Abstract
Background
Microbial cultures for diagnosis of neonatal sepsis have low sensitivity and reporting delay. Advances in molecular microbiology have fostered new molecular assays that are rapid and may improve neonatal outcomes.
Objectives
To assess the diagnostic accuracy of various molecular methods for the diagnosis of culture‐positive bacterial and fungal sepsis in neonates and to explore heterogeneity among studies by analyzing subgroups classified by gestational age and type of sepsis onset and compare molecular tests with one another.
Search methods
We performed the systematic review as recommended by the Cochrane Diagnostic Test Accuracy Working Group. On 19 January 2016, we searched electronic bibliographic databases (the Cochrane Library, PubMed (from 1966), Embase (from 1982), and CINAHL (from 1982)), conference proceedings of the Pediatric Academic Societies annual conference (from 1990), clinical trial registries (ClinicalTrials.gov, International Standard Randomised Controlled Trial Number (ISRCTN) registry, and World Health Organization (WHO) International Clinical Trials Platform (ICTRP) Search portal), and Science Citation Index. We contacted experts in the field for studies.
Selection criteria
We included studies that were prospective or retrospective, cohort or cross‐sectional design, which evaluated molecular assays (index test) in neonates with suspected sepsis (participants) in comparison with microbial cultures (reference standard).
Data collection and analysis
Two review authors independently assessed the methodologic quality of the studies and extracted data. We performed meta‐analyses using the bivariate and hierarchical summary receiver operating characteristic (HSROC) models and entered data into Review Manager 5.
Main results
Thirty‐five studies were eligible for inclusion and the summary estimate of sensitivity was 0.90 (95% confidence interval (CI) 0.82 to 0.95) and of specificity was 0.93 (95% CI 0.89 to 0.96) (moderate quality evidence). We explored heterogeneity by subgroup analyses of type of test, gestational age, type of sepsis onset, and prevalence of sepsis and we did not find sufficient explanations for the heterogeneity (moderate to very low quality evidence). Sensitivity analyses by including studies that analyzed blood samples and by good methodology revealed similar results (moderate quality evidence).
Authors' conclusions
Molecular assays have the advantage of producing rapid results and may perform well as 'add‐on' tests.
Plain language summary
Molecular tests to detect infections in newborn babies
Review question: Do molecular tests detect infection better than the standard culture methods for detecting infection in newborn babies?
Background
The current method of detecting infection (illness caused by germs) in newborn babies is to obtain blood or other body fluids (or both) and culture (grow) the bacteria (germs) in a laboratory. However, culture methods may miss some infections and take a long time to produce results (48 to 72 hours). Newer methods of detecting infection are based on detecting DNA (a molecule that carries the genetic instructions used in growth, development, functioning, and reproduction) from bacteria and other organisms that cause infections. Advances in microbiology have introduced new molecular tests for detecting infections. Molecular tests are rapid and may detect more infections compared to the traditional culture methods.
Study characteristics
We searched for evidence for the use of the molecular methods to detect infection in newborn babies. We found 35 studies that compared the new molecular methods to culture methods of the blood and spinal fluid to diagnose infection.
Study funding sources
None.
Key results
We found that the molecular methods may be very helpful additional tests because they provide rapid results.
Quality of evidence
Although there were some issues with selection of newborn babies for this review, overall the methods used by the studies were adequate. We rated the quality of the evidence as moderate to low.
Summary of findings
Summary of findings'. 'Summary of findings table.
| Groups | Number of studies |
Sensitivity (95% CI) |
Specificity (95% CI) |
Quality of evidence using GRADE | |
| All studies | ‐ | 35 | 0.90 (0.82 to 0.95) | 0.93 (0.89 to 0.96) | Moderate quality evidence* |
| Type of test | Broad‐range PCR | 9 | 0.97 (0.86 to 1.00) | 0.93 (0.77 to 0.98) | Moderate quality evidence* |
| Real‐time PCR | 9 | 0.86 (0.59 to 0.96) | 0.94 (0.90 to 0.97) | Moderate quality evidence* | |
| Post‐PCR processing | 5 | 0.97 (0.40 to 1.00) | 0.96 (0.93 to 0.98) | Low quality evidence** | |
| Multiplex PCR | 6 | 0.76 (0.60 to 0.88) | 0.81 (0.70 to 0.89) | Low quality evidence** | |
| Staphylococcal PCR* | 2 | ‐ | ‐ | Low quality evidence** | |
| Fungal PCR* | 4 | ‐ | ‐ | Low quality evidence** | |
| Type of sepsis | EOS* | 2 | ‐ | ‐ | Low quality evidence** |
| LOS | 10 | 0.79 (0.69 to 0.86) | 0.94 (0.85 to 0.98) | Low quality evidence** | |
| Mixed EOS and LOS | 23 | 0.94 (0.84 to 0.98) | 0.92 (0.87 to 0.95) | Moderate quality evidence* | |
| Gestational age | Preterm | 5 | 0.89 (0.75 to 0.96) | 0.87 (0.71 to 0.94) | Low quality evidence** |
| Mixed term and preterm | 30 | 0.90 (0.80 to 0.96) | 0.94 (0.90 to 0.96) | Moderate quality evidence* | |
| Prevalence | < 15% | 20 | 0.94 (0.80 to 0.99) | 0.95 (0.92 to 0.97) | Moderate quality evidence* |
| 15% to 30% | 8 | 0.85 (0.67 to 0.94) | 0.88 (0.79 to 0.94) | Low quality evidence** | |
| >30% | 7 | 0.87 (0.75 to 0.93) | 0.93 (0.64 to 0.99) | Low quality evidence** | |
| Specimen | Blood only | 32 | 0.92 (0.84 to 0.96) | 0.93 (0.89 to 0.95) | Low quality evidence** |
| Blood and CSF* | 3 | ‐ | ‐ | Moderate quality evidence* | |
| Quality | Good methodologic studies only | 22 | 0.90 (0.78 to 0.96) | 0.93 (0.88 to 0.96) | Moderate quality evidence* |
CI: confidence interval; CSF: cerebrospinal fluid; EOS: early‐onset sepsis; LOS: late‐onset sepsis; PCR: polymerase chain reaction.
Summary estimates of sensitivity and specificity were derived from meta‐analyses using the bivariate random‐effects model using statistical software STATA. Summary estimates for the subgroups are presented, where number of studies ≥ 4. *Summary estimates of sensitivity and specificity could not be calculated using STATA if number of studies ≤ 4.
GRADE rating of evidence: reasons for downgrading quality of evidence (Gopalakrishna 2014)
* Evidence downgraded one level for inconsistency of evidence.
** Evidence downgraded two levels for inconsistency and imprecision.
Background
Sepsis is a frequent life‐threatening event among neonates, particularly in very low birth weight infants (VLBW) (birth weight less than 1500 g) and is responsible for significant mortality and morbidity (Adams‐Chapman 2006; Stoll 2002; Stoll 2004). Early diagnosis of infections in newborns may improve clinical outcomes. Microbial cultures of blood or other sterile body fluids are the gold standard in the diagnosis of neonatal bacterial and fungal sepsis. Blood cultures are generally assumed to have low sensitivity in neonates for the following reasons: low degree of neonatal bacteremia or fungemia, small inoculation volumes in culture bottles, and the use of intrapartum antibiotics (Chiesa 2004; Schelonka 1996). In addition, results of the microbial culture are not available for at least 24 to 72 hours. Diagnostic capabilities of blood culture systems have improved since the early 2000s with the advent of automated continuous blood culture monitoring systems but still, subcultures for specific assays (e.g. biochemical) are ultimately needed for pathogen identification. New molecular methods for detection of infection may provide results earlier and improve neonatal outcomes.
Target condition being diagnosed
Neonatal bacterial and fungal sepsis is the target condition to be diagnosed and often described based on the age of the infant at the onset of infection. Early‐onset bacterial or fungal sepsis (sepsis in 72 hours of life or less) occurs in 1.5% to 1.9% of VLBW infants and late‐onset bacterial or fungal sepsis (sepsis onset after 72 hours of life) in about 20% of VLBW infants (Stoll 2002). Neonatal mortality in late‐onset sepsis (LOS) is approximately 18%, and in Gram‐negative infections as high as 36%. The incidence of LOS in neonates less than 33 weeks' postmenstrual age (PMA) in the Canadian neonatal network was 10% but varied from 0.61% to 14% in other studies (Canadian Neonatal Network 2014; Dong 2015). Sepsis increases neonatal morbidities including patent ductus arteriosus, need for intravascular access, need for parenteral nutrition, bronchopulmonary dysplasia, necrotizing enterocolitis and length of hospital stay. In addition, sepsis significantly impairs long‐term neurodevelopmental outcomes either by direct infection of the central nervous system or as a result of inflammatory injury (Adams‐Chapman 2006). In one large cohort study of more than 6000 extremely low birth weight infants (birth weight 1000 g or less), infected infants had a significantly higher incidence of adverse developmental outcomes at follow‐up, including cerebral palsy, lower Bayley's scores of infant development and visual impairment when compared to uninfected infants (Stoll 2004). Clinical signs and symptoms of neonatal sepsis are often nonspecific and early diagnosis and treatment may be critical to improve neonatal outcomes. Overdiagnosis of neonatal sepsis can lead to inappropriate antibiotic use that may foster antibiotic resistance.
Index test(s)
Advances in molecular microbiology have provided new molecular assays for the detection of infection. Molecular assays can be completed in less than 12 hours and may have better sensitivity than microbial cultures. In addition, the significant increase in workload related to bloodstream infections for the clinical microbiologic laboratory could potentially be offset by high‐throughput molecular assays coupled with automation (Rodriguez‐Creixems 2008). However, molecular assays do not provide information on antibiotic susceptibility.
Molecular pathogen detection methods are based on hybridization or amplification of pathogen DNA. Hybridization based methods (e.g. fluorescence in situ hybridization) have not yet been evaluated in the diagnosis of neonatal sepsis. However, neonatal studies have been conducted using amplification methods (e.g. polymerase chain reaction (PCR)) that amplify specific target regions in the microbial genome. Broad‐range PCR targets the 16S ribosomal ribonucleic acid (rRNA) gene, a ubiquitous gene that is preserved in all bacteria and comprises both conserved and variable regions (Woese 1987). The conserved regions are targeted by universal primers for identifying bacterial infection, and the variable regions by genus or species‐specific assays (Isaacman 1996; Relman 1999). Fungal PCRs target specific regions of the fungal genome (most commonly internal transcribed spacer regions of the rRNA). Amplified target regions may then be subjected to downstream applications such as sequencing or microarray/probe hybridization.
Amplification methods that have been evaluated in neonates for the diagnosis of sepsis can be grouped as follows.
Broad‐range conventional PCR assays: PCR amplification strategies targeting conserved regions such as 16S rRNA in bacteria.
Real‐time PCR, where amplification of the template is monitored in real time.
PCR followed by post‐PCR processing, such as sequencing or hybridization.
Multiplex‐PCR, where amplification is directed against multiple organisms in the same assay.
Species‐ or genus‐specific assays: staphylococcal, fungal PCR assays or other organism‐specific assays.
Clinical pathway
Neonates with clinical signs and symptoms of sepsis including lethargy, apnea, hypotension and oliguria are investigated for sepsis with a blood, cerebrospinal fluid (CSF) and urine cultures with or without markers of inflammation such as a white blood cell count, C‐reactive protein (CRP) or others. However, to prove an infection beyond doubt, cultures should be positive, which takes more than 24 hours and usually 48 hours. Also, the sensitivity of cultures has been questioned. An ideal diagnostic test for neonatal bacterial or fungal sepsis should be rapid, sensitive, specific, detect all organisms relevant to neonatal sepsis and not be affected by maternal antibiotics. The test should have high sensitivity so that infections are not missed and a negative test should reliably exclude sepsis so that no neonate is unnecessarily treated with antibiotics.
Alternative test(s)
Traditionally sepsis diagnosis is aided by abnormal white blood cell count (white blood cell less than 5000 cells/μL, sensitivity 0.2, specificity 0.96; white blood cell less than 1000 cells/μL, sensitivity 0.3 specificity 1.0), altered white cell indices, differential white cell count, elevation of immature white cells (I:T ratio greater than 0.20, sensitivity 0.55 and specificity 0.74) and low platelet count (less than 50 × 109/L, sensitivity 0.8 and specificity 0.99) (Hornik 2012). Serum biomarkers of infection consisting of acute‐phase proteins namely CRP (sensitivity 0.6 to 0.84, specificity 0.84 to 1.00), procalcitonin (sensitivity 0.77, specificity 0.62) or elevation of inflammatory cytokines; tumor necrosis factor (TNF)‐α (sensitivity 0.6 to 0.82, specificity 0.86 to 0.93) and interleukin (IL)‐6 (sensitivity 0.58 to 0.89, specificity 0.84 to 0.96) have also been used (Blommendahl 2002; Ng 1997; Ng 2012; Verboon‐Maciolek 2006). All sensitivities and specificities were calculated with culture as the reference standard. White cell indices and other serum biomarkers may aid in the diagnosis but not necessarily confirm infection.
Rationale
Blood cultures are generally assumed to have a relatively low sensitivity for the diagnosis of neonatal bacterial or fungal sepsis and results of the microbial culture are not available for at least 24 to 72 hours. Also, some cases of sepsis may be missed by cultures and a more sensitive diagnostic test such as a molecular test may be useful. Rapid advances in technology have led to molecular methods with rapid turnaround times, that may be more sensitive than culture and which may have an impact on current clinical practice. We will not be able to show that the molecular tests are more sensitive than culture, as culture is our reference standard. Still, culture is used in practice as a confirmation test (100% specificity) and thus knowing the relative performance of molecular tests compared to culture is very relevant. If a test misses too many culture‐positive samples, the test will not be implemented in practice. Alternative tests such as evaluation of acute phase reactants or cytokines are often used in conjunction with blood cultures but do not have sufficient diagnostic accuracy to replace microbial cultures as the reference standard. We have previously systematically reviewed molecular assays in the diagnosis of neonatal sepsis (Pammi 2011), but this is a rapidly advancing field. Optimization of the older molecular methods and development of newer methods may change the diagnostic accuracy of these tests and may change our clinical practice. In our view, a Cochrane Review is justified as new literature has accumulated since our last published review and will allow for updates as new studies are performed. We are not aware of any other systematic review on this topic in neonates although there are narrative reviews.
Objectives
To assess the diagnostic accuracy of various molecular methods for the diagnosis of culture‐positive bacterial and fungal sepsis in neonates and to explore heterogeneity among studies by analyzing subgroups classified by gestational age and type of sepsis onset and compare molecular tests with one another.
Methods
Criteria for considering studies for this review
Types of studies
We included prospective or retrospective, cohort or cross‐sectional studies that assessed the diagnostic accuracy of a molecular test in the clinical context of diagnosis of neonatal bacterial or fungal sepsis. We excluded studies that assessed the diagnostic accuracy of the test using only positive samples or healthy controls and not in the clinical context of suspected neonatal bacterial or fungal sepsis.
Participants
Neonates with clinically suspected bacterial or fungal sepsis. Clinical signs and symptoms of sepsis in neonates can be nonspecific and hence a high index of clinical suspicion is required for the diagnosis. Neonates are defined as a newborn of 28 days of age or less. We defined gestational age subgroups of preterm and term infants as:
preterm: neonates born at less than 37 completed weeks of gestation;
term: neonates born at 37 completed weeks of gestation or greater.
We made a post‐hoc decision to include data from studies that included infants aged more than 28 days if more than 50% of the study participants were under 28 days of age.
Index tests
We defined molecular assays as any assay that involves extraction and evaluation of nucleic acid from bacteria or fungi, and performed for the diagnosis of neonatal sepsis. The results of the index test were dichotomous; positive or negative. We assessed the results of the index test with the reference standard done at approximately same time. In the event of the index test identifying a different organism compared to the reference standard or identifying an organism when the reference standard was negative, we discussed among our author group as to whether we should discard or include as a false positive based on whether it was a contaminant or not. We analyzed subgroups of type of molecular assay namely broad‐range conventional PCR, real‐time PCR, PCR followed by post‐PCR processing, multiplex PCR, staphylococcal PCR and fungal PCR. New tests/methodology may arrive in the future as the technology advances and we will address this by subgroup analyses and using year of publication as a covariate in future meta‐analyses. We excluded molecular methods assessing infections other than those caused by bacteria or fungi (e.g. viruses or protozoa).
Target conditions
Neonatal bacterial or fungal sepsis, defined as a neonate with a positive culture of bacteria or fungi from the blood or CSF, or both. We analyzed subgroups of type of sepsis onset namely early‐onset sepsis (EOS) (72 hours of age or less) and LOS (greater than 72 hours of age).
Reference standards
The reference standard for the diagnosis of sepsis was microbial culture of blood or CSF, or both, for bacteria or fungi, or both. Microbial cultures are generally assumed to have low sensitivity but this decreased sensitivity has not been quantified. The low sensitivity of cultures in neonates may be due to the low degree of neonatal bacteremia or fungemia, small inoculation volumes in culture bottles and the use of intrapartum antibiotics. We documented the participant characteristics, risk factors and outcomes of people who were index test positive and reference standard negative to gain insight into the sensitivity of the reference standard. Alternative tests, such as evaluation of acute phase reactants or cytokines, are often used in conjunction with blood cultures but do not have sufficient diagnostic accuracy to replace microbial cultures as the reference standard.
Search methods for identification of studies
We used the standard search methods recommended by the Cochrane Neonatal Group and searched the literature on 19 January 2016. We applied no language restrictions in our search methods.
Electronic searches
Bibliographic databases: the Cochrane Library (2016, Issue 1), PubMed (from 1966), Embase (from 1982) and CINAHL (from 1982) using the search engines at Texas Medical Center library.
Abstract of conferences: proceedings of meetings of American Pediatric Society, Society for Pediatric Research and European Society for Pediatric Research (from 1990).
ClinicalTrials.gov (clinicaltrials.gov/), International Standard Randomised Controlled Trial Number (ISRCTN) registry (www.isrctn.com/), and the World Health Organization (WHO) International Clinical Trials Platform (ICTRP) Search portal (apps.who.int/trialsearch/).
Science Citation Index, Web of Science using subject search.
Our search strategies for PubMed and other databases including the platforms are outlined in Appendix 1. The search strategy was developed by discussion between the review author team, librarians and the Cochrane Neonatal Group's Trials Search Co‐ordinator.
Searching other resources
We screened reference lists of identified studies, relevant review articles and other publications held in our personal files. We also searched for ongoing and unpublished studies by contacting experts in this field.
Data collection and analysis
Selection of studies
Two review authors (MP, AF) screened all titles and abstracts identified by our search strategy for relevance to the inclusion criteria as detailed in Criteria for considering studies for this review. We retrieved full‐text articles of all identified articles that were deemed relevant to the review and evaluated them against our inclusion eligibility. We resolved disagreements by mutual discussion.
Data extraction and management
Two review authors (MP, AF) independently extracted the following data.
Author, year of publication and name of journal.
Study design including sample size, type of study (prospective or retrospective, cohort or cross‐sectional).
Study population characteristics and the clinical context in which the test was evaluated (e.g. suspected sepsis), and type of participant sample tested.
Type of reference standard, performance of the reference standard and whether evaluated manually or automated.
Index tests, performance of the index tests, type of assay, manufacturer, positivity thresholds, time between the performance of index and reference tests.
Information regarding quality assessment items of the Quality Assessment of Diagnostic Accuracy Studies‐2 (QUADAS‐2) tool (Assessment of methodological quality).
Data in two by two tables for calculation of diagnostic accuracy parameters.
Studies report number of neonates or episodes of sepsis as the unit of analysis. Some studies included neonates with more than one episode of sepsis. As the comparison here was between two tests, cultures versus molecular tests, we included the number of samples wherever possible for our analysis and most studies reported only one sample per participant which we analyzed as such. We compared the extracted data, and resolved discrepancies found upon comparison by mutual discussion. Data extracted from included studies are presented in Appendix 2.
Assessment of methodological quality
We assessed methodologic quality of each included study following guidance from the Cochrane Diagnostic Test Accuracy Working Group, which is adapted from the QUADAS‐2 tool (Whiting 2011). The four domains assessed for risk of bias are participant selection, index test, reference test, and flow and timing. Applicability concerns were assessed in the first three domains (participant selection, index test, reference test). In each domain, we answered the questions with 'Yes', 'No' or 'Unclear' and for each domain judged the risk of bias as 'Low', 'High' or 'Unclear' risk (Appendix 3).
Sources of bias in diagnostic accuracy studies that we assessed include those related to participants (spectrum bias and selection bias), the index test (information bias), the reference standard (misclassification bias, partial verification bias, differential verification bias, incorporation bias, disease progression bias and information bias) and data analysis (excluded data bias) (Appendix 3).
In addition, we decided post‐hoc to present quality of evidence using GRADE methodology recommended for diagnostic tests (Gopalakrishna 2014).
Statistical analysis and data synthesis
In our included studies, the reference standard and the index tests have dichotomous outcomes. We constructed two by two tables for all included studies and enumerated true positives, false positives, false negatives and true negatives. Any positive blood or CSF culture was considered a positive for the reference standard. Nine studies reported data from episodes of sepsis and hence more than one sample from some infants and other studies reported one episode of sepsis from one infant. We have meta‐analyzed data from both studies that reported as episodes of sepsis or as number of infants in this review with advice from our statistician.
As the results of the index tests were dichotomous without an explicit threshold, we used a bivariate random‐effects approach to estimate summary sensitivity and specificity for each index test type separately (Macaskill 2010; Reitsma 2009). The bivariate random‐effects approach enabled us to calculate the summary estimates of sensitivity and specificity, while dealing with the imprecision by which sensitivity and specificity have been measured within each study, variation beyond chance in sensitivity and specificity between studies and any correlation that may exist between sensitivity and specificity. We calculated summary estimates of sensitivity and specificity using 'xtmelogit' in the STATA software (Stata 2011) (Harbord 2007; Harbord 2008; Harbord 2009).
We generated forest plots with 95% confidence intervals (CIs) for sensitivity and specificity for each study using Review Manager 5 (RevMan 2014). We entered the relevant 'xtmelogit' STATA output in Review Manager 5 (RevMan 2014) for the creation of receiver operating characteristic (ROC) space, including summary estimates with 95% CIs and the summary curve.
Investigations of heterogeneity
Sepsis prevalence is higher in premature infants than in term infants because of their relative immunodeficiency, compromise in mucosal and skin integrity, need for intensive care and exposure to invasive procedures. The diagnostic accuracy parameters are likely to be influenced by prevalence of sepsis in term and preterm infants. Therefore, we investigated the effect of prevalence by including it as a covariate in the bivariate model. The same will be true for the onset of sepsis: prevalence rates and spectrum of organisms are different in late‐onset and early‐onset disease and may account for variation among studies. Therefore, we also included sepsis onset as a covariate in the models.
We compared the accuracy of different test types by comparing their summary estimates of sensitivity and specificity and the respective CIs. We did not report P values because the results are prone to confounding due to variations in participant characteristics and study methodology.
We used statistical tests using the 'xtmelogit' command in the statistical software STATA (Stata 2011) for evaluation of heterogeneity by subgroup and sensitivity analysis. We reported summary sensitivity and specificity for each subgroup in the subgroup analyses.
Sensitivity analyses
After performing analyses with data of all included studies, we performed sensitivity analysis to assess test accuracy in studies that evaluated blood samples only as well as studies that evaluated both blood and CSF samples, to test if inhibitors of PCRs in blood samples might influence our results. Furthermore, we investigated the effect of the potential sources of bias by removing biased studies from the total set of studies and re‐analyzing this new set.
Assessment of reporting bias
We used the Deeks' test to assess publication or reporting bias in this diagnostic test accuracy review (Reitsma 2009; Van Enst 2014).
Results
Results of the search
Our comprehensive search identified 932 studies of which we selected 47 relevant articles based on the title and abstract. We obtained the full publications whenever possible for the 47 relevant articles. Twelve articles were irrelevant to this review and discarded. Thirty‐five studies met the inclusion criteria assessing the diagnostic accuracy of molecular assays in neonatal sepsis. The inclusion process is detailed in the PRISMA flow diagram (Figure 1). Some studies did not include an upper limit for age and hence some infants were greater than 28 days of age (Chan 2009; Enomoto 2009; Esparcia 2011; Fujimori 2010; Jordan 2000; Lima 2007; Makhoul 2005; Makhoul 2006; Ohlin 2008; Ohlin 2012; Tirodker 2003; Torres‐Martos 2013). We made a post‐hoc decision that we would include studies where an upper age limit was not specified but more than 50% of the sample were from newborn to less than 28 days of age. Our decision was supported by the reasoning that LOS extends up to three months of age and participant characteristics are similar in the first two to three months of age. The included studies and their risk of bias are presented in Characteristics of included studies table and 12 excluded studies with reasons for exclusion are presented in the Characteristics of excluded studies table. We found no publication bias. Funnel plots were created with ln(DOR) on the x‐axis and the reciprocal of the effective sample size (ESS) on the y‐axis where 1/ESS = (1/(FP + TN) + 1/(TP + FN))1/2 (Figure 2). Then Deeks' test for publication bias was applied by computing Spearman's rank correlation (rs) for the association between ln(DOR) and 1/ESS. Asymmetry is not evident in the funnel plot, and Deeks' test did not indicate the presence of publication bias (rs = 0.012, p = 0.944).
1.
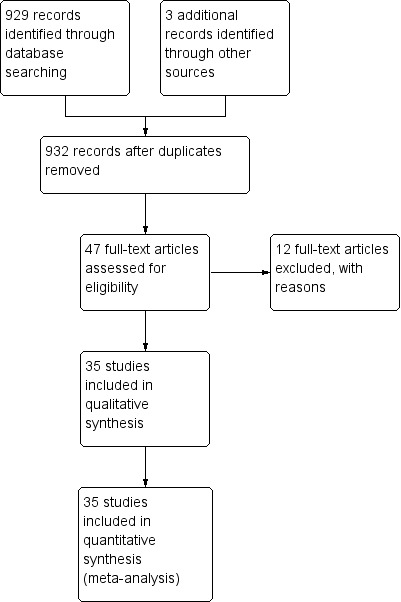
Study flow diagram.
2.
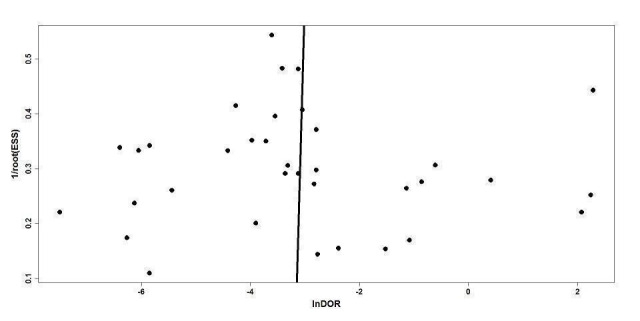
Deeks' funnel plot for publication bias.
Methodological quality of included studies
The results of the methodologic assessment of the studies included in the meta‐analyses are presented in Figure 3; Figure 4. Major risks for bias pertained to participant selection and blinding of index test. Applicability concerns pertained to selection of participants and blinding of the index test and blinding of the reference standard. All studies used an acceptable reference standard, avoided partial and differential verification, and avoided incorporation of the reference standard. Uninterpretable results and withdrawals were explained where applicable.
3.
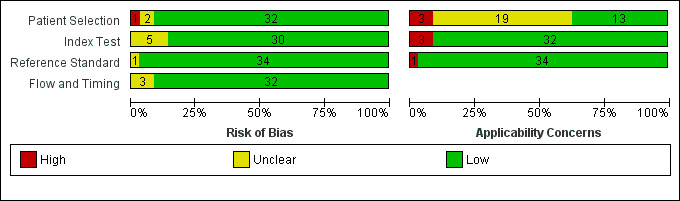
Risk of bias and applicability concerns graph: review authors' judgments about each domain presented as percentages across included studies.
4.
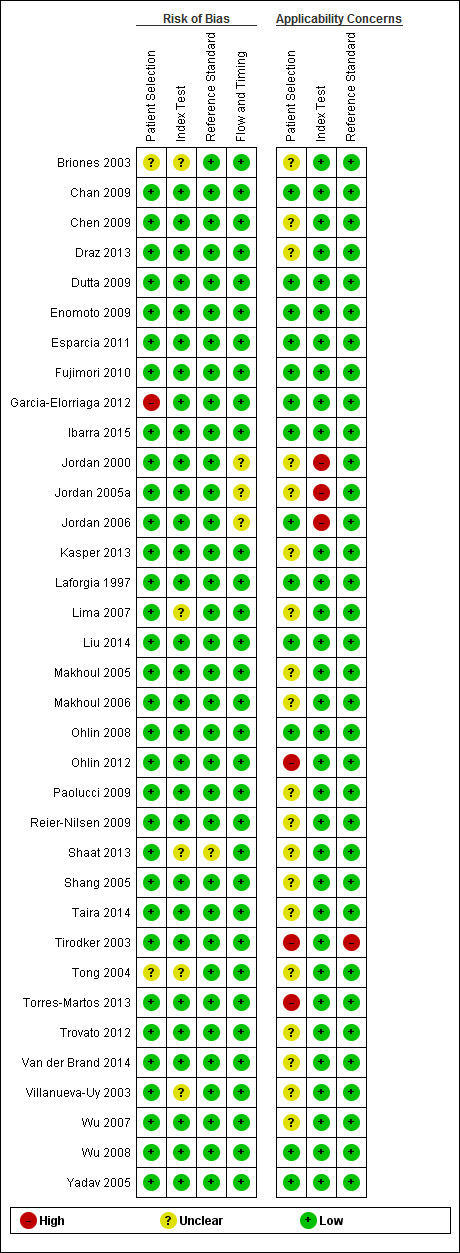
Risk of bias and applicability concerns summary: review authors' judgments about each domain for each included study.
Findings
Summary estimates of mean sensitivity for the 35 included studies were 0.90 (95% CI 0.82 to 0.95), while the mean specificity was 0.93 (95% CI 0.89 to 0.96) (moderate quality evidence) (Table 1). Forest plot (Figure 5) shows that sensitivity across studies ranged for 0.38 to 1.0 and specificity from 0.32 to 1.0. We also plotted the included studies in the ROC space to give a sense of distribution of sensitivity and specificity of the studies (Figure 6). Each study is represented by an oval symbol, with the width proportional to the inverse standard error of the specificity and the height to the inverse standard error of sensitivity.
5.
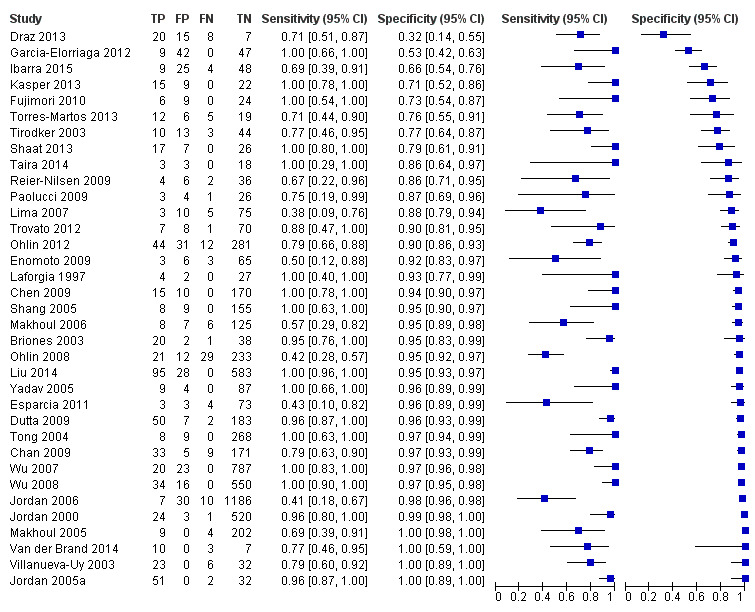
Forest plot of 1 All molecular tests. CI: confidence interval; FN: false negative; FP: false positive; TN: true negative; TP: true positive.
6.

Summary receiver operating characteristic plot of all molecular tests.
We explored heterogeneity by differentiating studies based on the type of molecular assay, onset of sepsis, gestational age and prevalence, and plotted the subgroups of studies in the ROC space (moderate to low quality evidence). Figure 7 represents the studies differentiated by the type of molecular assay in the ROC space. Summary estimates for real‐time PCR assays were sensitivity 0.86 (95% CI 0.59 to 0.96) and specificity 0.94 (95% CI 0.90 to 0.97). Broad‐range conventional PCR performed with sensitivity 0.97 (95% CI 0.86 to 1.00), specificity 0.93 (95% CI 0.77 to 0.98), tests with post‐PCR processing, sensitivity 0.97 (95% CI 0.40 to 1.00) and specificity 0.96 (95% CI 0.93 to 0.98) and multiplex PCR, sensitivity 0.76 (95% CI 0.60 to 0.88), specificity 0.81 (95% CI 0.70 to 0.89) (Table 1). Summary estimates of sensitivity and specificity for Staphylococcal PCR and fungal PCR were not possible as there four or fewer studies.
7.
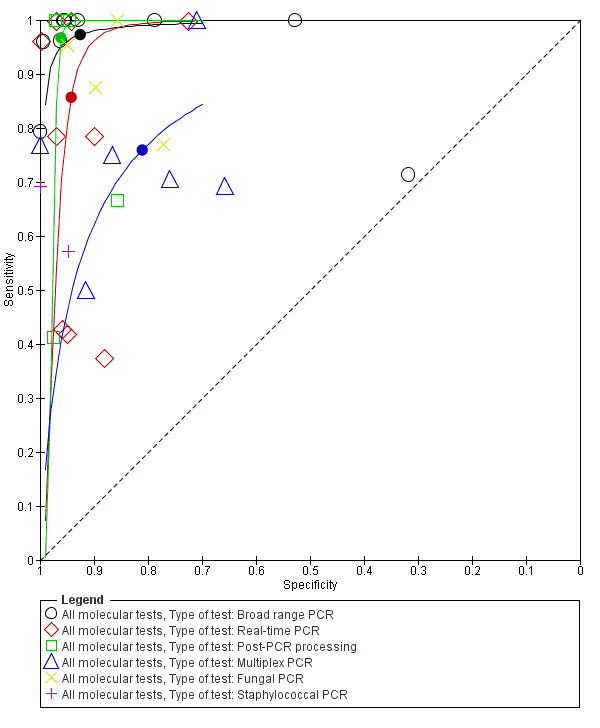
Summary receiver operating characteristic plot by type of molecular test. PCR: polymerase chain reaction.
Two studies reported on EOS, 10 on only LOS and 23 studies on both. Summary estimates for the molecular tests in the diagnosis of LOS were sensitivity 0.79 (95% CI 0.69 to 0.86), specificity 0.94 (95% CI 0.85 to 0.98) and mixed EOS and LOS were sensitivity 0.94 (95% CI 0.84 to 0.98), specificity 0.92 (95% CI 0.87 to 0.95) (Figure 8; Table 1).
8.
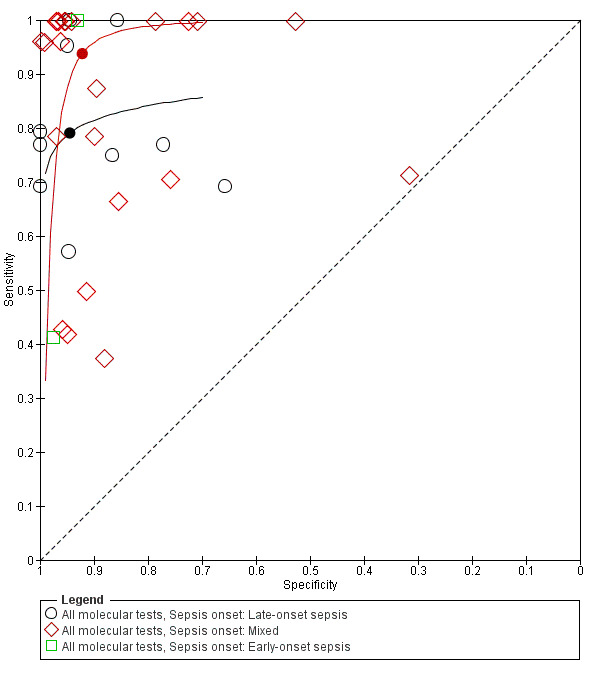
Summary receiver operating characteristic plot subgrouped by sepsis onset.
Five studies reported on testing on preterm infants only and 30 studies on a combination of preterm and term infants. Summary estimates for studies reporting on only preterm infants were sensitivity 0.89 (95% CI 0.75 to 0.96), specificity 0.87 (95% CI 0.71 to 0.94) and those for mixed term and preterm infants were sensitivity 0.90 (95% CI 0.80 to 0.96), specificity 0.94 (0.90 to 0.96) (Figure 9; Table 1).
9.

Summary receiver operating characteristic plot subgrouped by gestational age.
We categorized studies into three groups based on prevalence less than 15%, 15 % to 30% and greater than 30%. Summary estimates for 20 studies with a prevalence of less than 15% were sensitivity 0.94 (95% CI 0.80 to 0.99), specificity 0.95 (95% CI 0.92 to 0.97), with prevalence 15% to 30% were sensitivity 0.85 (95% CI 0.67 to 0.94), specificity 0.88 (95% CI 0.79 to 0.94) and those for studies with a sepsis prevalence greater than 30% were sensitivity 0.87 (95% CI 0.75 to 0.93), specificity 0.93 (95% CI 0.64 to 0.99) (moderate to low quality evidence) (Table 1; Figure 10; Figure 11).
10.
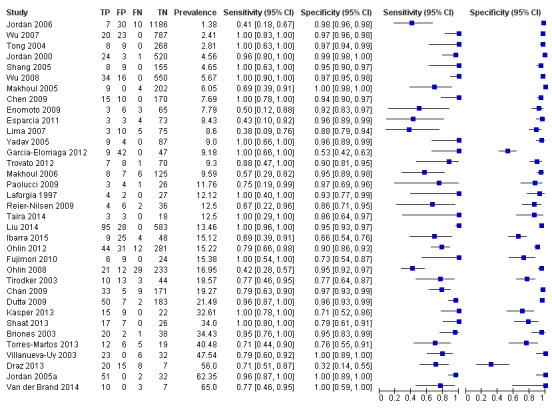
Forest plot of all molecular tests sorted in order of prevalence. CI: confidence interval; FN: false negative; FP: false positive; TN: true negative; TP: true positive.
11.
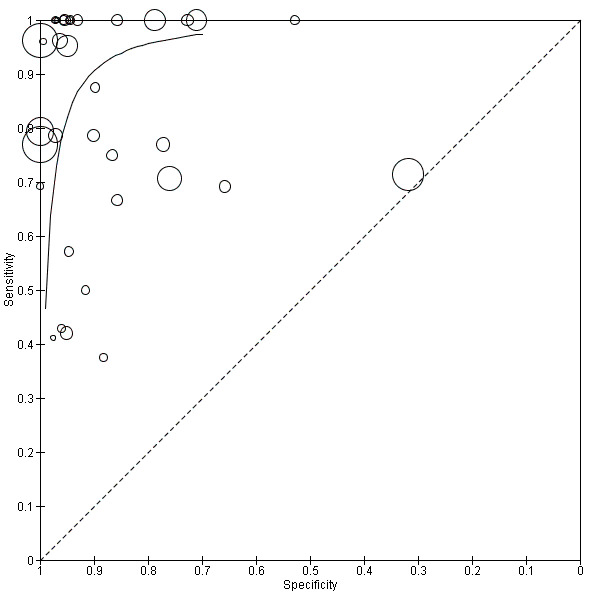
Summary receiver operating characteristic plot of all molecular tests where the size of the study symbol is directly proportional to the prevalence of sepsis in the study.
We performed sensitivity analyses using data from studies evaluating blood samples alone (not CSF) excluding three studies; the summary sensitivity was 0.92 (95% CI 0.84 to 0.96), specificity 0.93 (95% CI 0.89 to 0.95) (Figure 12; Table 1) (moderate quality evidence). Furthermore, we investigated the effect of the potential sources of bias by removing studies with unclear or high risk of bias or applicability concerns (13 studies) from the total set of studies and re‐analyzing this new set (22 studies) and found no differences in summary estimates; the summary sensitivity was 0.90 (95% CI 0.78 to 0.96), specificity 0.93 (95% CI 0.88 to 0.96) (moderate quality evidence) (Table 1).
12.
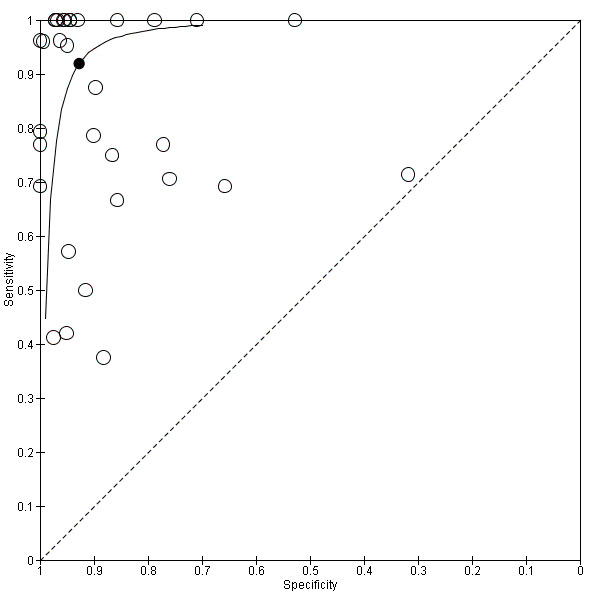
Summary receiver operating characteristic plot of studies that performed molecular tests on blood samples only.
Discussion
Summary of main results
Our search strategy identified 35 eligible studies and mean sensitivity of molecular tests in the diagnosis of neonatal sepsis was 0.90 (95% CI 0.82 to 0.95) and specificity was 0.93 (95% CI 0.89 to 0.96) and evidence was of moderate quality. We explored heterogeneity by subgroup analyses based on type of test, gestational age, type of sepsis onset and prevalence of neonatal sepsis (moderate to low quality evidence). We also performed sensitivity analysis by excluding studies which used both blood and CSF samples and excluding studies with high or uncertain risk of bias and applicability concerns.
Low sensitivity (less than 0.7 in nine studies) in some of the studies may be explained by the technicalities of the multiplex PCR assay, use of stored blood samples that were drawn by heel stick at a different time to the blood culture sample, participant characteristics and to Staphylococcus‐specific PCR. Jordon and colleagues commented that presence of white blood cells in the samples and hence human genomic DNA interference may have inhibited the PCR assay accounting for low sensitivity (Jordan 2006). However, 13 studies reported a sensitivity of 1.00 that did not conform to any particular type of test or participant population. In contrast, specificity was consistently higher than sensitivity and all except three of the included studies had specificity more than 0.70 (Draz 2013; Garcia‐Elorriaga 2012; Ibarra 2015). Primers used in the tests and differences in participant characteristics may accounted for low specificity. Four studies reported a specificity of 1.00 but with varying sensitivities and type of molecular assays (Jordan 2005a; Makhoul 2005; Van der Brand 2014; Villanueva‐Uy 2003).
We explored sources of heterogeneity by subgroup analyses based on type of test, gestational age, type of sepsis onset and prevalence. We found that studies evaluating molecular tests with post‐PCR processing, real‐time PCR and broad‐range conventional PCR plotted in the left upper corner of the ROC space and had higher sensitivity and specificity than multiplex PCR assay. Summary sensitivities from subgroups based on gestational age were similar with overlapping CIs and summary specificity was higher in studies that evaluated both preterm and term infants. In 10 studies that evaluated only LOS, the sensitivity was lower than the summary estimate for mixed EOS and LOS (0.79 (95% CI 0.66 to 0.87) versus 0.94 (95% CI 0.84 to 0.98)) but had higher specificity (0.94 (95% CI 0.85 to 0.98) versus 0.92 (95% CI 0.87 to 0.95)). But the wide 95% CIs precluded any delineation based on these subgroup analyses.
We categorized studies into three groups based on sepsis prevalence less than 15%, 15% to 30% and greater than 30%. Studies that evaluated molecular tests in a population with a sepsis prevalence less than 15% had higher sensitivity and specificity (sensitivity 0.94 (95% CI 0.80 to 0.99), specificity 0.95 (95% CI 0.92 to 0.97)) compared with studies in a higher sepsis prevalence population (Table 1). Variations in participant characteristics or test methodology may account for some of these differences.
We performed sensitivity analyses by type of samples used (blood or both blood and CSF, because inhibitors of PCRs may be present only in blood samples) and for studies evaluating blood samples alone (not CSF), the summary sensitivity was 0.92 (95% CI 0.84 to 0.96) and specificity 0.93 (95% CI 0.89 to 0.95) (Figure 12; Table 1). We also investigated the effect of the potential sources of bias by removing studies with unclear or high risk of bias or applicability concerns (13 studies) from the total set of studies and re‐analyzing this new set (22 studies) and found no differences (summary sensitivity 0.90 (95% CI 0.78 to 0.96), specificity 0.93 (95% CI 0.88 to 0.96)) (Figure 13; Table 1).
13.
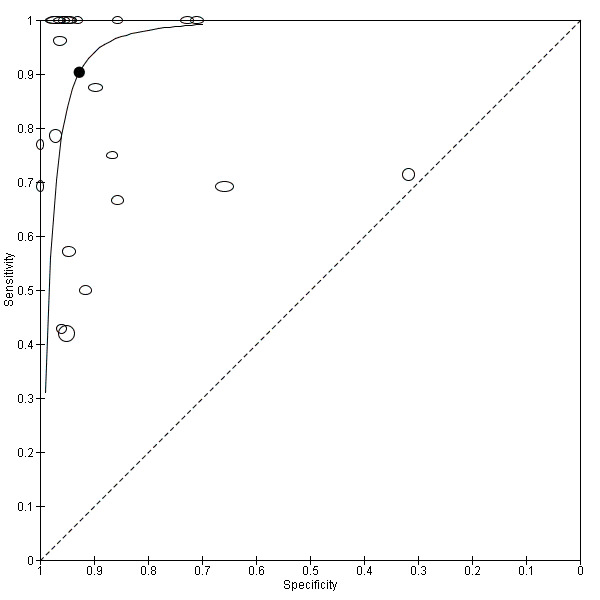
Summary receiver operating characteristic plot of molecular tests with good methodologic quality.
Other sources for variation of diagnostic test accuracy among studies evaluating molecular tests may be due to methods of DNA extraction or preprocessing the sample before DNA extraction (e.g. preincubation of the blood culture media before DNA extraction). Studies using whole blood DNA extraction had low sensitivity and preincubation of sample for five hours in tryptic soy broth increased sensitivity significantly. However, the methodologies of DNA extraction, samples from which DNA were extracted, varied considerably among the studies to make any meaningful comparisons.
New diagnostic tests can assume the following roles in a diagnostic pathway: replacement of the existing test, triage or 'add on' to the existing test (Bossuyt 2006). Our meta‐analysis estimated a mean sensitivity of 0.90 (95% CI 0.82 to 0.95) and a mean specificity 0.93 (95% CI 0.89 to 0.96) for molecular assays. The mean estimated sensitivity of molecular assays are better than other alternative tests used to diagnose sepsis such as platelet count, CRP, procalcitonin, TNF and IL‐6 while mean specificity was similar to these tests (Blommendahl 2002; Hornik 2012; Ng 1997; Ng 2012; Verboon‐Maciolek 2006). Theoretically, in 1000 VLBW neonates screened for EOS, where the prevalence was 2% (using the summary estimates of this review), we would miss two cases of sepsis and overtreat 69 neonates without sepsis. Similarly, in 1000 VLBW neonates screened for LOS (prevalence 10%), we would miss 10 culture‐positive cases and overtreat 63 neonates without sepsis. Thus, currently available molecular assays may not have sufficient diagnostic accuracy to replace microbial cultures. However, advancing technologies in molecular microbiology may bring forth newer assays with higher sensitivity and specificity, sufficient to replace microbial cultures in the diagnosis of neonatal sepsis.
In addition to test accuracy, it is important to consider management strategies for neonatal sepsis where molecular tests may be useful. Evidence to decision frameworks are recommended to assess how test results affect participant outcomes (Schünemann 2016; Trenti 2016). In the context of neonatal sepsis, molecular assays are unlikely to be used as a triage test that will select neonates who would undergo cultures. An unwanted delay in performing blood cultures may ensue and may postpone treatment. False negatives on the molecular tests will compromise neonatal safety. However, molecular assays have a faster turnaround time and may perform well as 'add‐on' tests where molecular assays may be performed concurrently with the gold standard (cultures). Results of molecular assays are available in six to eight hours and may help in optimizing clinical therapy. If the molecular test is negative, antibiotics may be discontinued if the test assay has high specificity and high negative predictive value. Decrease in antibiotic doses and decreased length of stay are potential advantages of such a strategy (Brozanski 2006). If the molecular test assay is positive (and if the assay has high sensitivity) then a case could be made for continuation of antibiotics. Molecular assays may theoretically diagnose sepsis in neonates exposed to antibiotics including maternal exposure to antibiotics in EOS, where cultures are negative and potentially decrease resource utilization. Combination of blood cultures with an 'add‐on' molecular test may improve sensitivity at the cost of specificity. Newer molecular assays that can identify the organism or detect antibiotic resistance can guide antibiotic therapy.
Jordan and colleagues and our group reviewed the methodology of molecular assays used in the diagnosis of neonatal sepsis without synthesizing data using meta‐analyses (Jordan 2010; Venkatesh 2010). Our group published one systematic review with meta‐analysis of 23 studies evaluating molecular assays in the diagnosis of neonatal sepsis (Pammi 2011). Overall, the summary estimates of sensitivity and specificity were similar with larger CIs and slightly higher specificity (sensitivity 0.90 (95% CI 0.78 to 0.95), specificity 0.96 (95% CI 0.94 to 0.97)). In our previous review, we were unable to analyze reasons for heterogeneity as data were not available, which we were able to do in this review.
Strengths and weaknesses of the review
Strengths: our systematic review was based on methodology recommended by the Cochrane Diagnostic Test Accuracy Working Group (Leeflang 2008). We performed a comprehensive search for all eligible studies using clinically relevant inclusion criteria. We used the bivariate random‐effects model for meta‐analyses of the included studies. We strived to explain the sources of heterogeneity by subgroup analyses using test type, gestational age of participants, type of sepsis onset and prevalence.
Weaknesses: evolution in methodology in the included studies over time (1997 to 2016) may account for variations in the diagnostic accuracy among studies. Unlike meta‐analyses of randomized controlled trials, heterogeneity is a well‐recognized problem in reviews of diagnostic test accuracy (Reitsma 2009). Despite our extensive search strategy, we may have missed potential studies, as diagnostic accuracy studies are poorly tagged in electronic databases. Publication bias in studies reporting diagnostic test accuracy has been poorly studied (Leeflang 2008). Poor reporting of study design, method of enrollment and participant characteristics may hamper methodologic assessment and external validity of the studies. Another limitation of our review might be that the reference standard (microbial cultures) is thought to be far from perfect. Interpretation of the accuracy of molecular assays is challenging given the assumed low sensitivity of the blood cultures. However, as our summary sensitivity of the molecular assays was poor (0.90) and the proportion of false positives was low, it does not seem to be the case.
Applicability of findings to the review question
Molecular assays have significant advantages when performed in conjunction with microbial cultures as an 'add‐on' test. The high specificity of molecular assay in LOS evaluation (0.94 (95% CI 0.85 to 0.98)) has the potential of decreasing antibiotic exposure by aiding physicians to make earlier decisions about discontinuation of antibiotics. Molecular assays, including PCR and hybridization methods, are feasible in neonates and have rapid detection times compared to blood cultures (six to eight hours versus 20 to 36 hours). Detection of pathogen DNA in the absence of viable organisms by culture and false‐negative results due to the presence of inhibitors may require careful interpretation. Molecular assays may have a significant impact on early diagnosis and treatment of neonatal sepsis. However, current molecular assays do not provide antibiotic susceptibility that may be important clinically. Microbiologic cultures detect most organisms causing neonatal sepsis, whereas molecular assays focused on fungi or a specific organism (Staphylococcus‐ or fungus‐specific PCR) do not. Costs, availability of equipment and technical skills in the microbiologic laboratory are important considerations that will impact applicability.
Authors' conclusions
Implications for practice.
The mean sensitivity of molecular assays in the diagnosis of clinically suspected neonatal sepsis was 0.90 (95% CI 0.82 to 0.95) and mean specificity was 0.93 (95% CI 0.89 to 0.96) (moderate quality evidence) and the diagnostic accuracy was variable among reported studies. Molecular tests for the diagnosis of sepsis may be useful 'add‐on' tests as they give rapid information that may aid clinical decisions regarding treatment. Our recommendations are based on moderate to low quality evidence. Optimization of existing assays or the development of new molecular assays in the future may improve diagnostic accuracy. Future molecular tests that may identify the pathogen and evaluate pathogen virulence and antibiotic susceptibility, in addition to diagnosis of sepsis may aid clinical management tremendously.
Implications for research.
Investigators evaluating current as well as future molecular tests should design their studies satisfying the items expounded in the QUADAS‐2 evaluation system, so that studies are of high methodologic quality and bias is minimal. Studies reporting diagnostic test accuracy should explicitly state the method of enrollment (prospective or retrospective), characteristics of the population assessed (such as gestational age, chronologic age range, birth weight, comorbidity), blinding of reference standard and index tests, and explanation of withdrawals. Details of the clinical setting and participant characteristics will help clinicians decide whether a diagnostic test is applicable in their population. Costs of the molecular assays need to be balanced with their ability to impact clinical outcomes before widespread acceptance in clinical practice.
What's new
| Date | Event | Description |
|---|---|---|
| 26 December 2016 | Amended | Revised based on suggestions from reviewers |
Acknowledgements
The Cochrane Neonatal Group has been funded in part with federal funds from the Eunice Kennedy Shriver National Institute of Child Health and Human Development National Institutes of Health, Department of Health and Human Services, USA, under Contract No. HHSN267200603418C. We acknowledge the help of Nha Huynh, librarian at the Texas Medical Center library in Houston, TX, USA and Milagros De Jesus, librarian at Texas Technical University Health Sciences Center, Lubbock, TX, USA for invaluable help with search strategy.
We acknowledge the help of Joe Hagan, statistician, Section of Neonatology at Baylor College of Medicine in creating the Deeks' funnel plot and checking the statistics for the hierarchical summary receiver operating characteristic (HSROC) and the bivariate model.
Appendices
Appendix 1. Search strategy
1. Our search strategy for PubMed below was developed by discussion between the author team and the Cochrane Neonatal Group's Trials Search coordinator. We adapted it for use in other databases. www‐ncbi‐nlm‐nih‐gov.ezproxyhost.library.tmc.edu/pubmed?otool=hamtmc
Search ((((((((((( "Sepsis/blood"[Mesh] OR "Sepsis/cerebrospinal fluid"[Mesh] OR "Sepsis/diagnosis"[Mesh] ))) OR neonatal sepsis) OR neonatal bacteremia) OR neonatal infections)) AND "Infant, Newborn"[Mesh])) AND ((((((((((polymerase chain reaction) OR pcr) OR real time pcr) OR multiplex pcr) OR molecular probes) OR nucleic acid amplification) OR hybridization) OR sequencing) OR pyrosequencing) OR genechip))) AND (((((diagnosis) OR detection) OR identification) OR rapid identification) OR diagnostic tool)
2. EMBASE search strategy (provided by Elsevier through TMC library)
#1 sepsis
#2 Infection
#3 bacteremia
#4 #1 OR #2 OR #3
#5 neonate
#6 newborn
#7 #5 OR #6
#8 diagnosis OR detection OR identification OR diagnostic
#9 PCR
#10 molecular AND methods
#11 nucleic AND acid AND amplification
#12 hybridization
#13 sequencing
#14 polymerase AND chain AND reaction
#15 #9 OR #10 OR #11 OR #12 OR #13 OR #14
#16 Human
#17 #4 AND #7 AND #8 AND #15 AND #16
3. CINAHL search strategy (platform EBSCO host)
#1 sepsis
#2 Infection
#3 bacteremia
#4 #1 OR #2 OR #3
#5 neonate
#6 newborn
#7 #5 OR #6
#8 diagnosis OR detection OR identification OR diagnostic
#9 PCR
#10 molecular AND methods
#11 nucleic AND acid AND amplification
#12 hybridization
#13 sequencing
#14 polymerase AND chain AND reaction
#15 #9 OR #10 OR #11 OR #12 OR #13 OR #14
#16 Human
#17 #4 AND #7 AND #8 AND #15 AND #16
4. Cochrane libraryhttp://www.cochranelibrary.com.ezproxyhost.library.tmc.edu/
Using advanced search and selecting Cochrane Reviews, other reviews, trials and methods
studies. Using search words, molecular, neonate, newborn, PCR and sepsis
5. Science citation index, platform‐Web of science
Searched using advanced search and subject search with search words, ‘molecular’, ‘neonate’,
‘newborn’, ‘PCR’, ‘nucleic acid’ ‘diagnostic’ and sepsis using BOOLEAN combination words.
Appendix 2. Data from included studies
| Ref | Method | Data | TP | FP | FN | TN | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Participants | Study period | Comments | |||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Briones 2003 | Fungal conventional PCR targeting ITS3 and ITS4 regions of the 5S rRNA. | Positive | 20 | 2 | 22 | 20 | 2 | 1 | 38 | 95.24 | 95.00 | 90.91 | 97.44 | Newborns > 3 days old suspected of sepsis. No information on demographics. |
Not mentioned. | Abstract. Same authors as Villanueva‐Uy and same number of cases but using different primers (bacterial vs fungal). |
| Negative | 1 | 38 | 39 | |||||||||||||
| 21 | 40 | 61 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Chan 2009 | RT‐PCR with universal primers and Gram‐specific probes Blood, peritoneal fluid and urine. |
Positive | 33 | 5 | 38 | 33 | 5 | 9 | 171 | 78.57 | 97.16 | 86.84 | 95.00 | Preterm infants < 37 wk GA, > 72 hr of age with signs and symptoms of systemic infection requiring full sepsis evaluation and antibiotic treatment. Interquartile range of age as reported in results suggested some infants > 28 days old. | Over 28‐month period from Mar 2006 to Jun 2008. | ‐ |
| Negative | 9 | 171 | 180 | |||||||||||||
| 42 | 176 | 218 | ||||||||||||||
| Reference std Blood and CSF Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Chen 2009 | Broad‐range 16S rRNA‐based real‐time FQ‐PCR. | Positive | 15 | 10 | 25 | 15 | 10 | 0 | 170 | 100.00 | 94.44 | 60.00 | 100.00 | Neonates admitted to the neonatal department and ICU of the Children's Hospital at Zhejiang University in China with suspected sepsis or meningitis. No information on demographics. |
Sept 2007 to Jun 2008. | Blood (n = 190) and CSF (n = 5) samples. Each sample tested for Cx and PCR. Not sure if blood drawn concurrently for Cx and PCR. Not blinded. |
| Negative | 0 | 170 | 170 | |||||||||||||
| 15 | 180 | 195 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Draz 2013 | Broad‐range 16S rDNA PCR. | Positive | 20 | 15 | 35 | 20 | 15 | 8 | 7 | 71.43 | 31.82 | 57.14 | 46.67 | Neonates with clinical or lab findings suggestive of sepsis. | May 2012 to Aug 2012. | The authors mentioned 6 samples were considered contaminated: 4 with Diphtheroid spp. and 2 with Candida. Appears these 6 were eventually considered as negative blood Cx. |
| Negative | 8 | 7 | 15 | |||||||||||||
| 28 | 22 | 50 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Dutta 2009 | Broad‐range conventional PCR after 5‐hr preamplification Cx. | Positive | 50 | 7 | 57 | 50 | 7 | 2 | 183 | 96.15 | 96.32 | 87.72 | 98.92 | Neonates who were clinically suspected to have an episode of sepsis with onset of ≥ 72 hr after cessation of antibiotics. | Not mentioned. | Aseptically collected and concurrent blood draw for PCR and Cx. Not blinded. |
| Negative | 2 | 183 | 185 | |||||||||||||
| 52 | 190 | 242 | ||||||||||||||
| Reference std Cx | ||||||||||||||||
| Positive | Negative | |||||||||||||||
| Enomoto 2009 | Multiplex PCR targeting 8 pathogens Also includes skin, BAL, mucus, CSF, urine and ascites. |
Positive | 3 | 5 | 8 | 3 | 5 | 3 | 66 | 50.00 | 92.96 | 37.50 | 95.65 | 130 clinical samples from 62 newborns with any suspicious infectious signs or infections and 50 cord bloods and blood after birth from healthy term infants without signs or history of infection. Total of 77 paired samples. |
Jun 2005 to Sept 2006. | In Table 2, number of positive PCR was 9 not 8 as in Table 3. Number of samples with no test was 8 unless pharyngeal mucus was included. Those doing Cx were blinded but no mention of those doing PCR. |
| Negative | 3 | 66 | 69 | |||||||||||||
| 6 | 71 | 77 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Esparcia 2011 | 16S RT‐PCR followed by microarray and sequencing. Includes CSF samples where PCR and Cx were performed. |
Positive | 3 | 3 | 6 | 3 | 3 | 4 | 73 | 42.86 | 96.05 | 50.00 | 94.81 | Newborn < 7 days old with suspected sepsis or early meningitis. | Nov 2005 to Jan 2007. | There were 105 samples from 83 newborns for EOS. In the paper, results referred to cases of EOS and not samples, hence n = 83. |
| Negative | 4 | 73 | 77 | |||||||||||||
| 7 | 76 | 83 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Fujimori 2010 | RT‐PCR. | Positive | 6 | 9 | 15 | 6 | 9 | 0 | 24 | 100.00 | 72.73 | 40.00 | 100.00 | Neonates admitted to NICU with suspected sepsis. Mean (SD) GA 34.8 ± 5.8 wk. 36 neonates with 39 episodes of neonatal sepsis. | Feb 2009 to Aug 2009. | Concurrent blood draw. Repeated samples taken in same episode were excluded. Not blinded. |
| Negative | 0 | 24 | 24 | |||||||||||||
| 9 | 33 | 39 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Garcia‐Elorriaga 2012 | Broad‐range PCR primer Note: authors' gold std was clinical Dx. |
Positive | 9 | 38 | 47 | 9 | 38 | 0 | 2 | 100.00 | 5.00 | 19.15 | 100.00 | Neonates aged ≤ 28 days admitted to NICU with clinical Dx of sepsis without antibiotic treatment or with maximum 48 hr antibiotic treatment or > 3 days' treatment but without response. | Aug 2005 to Jul 2006. | Calculation based on blood Cx of case only. Total positive Cx on table 2 = 33 but Table 4 = 23. Unsure where to add 2 positive catheter as it is unclear in table where PCR was done. |
| Negative | 0 | 2 | 2 | |||||||||||||
| 9 | 40 | 49 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Ibarra 2015 | LightCycler SeptiFast. | Positive | 9 | 25 | 34 | 9 | 25 | 4 | 48 | 69.23 | 65.75 | 26.47 | 92.31 | Neonates with suspected clinical sepsis and those presenting > 8 on NOSEP‐1 scale. 86 samples from 86 neonates included. Table 4 shows that neonates in the blood Cx group may be > 28 days old as it reported (mean ± SD) 23 ± 9.2 days. |
Not mentioned. | Concurrent samples for Cx and LightCycler SeptiFast. PPV and NPV reported were different (69% and 65%, respectively). |
| Negative | 4 | 48 | 52 | |||||||||||||
| 13 | 73 | 86 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Jordan 2000 | Broad‐range PCR and DNA blot analysis. | Positive | 24 | 3 | 27 | 24 | 3 | 1 | 520 | 96.00 | 99.43 | 88.89 | 99.81 | All infants admitted to NICU for sepsis evaluation. No information on demographics. |
Not mentioned. | Not blinded. Good technique. Eliminated contaminants. |
| Negative | 1 | 520 | 521 | |||||||||||||
| 25 | 523 | 548 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Jordan 2005a | 16S rRNA RT‐PCR. | Positive | 51 | 51 | 51 | 0 | 2 | 32 | 96.23 | 100.00 | 100.00 | 94.12 | Neonates admitted to NICU. No information on demographics. |
Not mentioned. | Calculation based on number of samples not cases. Numbers were derived from the paper that stated 53 were Cx positive and of the 53, 51 were also PCR positive and 2 that were PCR negative. 32 samples were Cx negative and PCR negative. No mention if blinded. |
|
| Negative | 2 | 32 | 34 | |||||||||||||
| 53 | 32 | 85 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Jordan 2006 | Conventional PCR based on 16S rRNA assay followed by pyrosequencing. | Positive | 7 | 30 | 37 | 7 | 30 | 10 | 1186 | 41.18 | 97.53 | 18.92 | 99.16 | Eligible infants had to be > 34 wk GA at time of birth, admitted to NICU within a few hours for EOS evaluation, and have both a blood Cx and CBC ordered. No details on demographics. | 1 Sept 2000 to 1 Apr 2004. | No mention if blinded. |
| Negative | 10 | 1186 | 1196 | |||||||||||||
| 17 | 1216 | 1233 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Kasper 2013 | Multiplex RT‐PCR (LightCycler) SeptiFast MGRADE system for detection of LOS. Targeted Gram‐positive and G‐negative organisms between 16S and 23S rRNA genes, and fungi by 18S and 5.8S rDNA. | Positive | 15 | 9 | 24 | 15 | 9 | 0 | 22 | 100.00 | 70.97 | 62.50 | 100.00 | 46 VLBW infants > 72 hr of life with suspected LOS. Details on demographic including day of life of sepsis evaluation were not mentioned. | Not mentioned. | ‐ |
| Negative | 0 | 22 | 22 | |||||||||||||
| 15 | 31 | 46 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Laforgia 1997 | Multiplex PCR. | Positive | 4 | 2 | 6 | 4 | 2 | 0 | 27 | 100.00 | 93.10 | 66.67 | 100.00 | 33 newborns at risk for EOS. | Jan to Sept 1996. | ‐ |
| Negative | 0 | 27 | 27 | |||||||||||||
| 4 | 29 | 33 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Lima 2007 | RT‐PCR using universal primer. | Positive | 3 | 10 | 13 | 3 | 10 | 5 | 75 | 37.50 | 88.24 | 23.08 | 93.75 | 93 samples for neonates with suspected sepsis. No information on demographics. |
Dec 2004 to Jun 2005. | Abstract. 93 blood samples. 3 were blood Cx and PCR‐positive. 5 were blood Cx positive, 10 were positive by molecular method. 4 samples not included as it was positive for human chromosomes. |
| Negative | 5 | 75 | 80 | |||||||||||||
| 8 | 85 | 93 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Liu 2014 | Broad‐range 16S rRNA gene PCR. | Positive | 95 | 28 | 133 | 95 | 28 | 0 | 583 | 100.00 | 95.42 | 77.24 | 100.00 | Neonates who had blood drawn for CBC and CRP. Infants were 4 hr to 28 days old. | 1 Sept to 31 Dec 2011. | ‐ |
| Negative | 0 | 583 | 583 | |||||||||||||
| 95 | 911 | 706 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Makhoul 2005 | Staphylococcal 16S rRNA PCR (both S. aureus and CONS). | Positive | 9 | 0 | 9 | 9 | 0 | 4 | 202 | 69.23 | 100.00 | 100.00 | 98.06 | Neonates hospitalized in the NICU with clinical signs suggestive of sepsis after 3 days of life. 124 neonates with 215 events. There was no mention of how many infants were > 28 days old. |
12‐month period. | Mean (± SD) GA 33.5 ± 4.4 (range 24 to 42 wk), mean birth weight 1962 ± 874 g (range 560 g to 3939 g), mean age at onset of presumed sepsis was 15.4 ± 17.3 days (range 4 to 96 days). Not blinded. |
| Negative | 4 | 202 | 206 | |||||||||||||
| 13 | 202 | 215 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Makhoul 2006 | Staphylococcal 16S rRNA PCR (both S. aureus and CONS). | Positive | 8 | 7 | 15 | 8 | 7 | 6 | 125 | 57.14 | 94.70 | 53.33 | 95.42 | Neonates with clinically suspected LOS beyond 3 days of life. No mention how many infants were > 28 days old. |
Not mentioned. | Mean age (± SD) at onset of presumed sepsis was 17.3 ± 18.7 days (range 4 to 105 days). Not mentioned if blinded. Discrepancy with published number and actual number (148 vs 146). |
| Negative | 6 | 125 | 131 | |||||||||||||
| 14 | 132 | 146 (?148) | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Ohlin 2008 | RT‐PCR 16S RNA. | Positive | 21 | 12 | 33 | 21 | 12 | 29 | 233 | 42.00 | 95.10 | 63.64 | 88.93 | Newborns < 28 days old admitted to NICU. n = 295 refers to samples from 288 infants. | 1995 to 2005. | Not blinded. |
| Negative | 29 | 233 | 262 | |||||||||||||
| 50 | 245 | 295 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Ohlin 2012 | Broad‐range 16S RT‐PCR. | Positive | 44 | 31 | 75 | 44 | 31 | 12 | 281 | 78.57 | 90.06 | 58.67 | 95.90 | Infants < 3 months of age subjected to blood Cx. total of 368 samples from 317 infants. | Oct 2007 to Nov 2009. | 34 samples were collected at postnatal age from 29 days to 3 months; however, no specific information on the blood Cx and PCR results of these samples. |
| Negative | 12 | 281 | 293 | |||||||||||||
| 56 | 312 | 368 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Paolucci 2009 | Commercial LightCycler SeptiFast System. | Positive | 3 | 4 | 7 | 3 | 4 | 1 | 26 | 75.00 | 86.67 | 42.86 | 96.30 | Newborns with suspected LOS. Age of infant at time of Dx not mentioned. |
Not mentioned. | Not blinded. |
| Negative | 1 | 26 | 27 | |||||||||||||
| 4 | 30 | 34 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Reier‐Nilsen 2009 | Broad‐range 16S rRNA PCR followed by sequencing of PCR products. | Positive | 4 | 6 | 10 | 4 | 6 | 2 | 36 | 66.67 | 85.70 | 40.00 | 94.70 | Infants with birth weight > 1000 g with suspected sepsis during first wk of life. | Not mentioned. | Prospective, non‐RCT. Sterile technique. Same blood draw for Cx and PCR. Blinded. Second table was used in article. Included all (n = 48) cases of suspected sepsis. |
| Negative | 2 | 36 | 38 | |||||||||||||
| 6 | 42 | 48 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Shaat 2013 | Broad‐range 16S rDNA PCR. | Positive | 17 | 7 | 24 | 17 | 7 | 0 | 26 | 100.00 | 78.79 | 70.83 | 100.00 | Newborns with clinically suspected sepsis. | Oct 2010 to Dec 2012. | GA ranged from 26 to 39 wk, mean (± SD) 32.44 ± 2.91 wk; however, age at Dx not mentioned. |
| Negative | 0 | 26 | 26 | |||||||||||||
| 17 | 33 | 50 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Shang 2005 | Broad‐range PCR with microarray hybridization. Positive specimens subjected to microarray hybridization. |
Positive | 8 | 9 | 17 | 8 | 9 | 0 | 155 | 100.00 | 94.51 | 47.06 | 100.00 | All neonates who developed clinical signs suggestive of sepsis after 3 days of life. | 1 Jan to 30 Jun 2004. | Authors did not provide additional characteristics of infants included in the study. Sensitivity was 94.51% (155/164). Not sure how the authors came up with 97.85%. Not blinded. |
| Negative | 0 | 155 | 155 | |||||||||||||
| 8 | 164 | 172 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Taira 2014 | Multiplex nested PCR for detection and identification of Candida species. | Positive | 8 | 5 | 13 | 8 | 5 | 0 | 41 | 100.00 | 89.13 | 61.54 | 100.00 | Information on neonates was based on correspondence with Dr Del Negro. | 18‐month period. | ‐ |
| Negative | 0 | 41 | 41 | |||||||||||||
| 8 | 46 | 54 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Tirodker 2003 | Fungal conventional PCR targeting 18S rRNA fungi. | Positive | 10 | 13 | 23 | 10 | 13 | 3 | 44 | 76.92 | 77.19 | 43.48 | 93.62 | 70 samples from 63 infants (46 from the NICU and 17 from PICU) with suspected clinical sepsis. | Nov 1999 to Nov 2000. | Study infants from NICU (46 infants) and PICU (17 infants). Neonatal specific data on blood Cx and PCR not available. Aseptic and concurrent blood sampling. Blinded. |
| Negative | 3 | 44 | 47 | |||||||||||||
| 13 | 57 | 70 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Tong 2004 | 16S rRNA‐based PCR followed by hybridization to chips with 18 probes. | Positive | 8 | 9 | 17 | 8 | 9 | 0 | 268 | 100.00 | 96.75 | 47.06 | 100.00 | Neonates with suspected sepsis. | Not mentioned. | Abstract only. No specific details provided for demographics. |
| Negative | 0 | 268 | 268 | |||||||||||||
| 8 | 277 | 285 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Torres‐Martos 2013 | LightCycler SeptiFast. | Positive | 12 | 6 | 18 | 12 | 6 | 5 | 19 | 70.59 | 76.00 | 66.67 | 79.17 | 42 blood samples from 35 infants with febrile episodes. Based on Table 1. Infants were 0 to 151 days. | Apr 2007 to Apr 2009. | Sensitivity, specificity, PPV and NPV values reported in paper were based on comparison on LightCycler SeptiFast with clinical Dx. |
| Negative | 5 | 9 | 24 | |||||||||||||
| 17 | 25 | 42 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Trovato 2012 | Fungus‐specific universal primers ITS1 and ITS2 used to amplify rDNA, the adjacent ITS1 and small portion of the 28S rDNA. | Positive | 7 | 8 | 15 | 7 | 8 | 1 | 70 | 87.50 | 89.74 | 46.67 | 98.59 | Neonates at high risk for invasive candidiasis. | Jan 2009 to Dec 2010. | No detailed information on demographics. |
| Negative | 1 | 70 | 71 | |||||||||||||
| 8 | 78 | 86 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Van der Brand 2014 | Multiplex RT‐PCR. | Positive | 10 | 0 | 10 | 10 | 0 | 3 | 7 | 76.92 | 100.00 | 100.00 | 70.00 | Preterm infants admitted to NICU and suspected to have LOS. No details on age of infants during evaluation for LOS. | Not mentioned. | ‐ |
| Negative | 3 | 7 | 10 | |||||||||||||
| 13 | 7 | 20 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Villanueva‐Uy 2003 | Broad‐range 16S rRNA conventional PCR. | Positive | 23 | 0 | 23 | 23 | 0 | 6 | 32 | 79.31 | 100.00 | 100.00 | 84.21 | Neonates > 3 days old with suspected sepsis. No information on upper age limit. |
Not mentioned. | Abstract. |
| Negative | 6 | 32 | 38 | |||||||||||||
| 29 | 32 | 61 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Wu 2007 | RT‐PCR 16S RNA. | Positive | 20 | 23 | 43 | 20 | 23 | 0 | 787 | 100.00 | 97.16 | 46.51 | 100.00 | Blood samples from cases of suspected septicemia. No mention of upper age limit. | Not mentioned. | Abstract only. |
| Negative | 0 | 787 | 787 | |||||||||||||
| 20 | 810 | 830 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Wu 2008 | RT‐PCR with Gram‐specific probes followed by sequencing. | Positive | 34 | 16 | 50 | 34 | 16 | 0 | 550 | 100.00 | 97.17 | 68.00 | 100.00 | Infants aged 1 to 28 days admitted to the neonatal ward or NICU for clinically suspected to have bacterial infection or to be susceptible to infection. | Jan 2005 to Jan 2007. | Not blinded but implied as Cx and PCR were done simultaneously. |
| Negative | 0 | 550 | 550 | |||||||||||||
| 34 | 566 | 600 | ||||||||||||||
| Reference std Blood Cx |
||||||||||||||||
| Positive | Negative | |||||||||||||||
| Yadav 2005 | Broad‐range 16S rRNA PCR. | Positive | 9 | 4 | 13 | 9 | 4 | 0 | 87 | 100.00 | 95.60 | 69.23 | 100.00 | Newborns with risk factor for sepsis. | Not mentioned. | Not blinded. |
| Negative | 0 | 87 | 87 | |||||||||||||
| 9 | 91 | 100 | ||||||||||||||
| BAL: bronchoalveolar lavage; CBC: complete blood count; CONS: coagulase‐negative staphylococci; CRP: C‐reactive protein; CSF: cerebrospinal fluid; Cx: culture; Dx: diagnosis; EOS: early‐onset sepsis; FP: false positive; FN: false negative; FQ‐PCR: quantitative fluorescence polymerase chain reaction; GA: gestational age; hr: hour; ICU: intensive care unit; LOS: late‐onset sepsis; NICU: neonatal intensive care unit; NPV: negative predictive value; PCR: polymerase chain reaction; PICU: pediatric intensive care unit; PPV: positive predictive value; RCT: randomized controlled trial; rRNA: ribosomal ribonucleic acid; RT‐PCR: real‐time polymerase chain reaction; SD: standard deviation; std: standard; TP: true positive; TN: true negative; wk: week. | ||||||||||||||||
Appendix 3. QUADAS‐2 methodologic assessment tool
QUADAS‐2 is structured so that four key domains are each rated in terms of the risk of bias and the concern regarding applicability to the research question (as defined above). Each key domain has a set of signaling questions to help reach the judgments regarding bias and applicability.
Domain 1: Participant selection
A. Risk of bias
Was a consecutive or random sample of participants enrolled?
YES: if the articles clearly stated that a consecutive or random samples was enrolled; NO: if it was clear that this was not the case (e.g. if a study included participants 'at the discretion of the clinician'); UNCLEAR: in other cases where it was not clear if consecutive or random samples were enrolled.
Was a case‐control design avoided?
YES: if the enrolled sample was a random or consecutive enrollment of neonates with suspected sepsis and not separate samples from sepsis‐positive participants and healthy controls; NO: if the enrolled samples consisted of sepsis‐confirmed cases and healthy controls; UNCLEAR: if the sampling regarding case‐control design was not clear.
Did the study avoid inappropriate exclusions?
Inappropriate exclusions included neonates whose mothers were treated with antibiotics, neonates from mothers infected with the human immunodeficiency virus (HIV), etc. YES: if inappropriate exclusions were not found in the included study, NO: if reasons for inappropriate exclusion were found. Unclear: if there was no description of the inclusion and exclusion criteria and inappropriate exclusion could not be ascertained.
Could the selection of participants have introduced bias?
LOW RISK: if all questions were scored "YES", or a maximum of one question with unclear.
HIGH RISK: if at least one question was scored as "NO".
UNCLEAR RISK: if at least two questions were scored as "UNCLEAR" and one as "NO".
B. Concerns regarding applicability
Was there concern that the included participants did not match the review question?
LOW CONCERN: if all included participants were neonates according to our definition and if they were suspected of sepsis.
HIGH CONCERN: if at least 10% of the included participants were not neonates or not suspected of sepsis.
UNCLEAR CONCERN: if it is unclear whether the study fulfilled either the criteria for low concern or for high concern.
Domain 2: Index test(s)
Describe the index test and how it was conducted and interpreted. If more than one index test was used, please complete for each test.
A. Risk of bias
Describe the index test and how it was conducted and interpreted
Were the index test results interpreted without knowledge of the results of the reference standard?
YES: if people performing the molecular assays were blinded to the results of blood or cerebrospinal fluid (or both) cultures or if the index test was performed and interpreted prior to the reference standard; NO: if people performing the molecular assays had knowledge of the results of blood or cerebrospinal fluid (or both) cultures; UNCLEAR: if the study did not explicitly describe how the index test was conducted and interpreted.
If a threshold was used, was it prespecified?
This signaling question is not applicable to the study as no thresholds were used in the conduct and interpretation of the index and the reference standards. Results of the tests were dichotomous and were reported as either positive or negative.
Could the conduct or interpretation of the index test have introduced bias?
LOW RISK: if the study was performed blinded to the results of the reference standard.
HIGH RISK: if there was prior knowledge of the results of the reference standard.
UNCLEAR RISK: if there was no clear description of how the tests were conducted and interpreted.
B. Concerns regarding applicability
Was there concern that the index test, its conduct, or interpretation differed from the review question?
LOW CONCERN: if the index test used for the diagnosis of sepsis was a molecular assay as defined in our protocol and if the index test was interpreted without the knowledge of the results of the reference standard.
HIGH CONCERN: if the index test used for the diagnosis of sepsis varied from what was defined in the protocol and if the index test was interpreted with knowledge of the results of the reference standard.
UNCLEAR CONCERN: if it was unclear whether the study fulfilled criteria for "low concern" or "high concern" or if the study provided limited information regarding the conduct and interpretation of the index test.
Domain 3: Reference standard
A. Risk of bias
Describe the reference standard and how it was conducted and interpreted
Was the reference standard likely to correctly classify the target condition?
YES: if the reference standard used was microbial culture of blood or cerebrospinal fluid (or both) in the diagnosis of neonatal sepsis. Microbial culture is currently the "gold standard" used in clinical practice in the diagnosis of neonatal sepsis; NO: if the test used as reference standard was a test other than microbial culture; UNCLEAR: if there was no description of the reference standard or if microbial cultures were used in combination with an "add‐on" test.
Were the reference standard results interpreted without knowledge of the results of the index test?
YES: if people evaluating the results of the microbial culture were blinded to the results of the molecular assays and if the reference standard was performed and interpreted prior to the index test; NO: if people evaluating the results of the microbial culture had knowledge of the results of the molecular assays; UNCLEAR: if the study did not explicitly describe how the reference standard was conducted and interpreted.
Could the reference standard, its conduct, or its interpretation have introduced bias?
LOW RISK: if the reference standard used met the definition described in the protocol, performed and evaluated without knowledge of the results of the index test.
HIGH RISK: if the reference standard did not meet the definition described in the protocol or was evaluated with the knowledge of the results of the index test.
UNCLEAR RISK: if there was no clear description of the reference standard used, how it was performed and interpreted in relation to the results of the index test.
B. Concerns regarding applicability
Was there concern that the target condition as defined by the reference standard did not match the review question?
LOW CONCERN: if the reference standard was microbial culture of blood or cerebrospinal fluid (or both) and if the target condition was suspected sepsis in a neonate as defined in our protocol.
HIGH CONCERN: if the reference standard was a test other than microbial culture of blood or cerebrospinal fluid (or both) and if the target condition included participants other than neonates or if the participants were not suspected of neonatal sepsis.
UNCLEAR CONCERN: if it was unclear whether the study fulfilled either the criteria for "low concern" or for "high concern".
Domain 4: Flow and timing
A. Risk of bias
Describe any participants who did not receive the index test(s) or reference standard (or both) or who were excluded from the 2 × 2 table (refer to flow diagram).
Describe the time interval and any interventions between index test(s) and reference standard.
Was there an appropriate interval between index test(s) and reference standard?
YES: if blood or cerebrospinal fluid (or both) samples used for both microbial culture and molecular assay were drawn concurrently at the same time during the workup for neonatal sepsis; NO: if blood or cerebrospinal fluid (or both) samples used for both microbial culture and molecular assay were drawn more than 6 hours apart for the workup of neonatal sepsis; UNCLEAR: if there was no description of how and when the samples for both the index text and the reference standard were collected.
Did all participants receive a reference standard?
YES: if all participants underwent microbial culture testing for their blood or cerebrospinal fluid (or both); NO: if at least 1 participant did not have the reference standard performed. UNCLEAR: if the study did not describe clearly which participants received the reference standard and which ones did not.
Did participants receive the same reference standard?
YES: if all participants underwent microbial culture testing for their blood or cerebrospinal fluid (or both); NO: if a different reference standard other than culture of blood or cerebrospinal fluid (or both) was used in at least 1 participant; UNCLEAR: if the study did not describe clearly what type of reference standard was used to diagnose a participant with neonatal sepsis.
Were all participants included in the analysis?
YES: if all enrolled participants with the target condition who underwent testing using the index test and reference standard were included in the analysis; NO: if all enrolled participants were not accounted in the analysis; UNCLEAR: if it was unclear from the study about the inclusion of all enrolled participants in the analysis.
Could the participant flow have introduced bias?
LOW CONCERN: if the answers to above questions were all "YES" which means that all participants enrolled in the study were subjected to the same reference standard and index test, clinical samples for testing were drawn concurrently from the same participant, and all participants were included in the final analysis.
HIGH CONCERN: if at least 2 questions had a "NO" answer.
UNCLEAR CONCERN: if at least 1 question had a "NO" answer or it was unclear whether the study fulfilled either the criteria for "low concern" or for "high concern".
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
| Test | No. of studies | No. of participants |
|---|---|---|
| 1 All molecular tests | 35 | 7339 |
| 2 Molecular tests: blood samples only | 32 | 6999 |
| 3 Molecular tests with good methodologic quality | 22 | 4150 |
1. Test.
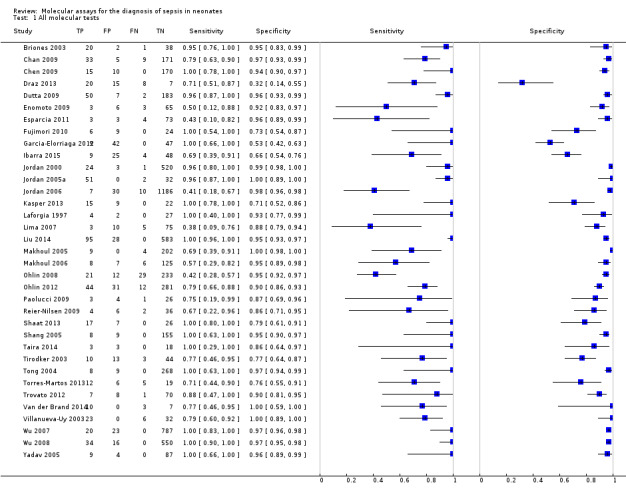
All molecular tests.
2. Test.
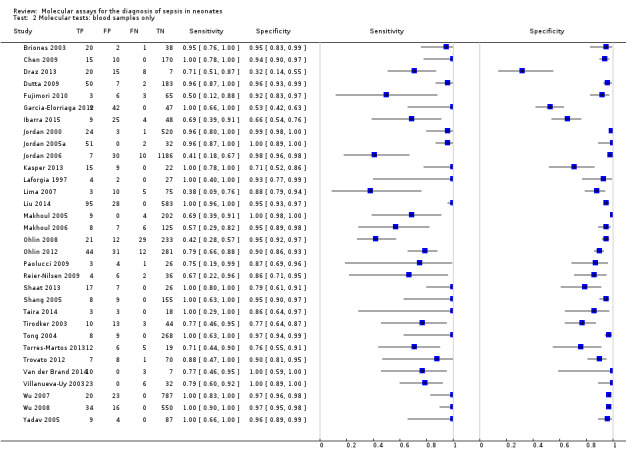
Molecular tests: blood samples only.
3. Test.
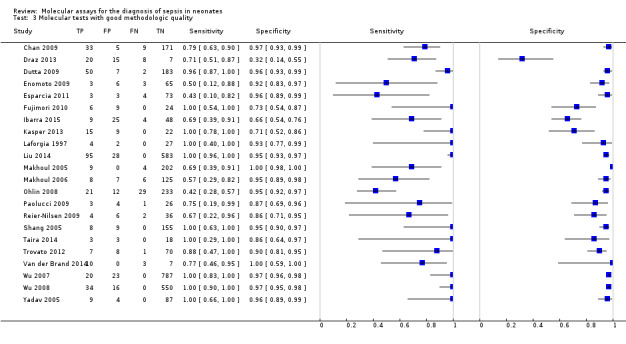
Molecular tests with good methodologic quality.
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Briones 2003.
| Study characteristics | |||
| Patient sampling | Participant sampling not clearly described. | ||
| Patient characteristics and setting | Newborns > 3 days old with suspected sepsis. No information on participant demographics or study period. | ||
| Index tests | PCR using universal candida DNA sequence. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples drawn at the same time. | ||
| Comparative | |||
| Notes | Data from conference abstract only. No information on participant demographics or study period. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Chan 2009.
| Study characteristics | |||
| Patient sampling | Participants were recruited consecutively. | ||
| Patient characteristics and setting | Preterm infants < 37 weeks and > 72 hours old with signs and symptoms of sepsis requiring antibiotic treatment. Interquartile range of age reported suggests some infants may have been > 28 days of age. Study period: March 2006 to June 2008 (28 months). | ||
| Index tests | Real‐time PCR using universal primers and Gram‐specific probes. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood, peritoneal fluid and CSF cultures. | ||
| Flow and timing | Index test and the reference standard performed at the same time. | ||
| Comparative | |||
| Notes | 15 samples were excluded due to insufficient amount of sample (n = 9) and mistakenly left in the refrigerator for > 72 hours (n = 6). Excluded samples not included in the analysis. Cycle threshold cut‐off values for positive PCR were defined. Interquartile range of age reported suggests some infants may have been > 28 days of age. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Chen 2009.
| Study characteristics | |||
| Patient sampling | Study did not classify whether participants were enrolled randomly or consecutively. Negative controls (n = 30) were not included in the analysis. | ||
| Patient characteristics and setting | Infants with suspected sepsis, admitted to the neonatal department and the intensive care unit of the Zhejiang University Children's University in China. It was unclear how many infants were < 28 days old as no participant demographics are available. Study period: September 2007 to June 2008. | ||
| Index tests | Broad‐range 16S rRNA‐based real‐time fluorescent PCR. | ||
| Target condition and reference standard(s) | Suspected sepsis and the reference standard were cultures of blood and CSF. | ||
| Flow and timing | Both index test and reference standard samples were drawn simultaneously. | ||
| Comparative | |||
| Notes | No participant demographics available and unclear if some infants were > 28 days of age. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Draz 2013.
| Study characteristics | |||
| Patient sampling | All neonates with suspected sepsis admitted during the period of May 2012 to August 2012 were enrolled. | ||
| Patient characteristics and setting | Neonates with suspected sepsis admitted to the NICU of Ain Shams University Hospitals. Study period: May 2012 to August 2012. Age range reported was 0 to 50 days. | ||
| Index tests | Broad‐range 16S rDNA PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood sample for culture and PCR were collected concurrently using standard sterile procedures. | ||
| Comparative | |||
| Notes | Participants were referred to as neonates although the age range reported was 0 to 50 days. Participants included both preterm and full‐term infants. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Dutta 2009.
| Study characteristics | |||
| Patient sampling | Not clearly reported. | ||
| Patient characteristics and setting | Neonates with suspected sepsis admitted to Level III NICU. Study period not mentioned. | ||
| Index tests | Broad‐range conventional PCR after 5‐hour preamplification culture. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for culture and PCR were drawn simultaneously. Reason for exclusion of participants were reported. | ||
| Comparative | |||
| Notes | Of the 64 participants that were excluded, 34 had malformations, 15 had < 12‐hour life expectancy and the remaining 15 had contaminated blood cultures. Study period not mentioned. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Enomoto 2009.
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met the inclusion criteria during the study period. Controls (n = 50) were not included in the analysis. | ||
| Patient characteristics and setting | Newborn participants with signs and history suggestive of sepsis admitted in the NICU at Kobe Hospital University from June 2005 to September 2006. | ||
| Index tests | Multiplex PCR targeting 8 common pathogens. | ||
| Target condition and reference standard(s) | Neonatal sepsis and bacterial culture of blood, skin, bronchoalveolar lavage, mucus, CSF, urine and ascitic fluid. | ||
| Flow and timing | Only 77 samples with paired specimen culture and PCR were included in the 2 × 2 table. Samples for culture and PCR were drawn simultaneously. | ||
| Comparative | |||
| Notes | Of the 6 specimens that were positive for PCR but negative for culture, 1 culture was positive for normal flora and was considered negative. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Esparcia 2011.
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met the inclusion criteria during the study period. | ||
| Patient characteristics and setting | Newborns < 7 days old with suspected sepsis or meningitis diagnosed at a participating hospital from November 2005 to January 2007. | ||
| Index tests | RT‐PCR targeting the 16S rRNA. | ||
| Target condition and reference standard(s) | Suspected early‐onset neonatal sepsis and blood and CSF cultures. | ||
| Flow and timing | Sample for PCR and culture were drawn concurrently. Samples for PCR were stored until DNA extraction. | ||
| Comparative | |||
| Notes | Analyzed only EOS in neonates and included 83 neonates. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Fujimori 2010.
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Neonates admitted to the NICU of Jutendo University Hospital or Jutendo Shizuoka Hospital from February to August 2009. Mean (SD) gestational age was 34.8 ± 5.8 weeks. There were 36 participants with 39 episodes of sepsis. | ||
| Index tests | RT‐PCR targeting 16S rRNA. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Whole blood collected concurrently for PCR and culture. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Garcia‐Elorriaga 2012.
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Neonates up to 28 days old admitted to the NICU from August 2005 to July 2006. | ||
| Index tests | Broad‐range PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Index test and reference standard sampling performed simultaneously. | ||
| Comparative | |||
| Notes | Only blood culture‐positive samples were included in the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| High | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Ibarra 2015.
| Study characteristics | |||
| Patient sampling | Participants who met the inclusion criteria were enrolled prospectively. | ||
| Patient characteristics and setting | Neonates with suspected clinical sepsis admitted to the Central South Hospital of Petroleos Mexicanos, the Gynecological‐Obstetrics Hospital number 4 of the Mexican Institute of Social Security, the Dalinde Hospital and the Monterrey Nuevo Leon University Hospital and National Institute of Perinatology. Study period not mentioned. | ||
| Index tests | LightCycler SeptiFast Test. | ||
| Target condition and reference standard(s) | Suspected neonatal sepsis and blood culture. | ||
| Flow and timing | Samples for blood culture and LightCycler SeptiFast were drawn concurrently. | ||
| Comparative | |||
| Notes | Study period not mentioned in the report. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Jordan 2000.
| Study characteristics | |||
| Patient sampling | All infants admitted to the NICU for sepsis evaluation. | ||
| Patient characteristics and setting | All infants admitted to the NICU for sepsis evaluation. No participant demographics available. | ||
| Index tests | Broad‐range conventional PCR and DNA dot‐blot hybridization. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Index test and reference standard were performed simultaneously. | ||
| Comparative | |||
| Notes | This was a feasibility study and blood sample for PCR was from discarded or unused sample sent to evaluate CBCs. It was not clear whether blood drawn for CBC was also done with the same aseptic technique as blood culture. Study period not mentioned. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Jordan 2005a.
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met inclusion criteria. | ||
| Patient characteristics and setting | Infant admitted to the NICU for sepsis evaluation that included at least blood culture and CBC. No demographic information or study period details available. | ||
| Index tests | Real‐time 16S rRNA PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood sample used for PCR was from discarded or unused samples sent for evaluation of CBC. Unclear whether blood drawn for CBC was done in an aseptic manner. | ||
| Comparative | |||
| Notes | Study was done to design a sample preparation protocol that would eliminate tryptic soy broth pre‐enrichment step and to convert conventional PCR assay to a real‐time PCR platform. The methodology here is real‐time PCR from whole blood without enrichment. So a different methodology fromJordan 2000paper and overlap is very unlikely. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Jordan 2006.
| Study characteristics | |||
| Patient sampling | All NICU admissions during the period of study were screened for eligibility. | ||
| Patient characteristics and setting | Infants > 34 weeks admitted to the NICU for suspected EOS from 1 September 2000 to 1 April 2004. | ||
| Index tests | Broad‐range conventional PCR followed by pyrosequencing. | ||
| Target condition and reference standard(s) | EOS in near‐term infants and blood culture. | ||
| Flow and timing | Samples for the index test and reference standard were collected simultaneously but PCR was evaluated from sample sent for CBC. Concerns about aseptic technique remain. | ||
| Comparative | |||
| Notes | Blood samples for PCR were from unused portion of the sample sent to evaluate CBC and were collected by venous, arteria or heel stick. The PCR was conventional PCR with enrichment with Trypticose soy before PCR just like the paperJordan 2000. The study period here was stated to be from September 2000.Jordan 2000paper was submitted for publication in 1999 as per the title page of the article and hence overlap ofJordan 2000andJordan 2006unlikely. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Kasper 2013.
| Study characteristics | |||
| Patient sampling | Neonates who met inclusion criteria were enrolled on admission. | ||
| Patient characteristics and setting | VLBW infants > 72 hours old. Participant demographics or study period not available. | ||
| Index tests | Multiplex real‐time PCR using Roche LightCycler SeptiFast MGRADE system. | ||
| Target condition and reference standard(s) | Neonates with suspected LOS and blood culture. | ||
| Flow and timing | Blood sample for PCR was collected during routine sepsis work‐up and before antibiotics. | ||
| Comparative | |||
| Notes | Participant demographics or study period not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Laforgia 1997.
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Newborn at risk for EOS from January to September 1996. Predefined major and minor criteria were used to classify participants "at risk" for sepsis. | ||
| Index tests | Broad‐range conventional PCR | ||
| Target condition and reference standard(s) | Neonatal EOS and blood culture. | ||
| Flow and timing | Blood samples for analyses were drawn concurrently. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Lima 2007.
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Neonates with suspected sepsis during the period of December 2004 to June 2005. Participant demographics not available. | ||
| Index tests | Real‐time PCR using universal primers. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | Recalculated sensitivity, specificity, PPV, and NPV as samples positive for PCR were also positive for human DNA and not bacterial DNA. Participant demographics not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Liu 2014.
| Study characteristics | |||
| Patient sampling | All neonates with suspected sepsis and had blood samples drawn for concomitant culture, CBC and CRP assay were included in the study. | ||
| Patient characteristics and setting | Neonates with suspected sepsis admitted to the NICU of the Women and Children's Hospital, the Children's Hospital and Tongji Hospital in Hubei Province from 1 September 2011 to 31 December 2011. Participants were from 4 hour to 28 days old. | ||
| Index tests | 16S rRNA gene PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Additional 0.5 mL to 1 mL EDTA blood sample was collected for PCR at the time of sepsis workup. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Makhoul 2005.
| Study characteristics | |||
| Patient sampling | Prospective enrollment of infants that met inclusion criteria during a 12‐month period. | ||
| Patient characteristics and setting | Neonates aged > 3 days, admitted to the NICU with suspected LOS. Gestational age range 24 to 42 weeks and range of age at enrollment was 4 to 96 days. Study period not mentioned although reported over 12 months. | ||
| Index tests | Staphylococcal 16S rRNA PCR (both Staphylococcus aureus and coagulase‐negative Staphylococcus). | ||
| Target condition and reference standard(s) | Neonatal LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | There were 32 culture‐positive samples for bacteria and fungi but only 13 were positive for staphylococci and this was incorporated into the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Makhoul 2006.
| Study characteristics | |||
| Patient sampling | Prospective enrollment of neonates that met the criteria for suspected LOS. | ||
| Patient characteristics and setting | Neonates aged > 3 days with suspected LOS. The age range of infants included were 4 to 105 days. Study period not available. | ||
| Index tests | Staphylococcal 16S rRNA PCR (both Staphylococcus aureus and coagulase‐negative Staphylococci). | ||
| Target condition and reference standard(s) | Neonates with suspected LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | The article mentioned 148 events of LOS but on further scrutiny there were on 146 events which were incorporated into the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Low | |||
Ohlin 2008.
| Study characteristics | |||
| Patient sampling | Newborn infant that met inclusion criteria for EOS and LOS admitted to the NICU during the period of 1999 to 2005. | ||
| Patient characteristics and setting | Newborn infants < 28 days old with suspected EOS or LOS admitted to Öbrero University from 1999 to 2005. | ||
| Index tests | Real‐time PCR targeting 16S rRNA. | ||
| Target condition and reference standard(s) | Neonates with suspected EOS or LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn simultaneously. | ||
| Comparative | |||
| Notes | PCR results from 1 sample that was positive for culture and PCR was considered uninterpretable as PCR result showed double sequence. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Ohlin 2012.
| Study characteristics | |||
| Patient sampling | All infants that met inclusion criteria were enrolled prospectively. | ||
| Patient characteristics and setting | All infants aged < 3 months who underwent sepsis evaluation and admitted to the NICU at 2 Swedish University Hospitals between October 2007 and November 2009. Of the participants enrolled in the study, 34 infants were > 28 days old. | ||
| Index tests | Broad‐range 16S real‐time PCR. | ||
| Target condition and reference standard(s) | Suspected sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn simultaneously. | ||
| Comparative | |||
| Notes | 16 participants were excluded due to lack of consent, 7 for being older than 3 months and 10 participants whose blood sample for PCR and culture were not drawn concurrently. Excluded participants were not included in the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Paolucci 2009.
| Study characteristics | |||
| Patient sampling | 34 newborns with LOS were enrolled in the study. | ||
| Patient characteristics and setting | Newborns > 3 days old with suspected LOS. Age of participants at enrollment and study period not available. | ||
| Index tests | Commercial real‐time PCR using LightCycler SeptiFast system (multiplex PCR). | ||
| Target condition and reference standard(s) | Neonatal LOS and blood culture. | ||
| Flow and timing | Blood samples for LightCycler SeptiFast and culture were simultaneously. | ||
| Comparative | |||
| Notes | Age of participants at enrollment and study period not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Reier‐Nilsen 2009.
| Study characteristics | |||
| Patient sampling | Prospective, non‐randomized enrollment of participants that met inclusion criteria. | ||
| Patient characteristics and setting | Infants with birth weight > 1000 g admitted to the NICU at Akershus University Hospital with suspected sepsis during the first week of life. Age at study enrollment and study period not mentioned. | ||
| Index tests | Broad‐range 16S rRNA PCR followed by sequencing. | ||
| Target condition and reference standard(s) | Suspected neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | PCR samples were stored until analysis. 4 infants were excluded from the study with 3 having incomplete registration and 1 with missing sample. 1 infant in the final analysis ended up with a diagnosis of asphyxia rather than sepsis. Age at study enrollment and study period not mentioned. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Shaat 2013.
| Study characteristics | |||
| Patient sampling | Neonates with clinically suspected sepsis. | ||
| Patient characteristics and setting | Neonates with suspected sepsis. The gestational age ranged from 26 to 39 weeks but age at enrollment not mentioned. Study period: October 2010 to December 2012. | ||
| Index tests | 16S rDNA PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for blood culture and PCR were done simultaneously. | ||
| Comparative | |||
| Notes | Age at enrollment not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Shang 2005.
| Study characteristics | |||
| Patient sampling | All infants that met inclusion criteria during a specified period of time. Controls were excluded from analysis. | ||
| Patient characteristics and setting | All neonates > 3 days old admitted to the neonatal ward or NICU who developed clinical signs of LOS during the period of 1 January 2004 to June 30, 2004. Other participant demographics not available. | ||
| Index tests | Broad‐range 16S rRNA PCR followed by microarray hybridization. | ||
| Target condition and reference standard(s) | Suspected neonatal LOS and blood culture. | ||
| Flow and timing | Unclear whether blood samples for PCR and blood culture were drawn simultaneously. | ||
| Comparative | |||
| Notes | Participant demographics not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Taira 2014.
| Study characteristics | |||
| Patient sampling | Consecutive enrollment of infants (24 were neonates) with signs of systemic inflammatory response syndrome and risk factors for candidemia. | ||
| Patient characteristics and setting | Infants who were admitted to the ICU of 2 pediatric hospital in Sao Paulo State, Brazil over an 18‐month period. Study period (month and year) or participant demographics not available. Author provided results for the 24 neonates. | ||
| Index tests | Multiplex nested PCR with specific primers designed to identify 7 Candida species | ||
| Target condition and reference standard(s) | Candidemia and blood culture. | ||
| Flow and timing | Blood sample for both culture and PCR were done concurrently. | ||
| Comparative | |||
| Notes | Data based on email communication with Dr. Del Negro. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Tirodker 2003.
| Study characteristics | |||
| Patient sampling | All infants with suspected sepsis in the NICU and PICU during the study period were considered for inclusion in the study. | ||
| Patient characteristics and setting | Infants admitted in the NICU (n = 46) and PICU (n = 17) with suspected sepsis during the period from November 1999 to November 2000. PCR and blood culture data separately for neonates not available. | ||
| Index tests | Fungal conventional PCR targeting 18S rRNA. | ||
| Target condition and reference standard(s) | Suspected sepsis and blood culture. | ||
| Flow and timing | Excess blood used for culture was used for PCR. | ||
| Comparative | |||
| Notes | PCR and blood culture data separately for neonates not available. It was unclear how many of the infants admitted in the PICU were neonates hence, not all infants may have met the target condition of neonatal sepsis defined in this study. PCR products were analyzed by 2 independent observers blinded to blood culture results and participant information. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Tong 2004.
| Study characteristics | |||
| Patient sampling | Study data derived from conference abstract only and hence limited. | ||
| Patient characteristics and setting | Neonates with suspected sepsis. No participant demographics or study period details available. | ||
| Index tests | 16S rRNA‐based PCR followed by hybridization to chips with 18 probes. | ||
| Target condition and reference standard(s) | Infants with suspected sepsis and blood culture. | ||
| Flow and timing | Possible simultaneous sampling for index test and reference standard. | ||
| Comparative | |||
| Notes | Limited information from abstract. No participant demographics or study period details available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Torres‐Martos 2013.
| Study characteristics | |||
| Patient sampling | Participants who met inclusion criteria were admitted consecutively. | ||
| Patient characteristics and setting | Infants with febrile episodes admitted to the NICU at the Hospital Universitario Virgen de las Nieves. Study period: April 2007 to April 2009. Participants enrolled in the study were both preterm and term infants; however, age of participants at the time of enrollment range from 0 to 151 days old. | ||
| Index tests | LightCycler SeptiFast Assay. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Sample for blood culture and LightCycler SeptiFast assay were collected at the same time. | ||
| Comparative | |||
| Notes | Participants enrolled in the study were both preterm and term infants; however, age of participants at the time of enrollment range from 0 to 151 days old. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Trovato 2012.
| Study characteristics | |||
| Patient sampling | Only participants with probable candidiasis were included in the study. | ||
| Patient characteristics and setting | Neonates at high risk for invasive candidiasis from Jan 2009 to Dec 2010. No information on participant demographics available. | ||
| Index tests | Detection of fungal DNA directly from lysis‐centrifugation blood culture. Fungus‐specific universal primer ITS1 and ITS2 were used to amplify 18S rDNA, the adjacent ITS1 and a small portion of the 28S rDNA region. | ||
| Target condition and reference standard(s) | Suspected neonatal candidiasis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture came from the same Isolator 1.5 microbial tubes. | ||
| Comparative | |||
| Notes | No information on participant demographics available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Van der Brand 2014.
| Study characteristics | |||
| Patient sampling | Consecutive enrollment of preterm infants with suspected LOS. | ||
| Patient characteristics and setting | Preterm infants with suspected LOS admitted to the NICU. Participant demographics or study period not mentioned. | ||
| Index tests | Multiplex real‐time PCR assay. | ||
| Target condition and reference standard(s) | LOS in neonates and blood culture. | ||
| Flow and timing | Blood samples for culture and PCR were drawn concurrently. | ||
| Comparative | |||
| Notes | Participant demographics or study period not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Villanueva‐Uy 2003.
| Study characteristics | |||
| Patient sampling | Limited information from abstract. | ||
| Patient characteristics and setting | Newborns aged > 3 days with suspected LOS. Participant demographics or study period data not available. | ||
| Index tests | Broad‐range 16S rRNA conventional PCR. | ||
| Target condition and reference standard(s) | Neonatal LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | Study data derived from abstract only. Participant demographics or study period data not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Wu 2007.
| Study characteristics | |||
| Patient sampling | Limited information from abstract. Controls not included in the analysis. | ||
| Patient characteristics and setting | Newborns with suspected sepsis admitted to the neonatal ward or NICU. Participant demographics or study period data not available. | ||
| Index tests | Real‐time PCR targeting 16S rRNA. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples were tested for routine culture and PCR separately. There was no mention if blood sample was drawn simultaneously. | ||
| Comparative | |||
| Notes | Abstract only. Participant demographics or study period data not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Wu 2008.
| Study characteristics | |||
| Patient sampling | Neonates who met inclusion criteria during the study period were enrolled. Controls were not included in the analysis. | ||
| Patient characteristics and setting | Neonates aged 1 to 28 days with suspected sepsis admitted to the neonatal ward and NICU of Zhejiang University Children's Hospital from January 2005 to January 2007. 108 of the participants were preterm infants. | ||
| Index tests | Real‐time PCR with Gram‐specific probes followed by sequencing. | ||
| Target condition and reference standard(s) | Suspected neonatal EOS and LOS and blood culture. | ||
| Flow and timing | PCR and culture were done simultaneously. Unclear if samples were concurrently. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Yadav 2005.
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met inclusion criteria. | ||
| Patient characteristics and setting | Infants < 7 days old with suspected sepsis admitted to a level II NICU. Study period details not available. | ||
| Index tests | Broad‐range 16S rRNA PCR. | ||
| Target condition and reference standard(s) | Suspected neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | Study period details not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
CBC: complete blood count; CSF: cerebrospinal fluid; EDTA: ethylenediaminetetraacetic acid; EOS: early‐onset sepsis; LOS: late‐onset sepsis; n: number of participants; NICU: neonatal intensive care unit; NPV: negative predictive value; PCR: polymerase chain reaction; PICU: pediatric intensive care unit; PPV: positive predictive value; rDNA: ribosomal DNA; rRNA: ribosomal ribonucleic acid; RT‐PCR: real‐time polymerase chain reaction; SD: standard deviation; VLBW: very low birth weight.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Chiba 2009 | All samples (CSF) were positive by culture for bacterial meningitis and not in the context of suspected infection. |
| Das 2015 | Urine instead of blood sample was used for broad‐range 16S rDNA in detecting neonatal septicemia. |
| de Zoysa 2012 | All samples investigated were culture negative samples and not in the context of suspected infection. |
| Golden 2004 | GBS fluorescent PCR not compared with the reference standard (all were culture negative samples). |
| Jones 2010 | Analyzed gastric aspirates by molecular methods for DNA load followed by sequencing and cultures. Neonates were suspected of sepsis but no details of blood cultures to diagnose sepsis were available. |
| Jordan 2005b | Culture‐positive specimens were examined for 16srRNA for PCR and sequencing. Not evaluated in the clinical context of suspected sepsis. |
| Jordan 2009 | Pyrosequencing used to identify bacteria from positive blood culture bottles. Not evaluated in the clinical context of suspected sepsis. |
| Lucignano 2011 | It is unclear how many participants included in the study were neonates. Attempt made to contact author for details. |
| Makhoul 2007 | Term neonates had risk factors of sepsis (maternal fever, unknown maternal GBS) but not suspected of having sepsis. Both blood cultures and PCR were negative in this cohort. |
| Shang 2001 | Culture‐positive specimens and healthy controls were evaluated and not in the clinical context of suspected sepsis. |
| Shen 2004 | No clinical specimens from neonates with suspected sepsis. Spiked samples were used. |
| Tschiedel 2012 | Non‐neonatal population. |
CSF: cerebrospinal fluid; GBS: group B streptococcus; PCR: polymerase chain reaction.
Differences between protocol and review
1. We decided post‐hoc to present quality of evidence using GRADE methodology recommended for diagnostic tests.
2. Some studies did not include an upper limit for age and hence some infants were over 28 days of age. We made a post‐hoc decision that we would include studies where an upper age limit was not specified but where more than 50% of the samples were from newborn less than 28 days of age. Our decision was supported by the reasoning that LOS extends up to three months of age and participant characteristics are similar in the first two to three months of age.
Contributions of authors
MP conceived the project, searched literature, extracted and analyzed data, and wrote the review.
AF participated in the design, searched literature, extracted data, performed the QUADAS evaluation of included studies and assisted in writing the review.
JV provided critical intellectual input and revised the review.
ML provided critical intellectual input and revised the review.
Sources of support
Internal sources
No sources of support supplied
External sources
-
Eunice Kennedy Shriver National Institute of Child Health and Human Development National Institutes of Health, Department of Health and Human Services, USA.
Editorial support of the Cochrane Neonatal Review Group has been funded with Federal funds from the Eunice Kennedy Shriver National Institute of Child Health and Human Development National Institutes of Health, Department of Health and Human Services, USA, under Contract No. HHSN275201100016C
Declarations of interest
Mohan Pammi, Angela Flores, James Versalovic and Mariska MG Leeflang have no financial or other conflicts of interest to disclose.
New
References
References to studies included in this review
Briones 2003 {published data only}
- Briones CR, Villanueva‐Uy ME, Uy HGTI. The use of polymerase chain reaction in neonatal candidemia. Pediatric Research 2003;53:396A. [Google Scholar]
Chan 2009 {published data only}
- Chan KY, Lam HS, Cheung HM, Chan AK, Li K, Fok TF, et al. Rapid identification and differentiation of Gram‐negative and Gram‐positive bacterial bloodstream infections by quantitative polymerase chain reaction in preterm infants. Critical Care Medicine 2009;37:2441‐7. [PUBMED: 19531943] [DOI] [PubMed] [Google Scholar]
Chen 2009 {published data only}
- Chen LH, Duan QJ, Cai MT, Wu YD, Shang SQ. Rapid diagnosis of sepsis and bacterial meningitis in children with real‐time fluorescent quantitative polymerase chain reaction amplification in the bacterial 16S rRNA gene. Clinical Pediatrics 2009;48(6):641‐7. [PUBMED: 19407210] [DOI] [PubMed] [Google Scholar]
Draz 2013 {published data only}
- Draz NI, Taha SE, Abou Shady NM, Abdel Ghany YS. Comparison of broad range 16S rDNA PCR to conventional blood culture for diagnosis of sepsis in the newborn. Egyptian Journal of Human Medical Genetics 2013;14:403‐11. [PUBMED: 19152691]19152691 [Google Scholar]
Dutta 2009 {published data only}
- Dutta S, Narang A, Chakraborty A, Ray P. Diagnosis of neonatal sepsis using universal primer polymerase chain reaction before and after starting antibiotic drug therapy. Archives of Pediatrics and Adolescent Medicine 2009;163(1):6‐11. [PUBMED: 19124696] [DOI] [PubMed] [Google Scholar]
Enomoto 2009 {published data only}
- Enomoto M, Morioka I, Morisawa T, Yokoyama N, Matsuo M. A novel diagnostic tool for detecting neonatal infections using multiplex polymerase chain reaction. Neonatology 2009;96(2):102‐8. [PUBMED: 19279393] [DOI] [PubMed] [Google Scholar]
Esparcia 2011 {published data only}
- Esparcia O, Montemayor M, Ginovart G, Pomar V, Soriano G, Pericas R, et al. Diagnostic accuracy of a 16S ribosomal DNA gene‐based molecular technique (RT‐PCR, microarray, and sequencing) for bacterial meningitis, early‐onset neonatal sepsis, and spontaneous bacterial peritonitis. Diagnostic Microbiology and Infectious Disease 2011;69(2):153‐60. [PUBMED: 21251558] [DOI] [PubMed] [Google Scholar]
Fujimori 2010 {published data only}
- Fujimori M, Hisata K, Nagata S, Matsunaga N, Komatsu M, Shoji H, et al. Efficacy of bacterial ribosomal RNA‐targeted reverse transcription‐quantitative PCR for detecting neonatal sepsis: a case control study. BMC Pediatrics 2010;10:53. [PUBMED: 20667142] [DOI] [PMC free article] [PubMed] [Google Scholar]
Garcia‐Elorriaga 2012 {published data only}
- Garcia‐Elorriaga G, Cortes‐Torres N, Ballesteros‐Del‐Olmo JC, Rey‐Pineda G, Gonzaez‐Bonilla C. The usefulness of the buffy coat smear and panbacterial polymerase chain reaction in early diagnosis of neonatal sepsis. Revista de Investigacion Clinica 2012;64(3):275‐83. [PUBMED: 23045950] [PubMed] [Google Scholar]
Ibarra 2015 {published data only}
- Ibarra JO, Valdez PT, Mendez EV, Rojas AL, Flores GL, Bocanegra AC, et al. Evaluation of the Light‐Cycler® SeptiFast Test in newborns with suspicion of nosocomial sepsis. Iran Journal of Pediatrics 2015;25(1):e253. [PUBMED: 26199693] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jordan 2000 {published data only}
- Jordan JA, Durso MB. Comparison of 16S rRNA gene PCR and BACTEC 9240 for detection of neonatal bacteremia. Journal of Clinical Microbiology 2000;38(7):2574‐8. [PUBMED: 10878046] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jordan 2005a {published data only}
- Jordan JA, Durso MB. Real‐time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis. Journal of Molecular Diagnostics 2005;7(5):575‐81. [PUBMED: 16258155] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jordan 2006 {published data only}
- Jordan JA, Durso MB, Butchko AR, Jones JG, Brozanski BS. Evaluating the near‐term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing. Journal of Molecular Diagnostics 2006;8(3):357‐63. [PUBMED: 16825509] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kasper 2013 {published data only}
- Kasper DC, Altiok I, Mechtler TP, Bohm J, Straub J, Langgartner M, et al. Molecular detection of late‐onset neonatal sepsis in premature infants using small blood volumes: proof‐of‐concept. Neonatology 2013;103(4):268‐73. [PUBMED: 23485823] [DOI] [PubMed] [Google Scholar]
Laforgia 1997 {published data only}
- Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Lolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatrica 1997;86(10):1097‐9. [PUBMED: 9350892] [DOI] [PubMed] [Google Scholar]
Lima 2007 {published data only}
- Lima V, Alpuche A, Noyola D, Soria R, Nieto K. Polymerase chain reaction technique in the diagnosis of neonatal sepsis: future gold standard?. Pediatric Academic Societies Annual Meeting; 2007 May 5‐8; Toronto (ON) 2007.
Liu 2014 {published data only}
- Liu CL, Ai HW, Wang WP, Chen L, Hu HB, Ye T, et al. Comparison of 16S rRNA gene PCR and blood culture for diagnosis of neonatal sepsis. Archives of Pediatrics 2014;21(2):162‐9. [PUBMED: 24388336] [DOI] [PubMed] [Google Scholar]
Makhoul 2005 {published data only}
- Makhoul I, Smolkin RT, Sujov P, Kassis I, Tamir A, Shalginov R, et al. PCR‐based diagnosis of neonatal staphylococcal bacteremias. Journal of Clinical Microbiology 2005;43(9):4823‐5. [PUBMED: 16145149] [DOI] [PMC free article] [PubMed] [Google Scholar]
Makhoul 2006 {published data only}
- Makhoul IR, Yacoub A, Smolkin T, Sujov P, Kassis I, Sprecher H. Values of C‐reactive protein, procalcitonin, and Staphylococcus‐specific PCR in neonatal late‐onset sepsis. Acta Paediatrica 2006;95(10):1218‐23. [PUBMED: 16982493] [DOI] [PubMed] [Google Scholar]
Ohlin 2008 {published data only}
- Ohlin A, Backman A, Bjorkqvist M, Molling P, Jurstrand M, Schollin J. Real‐time PCR of the 16S‐rRNA gene in the diagnosis of neonatal bacteraemia. Acta Paediatrica 2008;97(10):1376‐80. [PUBMED: 18624992] [DOI] [PubMed] [Google Scholar]
Ohlin 2012 {published data only}
- Ohlin A, Backman A, Ewald U, Schollin J, Bjorkqvist M. Diagnosis of neonatal sepsis by broad‐range 16S real‐time polymerase chain reaction. Neonatology 2012;10(4):241‐6. [PUBMED: 22205207] [DOI] [PubMed] [Google Scholar]
Paolucci 2009 {published data only}
- Paolucci M, Capretti MG, Dal Monte P, Corvaglia L, Landini MP, Varani S, et al. Laboratory diagnosis of late‐onset sepsis in newborns by multiplex real‐time PCR. Journal of Medical Microbiology 2009;58(Pt 4):533‐4. [PUBMED: 19273654] [DOI] [PubMed] [Google Scholar]
Reier‐Nilsen 2009 {published data only}
- Reier‐Nilsen T, Farstad T, Nakstad B, Lauvrak V, Steinbakk M. Comparison of broad range 16S rDNA PCR and conventional blood culture for diagnosis of sepsis in the newborn: a case control study. BMC Pediatrics 2009;9:5. [PUBMED: 19152691] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shaat 2013 {published data only}
- Shaat SS, Shazly SA, Badr Eldin MM, Barakat SS, Hashish MH. Role of polymerase chain reaction as an early diagnostic tool for neonatal bacterial sepsis. Journal of Egypt Public Health Association 2013;88(3):160‐4. [PUBMED: 24374951] [DOI] [PubMed] [Google Scholar]
Shang 2005 {published data only}
- Shang S, Chen G, Wu Y, Du L, Zhao Z. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatric Research 2005;58(1):143‐8. [PUBMED: 15985688] [DOI] [PubMed] [Google Scholar]
Taira 2014 {published data only}
- Taira CL, Okay TS, Delgado AF, Ceccon ME, Almeida MT, Negro GM. A multiplex nested PCR for the detection and identification of Candida species in blood samples of critically ill paediatric patients. BMC Infectious Diseases 2014;14:406. [PUBMED: 25047415] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tirodker 2003 {published data only}
- Tirodker UH, Nataro JP, Smith S, LasCasas L, Fairchild KD. Detection of fungemia by polymerase chain reaction in critically ill neonates and children. Journal of Perinatology 2003;23(2):117‐22. [PUBMED: 12673260] [DOI] [PubMed] [Google Scholar]
Tong 2004 {published data only}
- Tong MQ, Shang SQ, Wu YD, Zhao ZY. Rapid diagnosis of neonatal sepsis by 16SrRNA genes PCR amplification and genechip hybridization. Zhonghua Er Ke Za Zhi 2004;42(9):663‐7. [PUBMED: 15482666] [PubMed] [Google Scholar]
Torres‐Martos 2013 {published data only}
- Torres‐Martos E, Perez‐Ruiz M, Pedrosa‐Corral I, Pena‐Caballero M, Jimenez‐Valera MM, Perez‐Ramirez MD, et al. Evaluation of the LightCycler® SeptiFast test in newborns and infants with clinical suspicion of sepsis. Enfermedades Infecciosas Microbiologia Clinica 2013;31(6):375‐9. [PUBMED: 23137657] [DOI] [PubMed] [Google Scholar]
Trovato 2012 {published data only}
- Trovato L, Betta P, Romeo MG, Oliveri S. Detection of fungal DNA in lysis‐centrifugation blood culture for the diagnosis of invasive candidiasis in neonatal patients. Clinical Microbiology and Infection 2012;18(3):E63‐5. [PUBMED: 22192484] [DOI] [PubMed] [Google Scholar]
Van der Brand 2014 {published data only}
- Brand M, Peters RP, Catsburg A, Rubenjan A, Broeke FJ, Dungen FA, et al. Development of a multiplex real‐time PCR assay for the rapid diagnosis of neonatal late onset sepsis. Journal of Microbiological Methods 2014;106:8‐15. [PUBMED: 25102109] [DOI] [PubMed] [Google Scholar]
Villanueva‐Uy 2003 {published data only}
- Villanueva‐Uy ME, Briones CR, Uy HG. Application of polymerase chain reaction in late‐onset neonatal sepsis. Pediatric Research 2003;53:313A. [Google Scholar]
Wu 2007 {published data only}
- Wu YD, Shang SQ, Li JP, Yang ZQ, Zheng ZB, Du LZ, et al. A broad‐range 16S rRNA gene real‐time PCR assay for the diagnosis of neonatal septicemia. Zhonghua Er Ke Za Zhi 2007;45(6):446‐9. [PUBMED: 17880793] [PubMed] [Google Scholar]
Wu 2008 {published data only}
- Wu YD, Chen LH, Wu XJ, Shang SQ, Lou JT, Du LZ, et al. Gram stain‐specific‐probe‐based real‐time PCR for diagnosis and discrimination of bacterial neonatal sepsis. Journal of Clinical Microbiology 2008;46(8):2613‐9. [PUBMED: 18550744] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yadav 2005 {published data only}
- Yadav AK, Wilson CG, Prasad PL, Menon PK. Polymerase chain reaction in rapid diagnosis of neonatal sepsis. Indian Pediatrics 2005;42(7):681‐5. [PUBMED: 16085969] [PubMed] [Google Scholar]
References to studies excluded from this review
Chiba 2009 {published data only}
- Chiba N, Murayama SY, Morozumi M, Nakayama E, Okada T, Iwata S, et al. Rapid detection of eight causative pathogens for the diagnosis of bacterial meningitis by real‐time PCR. Journal of Infection and Chemotherapy 2009;15(2):92‐8. [PUBMED: 19396518] [DOI] [PubMed] [Google Scholar]
Das 2015 {published data only}
- Das BK, Suri S, Nath G, Prasad R. Urine nested polymerase chain reaction in neonatal septicemia. Journal of Tropical Pediatrics 2015;61(4):295‐300. [PUBMED: 26130622] [DOI] [PubMed] [Google Scholar]
de Zoysa 2012 {published data only}
- Zoysa A, Edwards K, Gharbia S, Underwood A, Charlett A, Efstratiou A. Non‐culture detection of Streptococcus agalactiae (Lancefield group B Streptococcus) in clinical samples by real‐time PCR. Journal of Medical Microbiology 2012;61(Pt 8):1086‐90. [PUBMED: 22740612] [DOI] [PubMed] [Google Scholar]
Golden 2004 {published data only}
- Golden SM, Stamilio DM, Faux BM, dela Cruz WP, Shoemaker CT, Blackmon CL, et al. Evaluation of a real‐time fluorescent PCR assay for rapid detection of Group B Streptococci in neonatal blood. Diagnostic Microbiology and Infectious Disease 2004;50(1):7‐13. [PUBMED: 15380273] [DOI] [PubMed] [Google Scholar]
Jones 2010 {published data only}
- Jones V, Wilks M, Johnson G, Warwick S, Hennessey E, Kempley S, et al. The use of molecular techniques for bacterial detection in the analysis of gastric aspirates collected from infants on the first day of life. Early Human Development 2010;86(3):167‐70. [PUBMED: 20223606] [DOI] [PubMed] [Google Scholar]
Jordan 2005b {published data only}
- Jordan JA, Butchko, AR, Durso MB. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. Journal of Molecular Diagnostics 2005;7(1):105‐10. [PUBMED: 15681481] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jordan 2009 {published data only}
- Jordan JA, Jones‐Laughner J, Durso MB. Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles. Journal of Clinical Microbiology 2009;47(2):368‐72. [PUBMED: 19091813] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lucignano 2011 {published data only}
- Lucignano B, Ranno S, Liesenfeld O, Pizzorno B, Putignani L, Bernaschi P, et al. Multiplex PCR allows rapid and accurate diagnosis of bloodstream infections in newborns and children with suspected sepsis. Journal of Clinical Microbiology 2011;49(6):2252‐8. [PUBMED: 21471340] [DOI] [PMC free article] [PubMed] [Google Scholar]
Makhoul 2007 {published data only}
- Makhoul IR, Sprecher H, Smolkin T, Sawaid R, Ben‐David S, Sujov P, et al. Approach to term neonates born after maternal intrapartum fever and unknown maternal group B Streptococcus status: value of serum C‐reactive protein and 16S rRNA gene PCR amplification. Pediatric Infectious Disease Journal 2007;26(11):1064‐6. [PUBMED: 17984819] [DOI] [PubMed] [Google Scholar]
Shang 2001 {published data only}
- Shang S, Chen Z, Yu X. Detection of bacterial DNA by PCR and reverse hybridization in the 16S rRNA gene with particular reference to neonatal septicemia. Acta Paediatrica 2001;90(2):179‐83. [PUBMED: 11236048] [DOI] [PubMed] [Google Scholar]
Shen 2004 {published data only}
- Shen DX, Du J, Feng ZC. Rapid diagnosis of common pathogenic bacteria infection in newborn infants by 16SrDNA oligonucleotide array. Zhonghua Er Ke Za Zhi 2004;42(9):668‐72. [PUBMED: 15482667] [PubMed] [Google Scholar]
Tschiedel 2012 {published data only}
- Tschiedel E, Steinmann J, Buer J, Onnebrink JG, Felderhoff‐Muser U, Rath PM, et al. Results and relevance of molecular detection of pathogens by SeptiFast ‐ a retrospective analysis in 75 critically ill children. Klinische Padiatrie 2012;224(1):12‐6. [PUBMED: 22258624] [DOI] [PubMed] [Google Scholar]
Additional references
Adams‐Chapman 2006
- Adams‐Chapman I, Stoll BJ. Neonatal infection and long‐term neurodevelopmental outcome in the preterm infant. Current Opinion in Infectious Diseases 2006;19(3):290‐7. [PUBMED: 16645492] [DOI] [PubMed] [Google Scholar]
Blommendahl 2002
- Blommendahl J, Janas M, Laine S, Miettinen A, Ashorn P. Comparison of procalcitonin with CRP and differential white blood cell count for diagnosis of culture‐proven neonatal sepsis. Scandinavian Journal of Infectious Diseases 2002;34(8):620‐2. [PUBMED: 12238581] [DOI] [PubMed] [Google Scholar]
Bossuyt 2006
- Bossuyt PM, Irwig L, Craig J, Glasziou P. Comparative accuracy: assessing new tests against existing diagnostic pathways. BMJ 2006;332(7549):1089‐92. [PUBMED: 16675820] [DOI] [PMC free article] [PubMed] [Google Scholar]
Brozanski 2006
- Brozanski BS, Jones JG, Krohn MJ, Jordan JA. Use of polymerase chain reaction as a diagnostic tool for neonatal sepsis can result in a decrease in use of antibiotics and total neonatal intensive care unit length of stay. Journal of Perinatology 2006;26(11):688‐692. [DOI] [PubMed] [Google Scholar]
Canadian Neonatal Network 2014
- Shah P, Yoon EW, Chan P, Members of the Annual Report Review Committee. Annual report of the Canadian Neonatal Network, 2004. www.canadianneonatalnetwork.org/Portal/LinkClick.aspx?fileticket=eGgxmMubxjk%3d&tabid=39 (accessed 18 February 2017).
Chiesa 2004
- Chiesa C, Panero A, Osborn JF, Simonetti AF, Pacifico L. Diagnosis of neonatal sepsis: a clinical and laboratory challenge. Clinical Chemistry 2004;50(2):279‐87. [PUBMED: 14752012] [DOI] [PubMed] [Google Scholar]
Dong 2015
- Dong Y, Speer CP. Late‐onset neonatal sepsis: recent developments. Archives of Disease in Childhood. Fetal and Neonatal Edition 2015;100(3):F257‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gopalakrishna 2014
- Gopalakrishna G, Mustafa RA, Davenport C, Scholten RJ, Hyde C, Brozek J, et al. Applying Grading of Recommendations Assessment, Development and Evaluation (GRADE) to diagnostic tests was challenging but doable. Journal of Clinical Epidemiology 2014;67(7):760‐8. [DOI] [PubMed] [Google Scholar]
Harbord 2007
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta‐analysis of diagnostic accuracy studies. Biostatistics 2007;8(2):239‐51. [PUBMED: 16698768] [DOI] [PubMed] [Google Scholar]
Harbord 2008
- Harbord RM, Whiting P, Sterne JA, Egger M, Deeks JJ, Shang A, et al. An empirical comparison of methods for meta‐analysis of diagnostic accuracy showed hierarchical models are necessary. Journal of Clinical Epidemiology 2008;61(11):1095‐103. [PUBMED: 19208372] [DOI] [PubMed] [Google Scholar]
Harbord 2009
- Harbord RM, Whiting P. Metandi: meta‐analysis of diagnostic accuracy using hierarchical logistic regression. Stata Journal 2009;9(2):211‐29. [Google Scholar]
Hornik 2012
- Hornik CP, Benjamin DK, Benjamin DKJ, Li J, Clark RH, Cohen‐Wolkowiez M, et al. Use of the complete blood cell count in late‐onset neonatal sepsis. Pediatric Infectious Disease Journal 2012;31(8):803‐7. [PUBMED: 22531232] [DOI] [PMC free article] [PubMed] [Google Scholar]
Isaacman 1996
- Isaacman DJ, Karasic RB, Reynolds EA, Kost SI. Effect of number of blood cultures and volume of blood on detection of bacteremia in children. Pediatrics 1996;128(2):190‐5. [PUBMED: 8636810] [DOI] [PubMed] [Google Scholar]
Jordan 2010
- Jordan JA. Molecular diagnosis of neonatal sepsis. Clinics in Perinatology 2010;37(2):411‐9. [PUBMED: 20569815] [DOI] [PubMed] [Google Scholar]
Leeflang 2008
- Leeflang MM, Deeks JJ, Gatsonis C, Bossuyt PM. Systematic reviews of diagnostic test accuracy. Annals of Internal Medicine 2008;149(12):889‐97. [PUBMED: 19075208] [DOI] [PMC free article] [PubMed] [Google Scholar]
Macaskill 2010
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: Analysing and presenting results. In: Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2010. Available from srdta.cochrane.org/.
Ng 1997
- Ng PC, Cheng SH, Chui KM, Fok TF, Wong MY, Wong W, et al. Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C‐reactive protein in preterm very low birthweight infants. Archives of Disease in Childhood Fetal and Neonatal Edition 1997;77(3):F221‐7. [PUBMED: 9462194] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ng 2012
- Ng PC, Lam HS. Biomarkers in neonatology: the next generation of tests. Neonatology 2012;102(2):145‐51. [PUBMED: 22759988] [DOI] [PubMed] [Google Scholar]
Pammi 2011
- Pammi M, Flores A, Leeflang M, Versalovic J. Molecular assays in the diagnosis of neonatal sepsis: a systematic review and meta‐analysis. Pediatrics 2011;128(4):e973‐85. [PUBMED: 21949139] [DOI] [PubMed] [Google Scholar]
Reitsma 2009
- Reitsma JB, Whiting P, Vlassov VV, Leeflang MMG, Deeks JJ. Chapter 9: Assessing methodological quality. In: Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2009. Available from srdta.cochrane.org/.
Relman 1999
- Relman DA. The search for unrecognized pathogens. Science 1999;284(5418):1308‐10. [PUBMED: 10334977] [DOI] [PubMed] [Google Scholar]
RevMan 2014 [Computer program]
- The Nordic Cochrane Centre, The Cochrane Collaboration. Review Manager (RevMan). Version 5.3. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Rodriguez‐Creixems 2008
- Rodriguez‐Creixems M, Alcala L, Munoz P, Cercenado E, Vicente T, Bouza E. Bloodstream infections: evolution and trends in the microbiology workload, incidence, and etiology, 1985‐2006. Medicine 2008;87(4):234‐49. [PUBMED: 18626306] [DOI] [PubMed] [Google Scholar]
Schelonka 1996
- Schelonka RL, Chai MK, Yoder BA, Hensley D, Brockett RM, Ascher DP. Volume of blood required to detect common neonatal pathogens. Journal of Pediatrics 1996;129(2):275‐8. [PUBMED: 8765627] [DOI] [PubMed] [Google Scholar]
Schünemann 2016
- Schünemann HJ, Mustafa R, Brozek J, Santesso N, Alonso‐Coello P, Guyatt G, et al. GRADE Working Group. GRADE Guidelines: 16. GRADE evidence to decision frameworks for tests in clinical practice and public health. Journal of Clinical Epidemiology 2016;76:89‐98. [DOI] [PubMed] [Google Scholar]
Stata 2011 [Computer program]
- StataCorp. LP. Stata Statistical Software: Release 11. College Station (TX): StataCorp. LP, 2011.
Stoll 2002
- Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late‐onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics 2002;110(2 Pt 1):285‐91. [PUBMED: 12165580] [DOI] [PubMed] [Google Scholar]
Stoll 2004
- Stoll BJ, Hansen NI, Adams‐Chapman I, Fanaroff AA, Hintz SR, Vohr B, et al. Neurodevelopmental and growth impairment among extremely low‐birth‐weight infants with neonatal infection. JAMA 2004;292(19):2357‐65. [PUBMED: 15547163] [DOI] [PubMed] [Google Scholar]
Trenti 2016
- Trenti T, Schunemann HJ, Plebani M. Developing GRADE outcome‐based recommendations about diagnostic tests: a key role in laboratory medicine policies. Clinical Chemistry and Laboratory Medicine 2016;54(4):535‐43. [DOI] [PubMed] [Google Scholar]
Van Enst 2014
- Enst WA, Ochodo E, Scholten RJPM, Hooft L, Leeflang MA. Investigation of publication bias in meta‐analyses of diagnostic test accuracy: a meta‐epidemiologic study. BMC Medical Research Methodology 2014;14:70. [PUBMED: 24884381] [DOI] [PMC free article] [PubMed] [Google Scholar]
Venkatesh 2010
- Venkatesh M, Flores A, Luna RA, Versalovic J. Molecular microbiological methods in the diagnosis of neonatal sepsis. Expert Reviews in Anti Infective Therapy 2010;8(9):1037‐48. [PUBMED: 20818947] [DOI] [PMC free article] [PubMed] [Google Scholar]
Verboon‐Maciolek 2006
- Verboon‐Maciolek MA, Thijsen SF, Hemels MA, Menses M, Loon AM, Krediet TG, et al. Inflammatory mediators for the diagnosis and treatment of sepsis in early infancy. Pediatric Research 2006;59(3):457‐61. [PUBMED: 16492989] [DOI] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] [DOI] [PubMed] [Google Scholar]
Woese 1987
- Woese CR. Bacterial evolution. Microbiology Reviews 1987;51(2):221‐71. [PUBMED: 2439888] [DOI] [PMC free article] [PubMed] [Google Scholar]


