ABSTRACT
The Anaphase-Promoting Complex/Cyclosome (APC/C) is an E3 ubiquitin ligase and a key regulator of cell cycle progression. By triggering the degradation of mitotic cyclins, APC/C controls cell cycle-dependent oscillations in cyclin-dependent kinase (CDK) activity. Thus, the dynamic activities of both APC/C and CDK sit at the core of the cell cycle oscillator. The APC/C controls a large number of substrates and is regulated through multiple mechanisms, including cofactor-dependent activation. These cofactors, Cdc20 and Cdh1, recognize substrates, while the specific E2 enzymes UBE2C/UbcH10 and UBE2S cooperate with APC/C to build K11-linked ubiquitin chains on substrates to target them for proteasomal degradation. However, whether deubiquitinating enzymes (DUBs) can antagonize APC/C substrate ubiquitination during mitosis has remained largely unknown. We recently demonstrated that Cezanne/OTUD7B is a cell cycle-regulated DUB that opposes the ubiquitination of APC/C substrates. Cezanne binds APC/C substrates, reverses their ubiquitination and protects them from degradation. Accordingly, Cezanne depletion accelerates APC/C substrate degradation, leading to errors in mitotic progression and formation of micronuclei. Moreover, Cezanne is significantly amplified and overexpressed in breast cancers. This suggests a potential role for APC/C antagonism in the pathogenesis of disease. APC/C contributes to chromosome segregation fidelity in mitosis raising the possibility that copy-number and expression changes in Cezanne observed in cancer contribute to the etiology of disease. Collectively, these observations identify a new player in cell cycle progression, define mechanisms of tempered APC/C substrate destruction and highlight the importance of this regulation in maintaining chromosome stability.
KEYWORDS: Mitosis, APC/C, Cezanne
The E3 ubiquitin ligase APC/C is a master cell cycle regulator
The cell cycle, or the process by which a cell gives rise to two identical daughter cells, is a fundamental aspect of biology. While the identification of many core cell cycle genes dates more than 40 years to genetic studies originally performed in budding yeast [1], it is still an area of extensive research, since alterations in cell cycle control lead to unrestrained cell proliferation and chromosome instability, both of which are hallmarks of cancers. Central to cell cycle progression is the activation of cyclin-dependent kinases (CDKs) by cyclins. The temporal activation of CDKs depends on the availability of a cyclin subunit to form a functional and active CDK-cyclin heterodimer. Several combinations of CDK-cyclin pairings allow cells to progress through particular stages of the cell cycle, and the temporal, sequential oscillations in cyclin abundance are instrumental to faithful and accurate cell division [2].
A groundbreaking discovery in the cell cycle field was the demonstration that the abundance of mitotic cyclins is regulated by the ubiquitin-proteasome system (UPS) [3]. The UPS relies on a three-step enzymatic cascade that, in humans, involves two ubiquitin-activating enzymes (E1), several dozen ubiquitin-conjugating enzymes (E2) and hundreds of ubiquitin ligases (E3). E3 ubiquitin ligases provide specificity in the UPS by designating substrates for ubiquitination. Conjugation of a single ubiquitin molecule onto a substrate lysine (mono-ubiquitination) is a non-proteolytic event that usually alters protein-protein interactions [4]. Protein degradation is triggered by conjugation of a ubiquitin chain onto a target protein (polyubiquitination), which is then degraded by the 26S proteasome. In a very simplistic way, polyubiquitination of a substrate occurs by directly conjugating ubiquitin onto a lysine residue in the target protein, and then adding ubiquitin molecules to the first and subsequent ubiquitins to form a chain. However, in reality, ubiquitin chain formation is much more complex: it is a highly ordered process, and we discuss some of the molecular mechanisms below.
Although first considered a non-specific disposal system for cells to get rid of abnormal proteins, the involvement of polyubiquitination in cell division completely changed this view [5]. A series of papers in the late 1980s and early 1990s demonstrated that mitotic cyclins are degraded by the UPS and that their degradation controls cell cycle in early embryonic systems [3,6]. This line of inquiry culminated in the simultaneous identification, by the laboratories of Avram Hershko and Mark Kirschner, in 1995, of the gigantic 1.2 MDa E3 ubiquitin ligase, the Anaphase Promoting Complex/Cyclosome (APC/C) [7,8]. Since APC/C controls degradation of the CDK-coactivators Cyclins A and B, it sits at the heart of the cell cycle. Moreover, by triggering the degradation of Cyclin B and Securin, APC/C literally “promotes anaphase” and therefore chromosome segregation, allowing cells to complete mitosis. The APC/C also contributes to G1 maintenance [9] by targeting a myriad of additional cell cycle proteins, including transcription factors, replication regulators, mitotic kinases and spindle assembly factors. This long list of substrates includes FoxM1, the Aurora kinases A and B, Geminin, Cdc6, Cyclin F, Nusap1 and others [10].
The tremendous importance of APC/C in cell division has led many research groups to dissect the molecular mechanisms underlying its mode of action and regulation. Some of the key features of the APC/C are its ability to recognize short, linear sequence motifs, also known as degrons. The APC/C degrons, termed D and KEN boxes, are bound by its co‐activator subunits Cdc20 and Cdh1/Fzr1, primarily during mitosis and G1, respectively [11]. Another important aspect of APC/C-mediated ubiquitination resides in the type of ubiquitin chains that are built on substrates. Historically, it was believed that proteolytic signals consist of four ubiquitin molecules linked to each other through lysine 48 in ubiquitin (hereafter referred to as K48-linked chains) and attached to a target protein [12]. However, ubiquitin contains six additional lysine residues and an amino-terminal methionine, all of which can be used to generate distinct ubiquitin chain linkages with differing topologies [13]. Using ubiquitin mutants harboring point mutations in each lysine residue, it was found that the APC/C triggers the formation of K11-linked ubiquitin chains [14]. This specific linkage is made by the sequential action of the APC/C with two specific cell cycle-regulated E2 enzymes, UBE2C/UbcH10 and UBE2S [15,16]. UBE2C primes substrates by decorating them with single ubiquitin molecules on lysine residues within the target, leading to multi-monoubiquitination of the substrate [17]. A second E2, UBE2S, extends these modifications by building K11-linked ubiquitin chains [18,19]. Therefore, the molecular signature of the APC/C consists of K11-linked ubiquitin chains on substrates whose abundance is cell cycle regulated and which coincides with APC/C activity [20].
Deubiquitinating enzymes, the other side of protein ubiquitination
Analogous to other post-translational modifications, ubiquitin conjugation is a reversible process. This important yet understudied aspect of protein ubiquitination is mediated by deubiquitinating enzymes (DUBs). Approximately 100 DUBs are encoded in our genome, and they are classified into seven families based on the architecture of their catalytic domains: ubiquitin-specific proteases (USPs), ovarian tumor proteases (OTUs), ubiquitin carboxy-terminal hydrolases (UCHs), the Machado–Joseph disease proteases (MJDs), zinc-dependent metalloproteases (JAMMs) and the recently identified motif interacting with ubiquitin-containing novel DUB family (MINDY) and ZUP1 [21–23]. In contrast to our understanding of E3 ubiquitin ligase functions and mechanisms of action, the role of DUBs in crucial cellular processes is still in its infancy. However, a growing body of evidence suggests that DUBs are important regulators of cell physiology, whose misregulation contributes to diseases. A notable example is the DUB USP7/HAUSP, which regulates signaling through the Mdm2-p53 tumor suppressor pathway [24]. By deubiquitinating and stabilizing Mdm2, USP7 leads to p53 degradation and is therefore closely linked to genome stability and checkpoint function. Since its inhibition stabilizes p53, USP7 represents an attractive therapeutic target in cancer [25,26].
Because of the promising roles and regulation of DUBs, several groups have characterized the molecular features of these enzymes. David Komander and colleagues uncovered an unexpected feature of the OTU class of DUBs [27]. This family is comprised of 16 members, and by using di-ubiquitin probes recapitulating all eight different linkages of ubiquitin chains, they observed that OTU DUBs harbor linkage specificity. One of them, OTUD7B/Cezanne (hereafter referred to as Cezanne) showed a preference for K11 linkages [28]. Structural studies from the same group characterized the molecular basis for this specificity [28], but remarkably, the significance and function of this feature in human cells remained elusive. The importance of this specificity was further complicated by additional reports on a K11-independent function of Cezanne in different biological processes, such as mTORC2 and NF-kappa-B signaling [29,30].
Cezanne is a cell cycle-regulated DUB antagonizing APC/C activity
In order to address this discrepancy, we examined Cezanne function and abundance in actively dividing cells [31]. Since K11-linked ubiquitin chains are (i) important regulators of cell division and (ii) the product of cell cycle-regulated enzymes like UBE2S and the APC/C, we hypothesized that Cezanne could also be a cell cycle-regulated protein. Using a combination of immunofluorescence imaging and immunoblot analysis of synchronized cells, we found that Cezanne is cell cycle regulated in several cell lines. Its abundance strongly increases during mitosis and decreases as cells exit mitosis and progress through G1. Importantly, Cezanne increases at the mRNA levels in mitotic cells, indicating a role for gene expression dynamics in controlling its oscillating expression during the cell cycle. Therefore, Cezanne abundance coincides with the timing of APC/C activation during mitosis. However, while the APC/C stays active through G1 [9], Cezanne protein levels strongly decrease after mitotic exit. Thus, we explored the possibility that Cezanne exerts its function in mitosis by interacting with established cell cycle regulators which are targeted for degradation by the APC/C in late mitosis. By combining co-immunoprecipitation and in vitro binding assays using bacterially purified proteins, we found that Cezanne interacts with the transcription factor FoxM1, the Aurora A kinase and Cyclin B. It is important to note that in these assays, the interaction observed between Cezanne and these substrates is ubiquitin independent. Since Cezanne is able to bind directly to substrates [31], and also can directly interact with K11-linked ubiquitin chains, this raises the interesting possibility that Cezanne engages substrates through simultaneous, multi-valent interactions with both targets and ubiquitin chains. Furthermore, these interactions appeared to be cell cycle regulated, indicating that currently unknown post-translational mechanisms determine the ability of Cezanne to recognize substrates. This could include modifications of substrates or Cezanne itself, allosteric changes in either Cezanne or its targets that facilitate enzyme-substrate binding, or the use of adaptor proteins that increase the affinity of Cezanne for substrates. Addressing these questions represents an important area of future research. Moreover, we found that Cezanne also interacts with Cdc20 and Cdh1, the co-activating subunits of the APC/C, again by co-immunoprecipitation and in vitro binding assays. However, the molecular basis and importance of this binding is currently unknown.
Based on these results, we tested whether Cezanne can deubiquitinate APC/C substrates and antagonize APC/C-mediated degradation. We used ubiquitination assays where the APC/C is isolated from mitotic cells and ubiquitin conjugation is reconstituted by adding E1, E2, ubiquitin, ATP and a recombinant substrate of interest, Cyclin B. This approach allowed us to observe that when Cezanne was added to the reaction, the extent of polyubiquitinated Cyclin B was strongly diminished and importantly, this effect was completely dependent on Cezanne catalytic activity. Moreover, we demonstrated that Cezanne specifically removes K11-linked ubiquitin chains from two APC/C substrates, Cyclin B and Securin. This was made possible by using recombinant APC/C complexes isolated from insect cells, together with distinct sets of E2 enzymes, which allowed us to compare the activity of Cezanne towards K11, K48 or K63-linked ubiquitin chains conjugated to the same substrate. Cezanne removed K11-linked chains from both Cyclin B and Securin, whereas K48 and K63-linked chains were left untouched. Finally, having established that Cezanne can efficiently deubiquitinate APC/C substrates, we looked at the impact of Cezanne on their rates of degradation. By first using a synchronized cellular extract in which we can monitor APC/C-dependent degradation of proteins over time, we found that adding Cezanne to the reaction reduced the rate of substrate turnover. Conversely, in a complementary assay, we observed that substrate degradation was accelerated in Cezanne-depleted cells compared to controls cells when both populations were released from a mitotic block.
We next examined the impact of Cezanne depletion on mitotic progression. Previous studies have shown that restraining APC/C activity results in slower mitotic progression as well as fewer mitotic errors [32,33]. Hence, we hypothesized that we would observe the opposite effect when knocking down Cezanne. Indeed, we observed a significant increase in micronuclei formation as well as lagging chromosomes in Cezanne-depleted cells. Remarkably, these effects were completely rescued by UBE2S co-depletion, which is responsible for the APC/C-dependent formation of K11-linked ubiquitin chains on substrates. Therefore, these observations suggest that by restraining APC/C activity in mitosis, Cezanne allows proper mitotic progression and faithful chromosome segregation. We hypothesized that a long-term effect of mitotic defects should impact cell proliferation, since improper chromosome segregation leads to aneuploidy [34]. Indeed, we observed that Cezanne depletion led to slower proliferation rates of several cancer cell lines, with slower progression into S-phase. Rather than directly controlling S-phase progression, we anticipate that the newly reported role of Cezanne during mitosis, which prevents micronuclei formation and lagging chromosomes, allow cells to progress through the cell cycle correctly. Thus, we speculate that several rounds of cell division without Cezanne should lead to accumulation of mitotic defects, which would ultimately be detrimental for their proliferation rate, therefore explaining the decrease of cells into S-phase. Experimentally addressing this question represents an important area of future study.
Perspectives
In summary, our recent study describes a novel, DUB-dependent mechanism which restrains APC/C activity during mitosis, thereby contributing to substrate degradation kinetics and proper mitotic progression [31] (Figure 1). We envision that Cezanne could be involved in a phenomenon referred to as substrates ordering, meaning that different APC/C substrates are degraded at different rates as cells exit mitosis and progress through G1-phase. Several mechanisms likely contribute to substrate ordering. One such mechanism is substrate processivity, that is, the number of ubiquitin molecules added to a substrate in a single binding event with APC/C. This concept has been shown experimentally, where Cyclin B becomes highly poly-ubiquitinated in a single APC/C encounter, whereas more distributive substrates need several rounds of binding before being adequately ubiquitinated to trigger their destruction [35]. This has been explained by different affinities of substrates for the APC/C. Therefore, substrate ordering has been thought to be a property intrinsic to the APC/C, and while it has been proposed that DUBs could play a role in this process, our work is the first to identify and describe an enzyme involved in this regulation.
Figure 1.
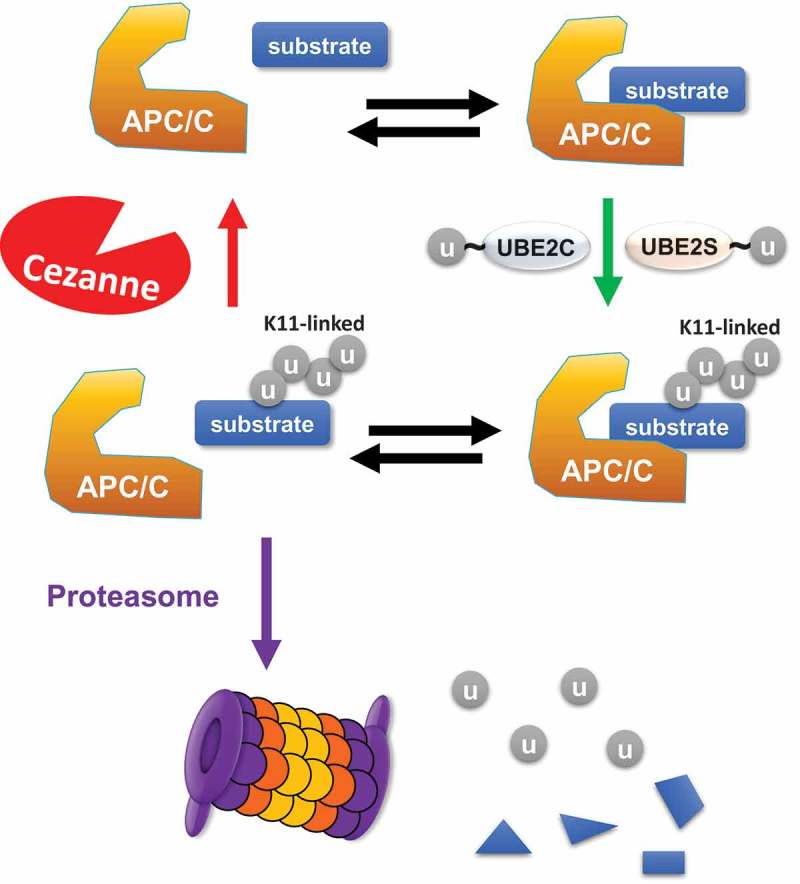
Cezanne is a DUB antagonizing APC/C ubiquitination and degradation.
APC/C recognizes and interacts with a cell cycle substrate. Following recruitment of the APC/C specific, ubiquitin charged E2s, UB2C and UBE2S, the substrate gets ubiquitinated with a K11-linked ubiquitin chain. After dissociation from APC/C, the ubiquitinated substrate can be either degraded by the proteasome or deubiquitinated by Cezanne, which therefore contributes to the kinetics of substrate degradation.
An intuitive way of picturing how Cezanne could alter the kinetics of substrate degradation is through the successive rounds of binding and dissociation of target proteins from E3 ligases (Figure 1). For instance, once a protein gets ubiquitinated it must dissociate from its E3 ligase in order to be recognized by the proteasome to be degraded. In this scenario, if a DUB “catches” the substrate before proteasomal recognition, this would lead to its deubiquitination and thus alter its degradation kinetics (Figure 1). Therefore, the affinity and processivity of DUBs like Cezanne for their substrate would influence the kinetics of substrate degradation, in the same way that affinity and processivity of APC/C for substrates influences degradation. Alternatively, since several DUB-E3 ligase interactions have been identified by mass spectrometry-based immunoprecipitation proteomics studies [36], DUBs could already be in complex with E3 ligases to contribute to the kinetics of substrate ubiquitination. This is an attractive hypothesis since we observed that Cezanne can bind the APC/C co-activating sub-units Cdc20 and Cdh1. However, Cezanne was not found in proteomics studies looking at the APC/C interactome [15] and Cezanne did not control the degradation kinetics of all APC/C substrates that we examined. In particular, we analyzed the stability of substrates such as Aurora B and Geminin following Cezanne depletion and failed to observe any effect on their rates of degradation. Since these are APC/C substrates not influenced by Cezanne, we anticipate that it might not be the only enzyme involved in substrate degradation at mitotic exit, and that other DUBs could serve a similar purpose for different sets of substrates. This would also explain why Cezanne has not been found as being essential in genome-wide screens looking for lethal genes. If Cezanne is the first of several DUBs which have a similar function in restraining proteolysis at mitotic exit, it is reasonable to predict that there might be redundancy among DUBs and that simultaneous loss would be required to cause lethality. According to data from the Komander lab, other OTU DUBs show some preferences towards K11-linkages, although none of them has a comparable activity to Cezanne [27]. However, such enzymes would represent high priority candidates to test in order to find additional DUBs with a function similar to the one we describe for Cezanne.
Alternatively, different APC/C substrates could be decorated with ubiquitin chains of distinct linkages. Indeed, it is important to add that the notion of APC/C being a K11-linked ubiquitin chain specific enzyme has evolved during the last couple of years. It has been shown previously that the APC/C also builds K48-linked ubiquitin chains on substrates, although to a much lesser extent than K11-linked chains [37]. Recently, the generation of bispecific antibodies allowed the detection of K11/K48-branched chains generated by the APC/C [38]. A branched chain refers to a ubiquitin molecule that can be simultaneously modified with two ubiquitin molecules on both lysines 11 and 48. This atypical linkage was first described using engineered ubiquitin, substrates and enzymes in in vitro assays, showing that the APC/C can assemble such linkages in a test tube [39]. Therefore, in light of these recent data, it is fair to assume that a subset of substrates could be preferentially ubiquitinated with K11-linked chains and sensitive to Cezanne regulation, whereas others could be preferentially ubiquitinated with K48-linked or K11/K48-branched chains and unaffected by Cezanne. This reinforces the notion that additional DUBs could serve a similar purpose as Cezanne in controlling the rates of APC/C substrate degradation during cell cycle progression. Testing this possibility, and identifying such enzymes would be critical to determine the diversity of ubiquitin chain topologies conjugated to cell cycle regulators. Ultimately, performing screens without focusing on the previously reported DUB specificity might be of interest too, since a recent study elegantly demonstrated that post-translational modifications can alter DUB linkage specificity [40]. Therefore, we expect that identifying additional players contributing to the kinetics of substrates degradation and antagonizing APC/C activity might be the subject of exciting research in the future.
Other important questions raised by our study are the underlying mechanisms controlling both Cezanne enzymatic activity and abundance. We have ongoing efforts to determine if Cezanne activity can be altered during mitosis or in G1. The K11-specificity of Cezanne has frequently been studied using recombinant protein in a cell-free system. It is important to note that when produced in bacteria, recombinant proteins do not get post-translational modifications compared to other systems such as insect cells. This is of particular importance in light of studies showing that phosphorylation can alter both activity and specificity of DUBs [40,41]. Nevertheless, we observed that the activity of Cezanne could be slowed down when recombinant Cezanne was incubated in extracts of mitotic but not asynchronous cells, and similarly when Cezanne was expressed and immunoprecipitated from mitotic but not asynchronous cells (unpublished results). This suggests that Cezanne activity tracks that of the APC/C, in the sense that its activity is low in early mitosis and presumably increases as cells progress through mitosis. The regulation of APC/C activity in early mitosis is suppressed by the spindle assembly checkpoint (SAC), which promotes the assembly of a mitotic checkpoint complex (MCC) that binds the APC/C and prevents its activation [42]. Once all chromosomes become properly attached to the mitotic spindle, disassembly of the MCC leads to APC/C activation and anaphase onset. Whether a similar mechanism exists for regulating Cezanne activation, and whether it depends on the SAC, will be of tremendous importance to deepen our understanding of its enzymatic activity. Another important aspect of Cezanne function is the control of its abundance during cell cycle progression. A key finding of our study is that Cezanne is a new cell cycle protein, whose regulation occurs at the transcriptional level and presumably also at the post-translational level. Identifying which transcription factor(s) and mechanisms contribute to this regulation would help to further define its contribution to cell cycle progression. An attractive possibility for how cells would get rid of Cezanne after mitosis is through APC/C-mediated degradation. Although Cezanne is the first DUB identified to counteract APC/C activity in mitosis, the Dixit lab identified USP37 as a DUB antagonizing the APC/C to stabilize Cyclin A at the end of G1 [43]. In that study, the authors showed that USP37 is a substrate of the APC/C in mitosis, which turns into a negative regulator of APC/C-mediated ubiquitination through Cdk2-dependent phosphorylation and activation to allow Cyclin A accumulation and progression into S-phase. In light of such a mechanism, we envision that Cezanne activity could be repressed in early G1 by a yet to be characterized process, allowing the APC/C to ubiquitinate and destroy it. This inactivation step would be mandatory since we observed that Cezanne can also deubiquitinate itself. Moreover, sequence analysis of Cezanne shows that it contains one putative KEN box as well as several potential D boxes, the latter being well conserved between humans and mice. Since our study is the first to point to a role of Cezanne in cell cycle control, characterizing the mechanisms regulating it in the context of cell division is of tremendous importance to establish Cezanne as a new cell cycle DUB.
Finally, transcriptional and DNA copy-number changes identified in large cohorts of patient tumors through TCGA show that Cezanne is amplified and/or overexpressed in roughly one-third of breast tumors [44]. Cezanne is localized in an amplicon on chromosome 1q, and remarkably, this region lacks a clear oncogenic driver [45]. This could suggest ian additional and potential role in genome maintenance and cancer cell proliferation. Since Cezanne is involved in progression through mitosis, its overexpression in cancers could negatively impact chromosome segregation and cause aneuploidy. Furthermore, since APC/C also plays a key role in restraining G1/S, it is possible that aberrant expression of Cezanne could weaken the proliferative barrier to S-phase entry. In addition to its role in mitotic progression, Cezanne has been shown to activate mTORC2 signaling by deubiquitinating GβL, thus facilitating tumorigenesis [29]. Therefore, this suggests that Cezanne might be involved in several proliferative pathways, which could explain the observation that some cancer cells show amplification of its mRNA. Moreover, it is noteworthy to add that the amplicon on chromosome 1q contains another DUB called USP21 [46]. In the past few years, a growing body of evidence has suggested that USP21 plays an important role in tumor growth [47]. Our unpublished data also suggest additional roles for USP21 in cell cycle. Taken together, this suggests that inhibition of DUBs could be a promising way of treating cancers that lack efficient therapies. Indeed, there is a growing interest in developing inhibitors targeting components of the UPS, with the proteasome inhibitor bortezomib/Velcade being the only drug currently approved for the treatment of multiple myeloma and mantle cell lymphoma. Pharmacological inhibition of DUBs is starting to draw attention from pharmaceutical companies since their catalytic activity can be targeted by small molecules [25,26]. Therefore, understanding the contribution of DUBs, such as Cezanne and others in tumor formation and maintenance, could pave the way to the development of new drugs to improve cancer treatment.
In conclusion, our molecular understanding of mitotic progression is now increasingly more refined. Although the APC/C is one of the most-studied and best-understood ubiquitin ligases, we keep unraveling additional layers of regulation and complexity in the mechanisms governing its function. Our finding that Cezanne is a newly identified player in the APC/C network, whose DUB activity contributes to proper kinetics of substrate degradation and mitotic progression, illustrates the need to continue defining the regulatory apparatus surrounding this critical ubiquitin ligase, whose physical size among E3s is only matched by its key function in cell cycle control.
Funding Statement
This work was supported by start-up funds from the University Cancer Research Fund to MJE, as well as grants to MJE from the Susan G. Komen Foundation [CCR14298820], the American Cancer Society [RSG-18-220-01-TBG], Jimmy-V Foundation and National Institute of Health [R01GM120309].
Acknowledgments
Special thanks to the Emanuele laboratory for feedback on this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Hartwell LH, Culotti J, Pringle JR, et al. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. [DOI] [PubMed] [Google Scholar]
- [2].Morgan DO. The cell cycle: principles of control. Sunderland (MA): New Science Press in association with Oxford University Press; 2007. [Google Scholar]
- [3].Glotzer M, Murray AW, Kirschner MW.. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. [DOI] [PubMed] [Google Scholar]
- [4].Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem [Internet] 2012;81:291–322. [DOI] [PubMed] [Google Scholar]
- [5].Hershko A, Ciechanover A, Varshavsky A. The ubiquitin system. Nat Med. 2000;6:1073–1081. [DOI] [PubMed] [Google Scholar]
- [6].Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. [DOI] [PubMed] [Google Scholar]
- [7].King RW, Peters JM, Tugendreich S, et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. [DOI] [PubMed] [Google Scholar]
- [8].Sudakin V, Ganoth D, Dahan A, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kernan J, Bonacci T, Emanuele MJ. Who guards the guardian? Mechanisms that restrain APC/C during the cell cycle. Biochim Biophys Acta Mol Cell Res. 2018;1865:1924–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choudhury R, Bonacci T, Arceci A, et al. APC/C and SCFcyclin FConstitute a reciprocal feedback circuit controlling S-Phase entry. Cell Rep [Internet] 2016;16:3359–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Alfieri C, Zhang S, Barford D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 2017;7:170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Thrower JS. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Publ Gr. 2016;18:579–586. [DOI] [PubMed] [Google Scholar]
- [14].Jin L, Williamson A, Banerjee S, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Williamson A, Wickliffe KE, Mellone BG, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci [Internet] 2009;106:18213–18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu T, Merbl Y, Huo Y, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci [Internet] 2010;107:1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dimova NV, Hathaway NA, Lee B, et al. APC / C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Publ Gr [Internet] 2012;14:168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brown NG, Watson ER, Weissmann F, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: ring repurposing for ubiquitin chain assembly. Mol Cell [Internet] 2014;56:246–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brown NG, VanderLinden R, Watson ER, et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell [Internet] 2016;165:1440–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Matsumoto ML, Wickliffe KE, Dong KC, et al. Resource K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell [Internet] 2010;39:477–484. [DOI] [PubMed] [Google Scholar]
- [21].Nijman SMB, Luna-Vargas MPA, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. [DOI] [PubMed] [Google Scholar]
- [22].Abdul Rehman SA, Kristariyanto YA, Choi SY, et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell [Internet] 2016;63:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kwasna D, Arif S, Rehman A, et al. Distinct deubiquitinase class important for genome article discovery and characterization of ZUFSP / ZUP1, a distinct deubiquitinase class important for genome stability. Mol Cell [Internet] 2018;70:150–164.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tavana O, Gu W. Modulation of the p 53 / MDM 2 interplay by HAUSP inhibitors. J Mol Cell Biol. 2018;9:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kategaya L, Di Lello P, Rougé L, et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534. [DOI] [PubMed] [Google Scholar]
- [26].Turnbull AP, Ioannidis S, Krajewski WW, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mevissen TET, Hospenthal MK, Geurink PP, et al. XOTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell [Internet] 2013;154:169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mevissen TET, Kulathu Y, Mulder MPC, et al. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature [Internet] 2016;538:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang B, Jie Z, Joo D, et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545:365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moniz S, Bandarra D, Biddlestone J, et al. Cezanne regulates E2F1-dependent HIF2α expression. J Cell Sci. 2015;128:3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bonacci T, Suzuki A, Grant GD, et al. Cezanne/OTUD7B is a cell cycle‐regulated deubiquitinase that antagonizes the degradation of APC/C substrates. Embo J. 2018;37:e98701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zeng X, Sigoillot F, Gaur S, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sansregret L, Patterson JO, Dewhurst S, et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov. 2017;7:218–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Funk LC, Zasadil LM, Weaver BA. Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev Cell [Internet] 2016;39:638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. [DOI] [PubMed] [Google Scholar]
- [36].Sowa ME, Bennett EJ, Gygi SP, et al. Resource defining the human deubiquitinating enzyme interaction landscape. Cell [Internet] 2009;138:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kirkpatrick DS, Hathaway NA, Hanna J, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700. [DOI] [PubMed] [Google Scholar]
- [38].Yau RG, Doerner K, Castellanos ER, et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell [Internet] 2017;171:918–933.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell [Internet] 2014;157:910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhao Y, Mudge MC, Soll JM, et al. OTUD4 is a phospho-activated K63 deubiquitinase that regulates MyD88-dependent signaling. Mol Cell [Internet] 2018;69:505–515.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Huang OW, Ma X, Yin J, et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012;19:171–175. [DOI] [PubMed] [Google Scholar]
- [42].Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol [Internet] 2015;25:R1002–18. [DOI] [PubMed] [Google Scholar]
- [43].Huang X, Summers MK, Pham V, et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011;42:511–523. [DOI] [PubMed] [Google Scholar]
- [44].Cancer T, Atlas G. Comprehensive molecular portraits of human breast tumours. Nature. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Silva GO, He X, Parker JS, et al. Cross-species DNA copy number analyses identifies multiple 1q21-q23 subtype-specific driver genes for breast cancer. Breast Cancer Res Treat. 2015;152:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu J, Kruswick A, Dang H, et al. Ubiquitin-specific protease 21 stabilizes BRCA2 to control DNA repair and tumor growth. Nat Commun [Internet] 2017;8 DOI: 10.1038/s41467-017-00206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]


