct binding of the TRIM5α SPRY domain to the capsid surface, but isolated SPRY domains typically interact weakly with the divergent surfaces of different retroviral capsids. This limitation is overcome by avidity, which arises because TRIM5α assembles into hexagonal lattices that are complementary to the conserved retroviral capsid lattice (Ganser-Pornillos et al., 2011) (Fig. 1A). The TRIM5α hexagonal lattice comprises stable antiparallel dimers of the coiled-coil domain (hexagon edges) and trimers of the B-box/RING domains (hexagon vertices) (Fig. 1B). ‘Caging’ of viral capsids by the TRIM5α lattice inhibits viral infectivity, but this is only half the story. A new report (Fletcher et al., 2018) now explains the Ub-dependent reactions that induce: 1) rapid TRIM5α turnover, which likely prevents unwanted immune activation; 2) capsid dissociation, which blocks reverse transcription and augments restriction; and 3) production of anti-viral interferons and cytokines, which alerts the immune system to invasion.
Figure 1.
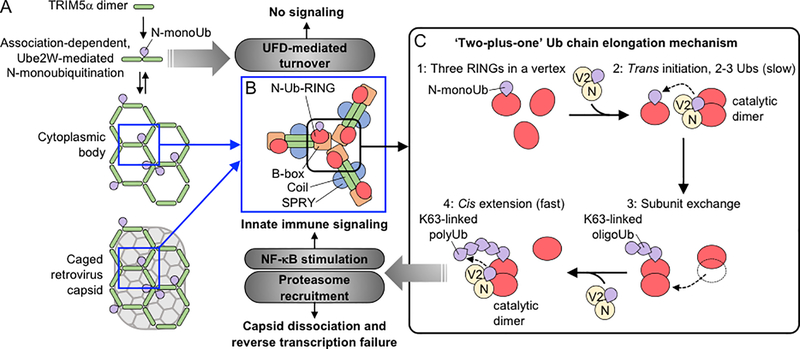
Models for TRIM5α assembly-dependent activation of different ubiquitin (Ub)-dependent processes. (A) The basal oligomeric state of TRIM5α is an antiparallel dimer. Under steady-state conditions, B-box dependent association of the TRIM5α dimers can lead to Ube2W recruitment, resulting in the formation of N-Ub-TRIM5α, which is turned over by the ubiquitin fusion degradation (UFD) machinery (Fletcher et al., 2018). This process ordinarily maintains TRIM5α at a low steady-state level, but if TRIM5α levels increase sufficiently then the protein can form larger cytoplasmic bodies. We speculate that the oligomeric plasticity of the TRIM5α B-box (Keown et al., 2016; Wagner et al., 2016) may contribute to the formation of these different species because the B-box can dimerize, as might be required for efficient Ube2W-mediated N-monoubiquitination, and can trimerize, as would be required to form the vertices of a hexagonal cytoplasmic body. Finally, the introduction of a susceptible retroviral capsid induces the TRIM5α subunits to assemble into a hexagonal lattice on the capsid surface (Ganser-Pornillos, 2011). (B) Each vertex in a TRIM5α hexagonal lattice is mediated by a B-box domain trimer (Wagner et al., 2016), which also clusters three RING domains. (C) Hexagonal lattice formation brings three TRIM5α RING domains together at each vertex (step 1) and drives a dynamic ‘two-plus-one’ K63-linked Ub chain elongation mechanism (Fletcher et al., 2018). Two RING subunits initially form a catalytic dimer and cooperate with Ube2N/V2 to attach 2–3 more Ub moieties in trans to the anchoring N-monoUb on the third RING (step 2). The substrate RING then exchanges into the original RING dimer (step 3) and can now collaborate with Ube2N/V2 to further extend the K63-linked chains in cis (step 4). The creation of N-anchored, K63-linked Ub chains on TRIM5α then leads to proteasome recruitment and NF-κB activation (Fletcher et al., 2018).
Like other RING domains, the TRIM5α RING catalyzes ubiquitination reactions. Some TRIM5α restriction activities are Ub-dependent, whereas others are not. For example, proteasome inhibition relieves the TRIM5α block to reverse transcription, but does not restore viral infectivity (Wu et al., 2006), whereas immune signaling is strictly Ub-dependent (Pertel et al., 2011). However, the relevant substrates, Ub chain compositions, and functional consequences of different Ub linkages have not been clear. The role of TRIM5α RING oligomerization has also been puzzling because three RING domains reside at each lattice vertex (Fig. 1B) but isolated RING domains can dimerize, and dimerization is required to bind ubiquitin-conjugated E2 enzymes (Yudina et al., 2015). To clarify these issues, Fletcher et al. dissected the contributions of distinct RING interactions to different TRIM5α activities. They generated human TRIM5α RING mutations that impaired either dimerization or binding to E2-conjugating enzymes, and tested the effects on infectivity of a restriction-sensitive N-tropic murine leukemia virus. As expected, deletion of the RING domain attenuated (but did not entirely eliminate) the infectivity block. Nicely, E2 binding mutants inhibited viral infectivity as potently as wild-type TRIM5α, whereas RING dimerization mutants reduced this activity, just like the RING deletion mutant. Consistent with previous findings, these experiments demonstrated that the RING domain helps reduce viral infectivity by contributing to TRIM lattice assembly, but this activity does not require ubiquitination activity.
Fletcher et al. next examined the connections between TRIM5α RING domain interactions, Ub conjugation, higher-order assembly, and protein turnover in cells. Even in the absence of a viral invader, TRIM5α can associate into punctate ‘cytoplasmic bodies’ (CB) (Fig. 1A). The size of these dynamic puncta increases with proteasome inhibition, implying a role for the ubiquitin-proteasome system (Diaz-Griffero et al., 2006; Wu et al., 2006). The authors found that their TRIM5α RING mutants displayed a range of punctate phenotypes, with the RING dimerization mutants phenocopying the CB enlargement seen for proteasome inhibition and RING deletion (Diaz-Griffero et al., 2006; Wu et al., 2006). Cycloheximide chase experiments further showed that the RING mutants failed to turn over efficiently, implying that increased puncta size reflects impaired protein degradation (Diaz-Griffero et al., 2006; Wu et al., 2006). Thus, the RING domain is not required for CB formation, and Ub conjugation regulates TRIM5α turnover and CB growth.
Fletcher et al. then examined how TRIM5α activates innate immune signaling. Like others (Pertel et al., 2011), they found that TRIM5α recognition of an incoming capsid potently induced expression of cytokine and other interferon-inducible genes. This occurred through the NF-κB pathway, via phosphorylation of IkBa and nuclear translocation of p65. To examine the relationship between TRIM5α assembly, ubiquitination and signaling, the authors took advantage of the observation that TRIM5α overexpression activates NF-κB signaling, even in the absence of virus. RING-E2 interface mutants failed to stimulate NF-κB activity, confirming that signaling is Ub-dependent. They also found that the degree of stimulation followed a bell-shaped curve: first increasing with increasing TRIM5α protein expression, but then dropping at higher TRIM5α levels. This behavior correlated with CB morphology and abundance, and the authors concluded that TRIM5α can form two different types of puncta – signaling-competent CB assemblies, which predominate at lower protein concentrations (and presumably correspond to hexagonal TRIM5α arrays), and non-signaling protein aggregates, which predominate at higher protein concentrations. The aggregates have the characteristics of ‘aggresomes’ (Diaz-Griffero et al., 2006) and may be turned over by autophagy, which could help to explain previous links between autophagy and TRIM5α turnover (Mandell et al., 2014). The resulting model is that CB TRIM5α assemblies (but not aggregates) form Ub conjugates that stimulate immune activation, and the activation can be self-limiting (albeit shown only at superphysiological TRIM5α levels). Interestingly, TRIM1, TRIM32, and TRIM38 also showed this bell-curve behavior, indicating that similar signaling mechanisms may operate in other TRIM family members with innate immune functions.
Importantly, Fletcher et al. also analyzed the different Ub conjugates formed by TRIM5α. TRIM5α uses at least two different E2 enzymes (Fletcher et al., 2015): Ube2W, which preferentially attaches monoUb to free substrate N-termini rather than lysine sidechains, and Ube2N/V2, which can generate unanchored K63-linked polyUb chains or can build K63-linked chains onto anchoring monoUb substrates. N-terminal Ub modification of TRIM5α in cells was previously discounted because TRIM5α N-termini can also be modified by competing N-acetylation reactions (Fletcher et al., 2015). However, the current report presents several lines of evidence that free TRIM5α N-termini are present in cells and are modified by Ube2W to generate N-Ub-TRIM conjugates (although Ube2W-dependent lysine-attached chains may also be present and the relative ratio of N-acetylated vs. free N-termini at steady state is not yet known).
This discovery allowed the authors to bypass the requirement for Ube2W by using genetically-encoded Ub-TRIM5α fusion constructs. Remarkably, they found that adding a Ub moiety directly to the TRIM5α N-terminus: 1) reduced CB formation, 2) abrogated inhibition of viral infectivity, and 3) strongly stimulated NF-κB activity, implying that Ub-TRIM5α is ‘on-pathway’ toward immune signaling. The first two results were attributed to the enhancement of proteasome recruitment and Ub-TRIM5α degradation prior to assembly. Strongly supporting this idea, proteasome inhibition via MG132 treatment rescued the ability of Ub-TRIM5α to form CB and to inhibit viral infectivity. Mutation of all of the Ub lysines except K63 in the Ub-TRIM5α construct also restored these activities, implying that degradation of Ub-TRIM5α requires addition of non-K63 Ub linkages. To integrate these observations, the authors propose that under steady-state conditions, Ube2W-mediated ubiquitination of the protein’s N-terminus results in rapid TRIM5α turnover because the Ub-TRIM5α conjugates are recognized by the Ub fusion degradation (UFD) machinery, elongated with non-K63-linked chains, and degraded by the proteasome (Fig. 1). However, if these Ub-TRIM5α conjugates are allowed to accumulate and assemble into a hexagonal lattice, then Ube2N/V2 attaches K63-linked Ub chains onto the anchoring N-monoUb moiety and the resulting TRIM5α-linked Ub chains signal to activate NF-κB.
In a final clever set of experiments, the authors dissected the molecular basis of assembly-induced ubiquitination. The TRIM hexagonal lattice clusters three RING domains at the trivalent B-box vertices (Fig. 1B). Fletcher et al. found that this architecture supports K63-linked Ub chain formation, in a surprisingly complex multistep reaction (Fig. 1C) in which a RING dimer initially collaborates with Ube2N/V2 to add 2–3 K63-linked Ub molecules onto the anchoring N-monoUb of the third RING domain in trans. The ubiquitinated RING substrate then exchanges into the dimer and further rapid chain elongation occurs in cis. This dynamic ‘two-plus-one’ ubiquitination model nicely explains how assembly of the TRIM hexagonal lattice on a retroviral capsid promotes efficient self-ubiquitination. In effect, the cascade of capsid-induced TRIM5α assembly, assembly-dependent Ub conjugation, and required trivalent RING clustering can be thought of as an elaborate ‘proofreading’ mechanism that tests for an invading viral capsid and for TRIM5α assembly before producing an inflammatory signal.
Like all good science, this study will stimulate new pursuits, including: 1) further characterizing the role of the UFD system in basal TRIM5α turnover and testing whether non-proteasomal mechanisms such as autophagy may also contribute, 2) testing whether more elaborate signals such as branched K63/K48 chains recruit proteasomes to TRIM5α-bound retroviral capsids, and if so, whether another E2 enzyme is required to add the K48-linked chains, 3) determining whether the interplay between TRIM5α N-acetylation and N-ubiquitination can regulate restriction, 4) learning precisely what factors bias N-Ub-TRIM5α toward non-signaling UFD-mediated degradation vs. K63-linked Ub-mediated restriction and NF-κB stimulation, and 5) dissecting the mechanistic details of downstream capsid inactivation and signaling. These issues highlight how TRIM5α uses the full sophistication of ubiquitin signaling to activate multiple defense mechanisms, specifically in response to retroviral invasion.
Acknowledgements:
We thank Devin Christensen and Barbie Ganser-Pornillos for helpful discussions. Work on TRIM5 in our laboratories are funded by NIH grants R01-GM112508 (O.P.) and P50-GM082545 (W.I.S.).
References:
- Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, and Sodroski J (2006). Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349, 300–315. [DOI] [PubMed] [Google Scholar]
- Fletcher AJ, Christensen DE, Nelson C, Tan CP, Schaller T, Lehner PJ, Sundquist WI, and Towers GJ (2015). TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J 34, 2078–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AJ, Vaysburd M, Maslen S, Zeng J, Skehel JM, Towers GJ, and James LC (2018). Trivalent RING assembly activates TRIM5 ubiquitination and innate immune signalling. Cell Host Microbe X, X-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, and Yeager M (2011). Hexagonal assembly of a restricting TRIM5α protein. Proc Natl Acad Sci U S A 108, 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown JR, Yang JX, Douglas J, and Goldstone DC (2016). Characterization of assembly and ubiquitylation by the RBCC motif of TRIM5α. Sci Rep 6, 26837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell MA, Kimura T, Jain A, Johansen T, and Deretic V (2014). TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy 10, 2387–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. (2011). TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JM, Roganowicz MD, Skorupka K, Alam SL, Christensen D, Doss G, Wan Y, Frank GA, Ganser-Pornillos BK, Sundquist WI, et al. (2016). Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α. eLife 5, e16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, and Hope TJ (2006). Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A 103, 7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudina Z, Roa A, Johnson R, Biris N, de Souza Aranha Vieira DA, Tsiperson V, Reszka N, Taylor AB, Hart PJ, Demeler B, et al. (2015). RING dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep 12, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]