Abstract
Complement factor B (cfB) is an essential component of alternative pathway (AP) and plays an important role in the pathogenesis of polymicrobial sepsis. However, the mechanism leading to cfB production and AP activation during sepsis remains poorly understood. In this study, we found that the plasma cell-free RNA was significantly increased following cecal ligation and puncture (CLP), an animal model of polymicrobial sepsis, and was closely associated with sepsis severity. qRT-PCR and miRNA array analysis revealed an increase in bacterial RNA and multiple host microRNAs (miR-145, miR-146a, miR-122, miR-210) in the blood following CLP. Treatment with tissue RNA or synthetic miRNA mimics (miR-145, miR-146a, miR-122, miR-34a) induced a marked increase in cfB production in cardiomyocytes or macrophages. The newly synthesized cfB released into medium was biologically active as it participated in the AP activation initiated by cobra venom factor. Genetic deletion of TLR7 or MyD88, but not TLR3, and inhibition of the MAP kinases (JNK and p38) or NF-κB abolished miR-146a-induced cfB production. In vivo, CLP led to a significant increase in splenic cfB expression, which was correlated with the plasma RNA or miRNA levels. Peritoneal injection of RNA or miR-146a led to a rise in cfB expression in the peritoneal space, which was attenuated in MyD88-KO or TLR7-KO mice, respectively. These findings demonstrate that host cellular RNA and specific miRNAs are released into the circulation during polymicrobial sepsis and may function as extracellular mediators capable of promoting cfB production and AP activation through the specific TLR7 and MyD88 signaling.
Keywords: RNA, microRNA, complement factor B, Toll-like receptors, sepsis
INTRODUCTION
During sepsis, invading pathogens as well as endogenous molecules released from injured tissues interact with host cells and cause massive activation of innate immune responses including immune cell activation, cytokine production, and complement system activation (1). Complement is a part of the innate immune system and functions against invading pathogen (2). It can also promote cytokine and reactive oxygen species (ROS) production that ultimately leads to organ injury in severe sepsis (3–6). Complement factor B (cfB) is an essential component of the alternative pathway (AP) and amplifies complement activation (7). We have recently reported that cfB is up-regulated in serum and in major organs including the heart and kidney following cecal ligation and puncture (CLP) procedure, and may contribute to the pathogenesis of polymicrobial sepsis (3). Systemic cfB deficiency improves the animal survival and cardiac function and attenuates acute kidney injury in severe sepsis. Moreover, others have demonstrated that locally synthesized rather than serum complements are of major importance in tissue injury (8–11). While TLRs are known to regulate cfB production (3, 12), the mechanisms leading to the critical cfB up-regulation and AP activation in severe sepsis remain incompletely understood.
During bacterial infection, nucleic acids are released into the extracellular space from invading pathogens and injured tissues (13). These include various types of RNA such as transfer RNA, ribosomal RNA, messenger RNA and microRNA (14). These extracellular RNAs may provoke inflammation and induce tissue injury. For example, in a sterile model of tissue injury, eliminating extracellular RNA by systemic delivery of RNase reduces myocardial inflammation and injury following a transient ischemia (15, 16). Extracellular RNA induces a potent inflammatory response of cardiovascular system by enhancing leukocyte recruitment (15, 17) and cytokine production (15, 18).
MicroRNAs (miRNAs) are a group of small noncoding RNAs. The primary role of miRNA is to regulate gene expression inside the cell at the post-transcriptional level by binding to the 3′ untranslated region of the target mRNAs. Thus, by affecting protein translation, miRNAs act as regulators (repressors) of a wide range of biological processes and play a dominant role in health and diseases (19–21). miRNA may also function as cell-to-cell communicators. miRNA can be actively secreted or passively released from cells (22) and in different pathological conditions such as ischemia (15) and sepsis (14). miRNAs are circulating in the blood in various forms, such as exosomes, microvesicles, HDL, and RNA binding proteins that protect them from RNase digestion and thus is highly stable in the blood. These miRNAs reportedly act on the target cells and play an important role in the pathogenesis of diseases (23).
In the current study, we tested the hypothesis that endogenous tissue RNA including various miRNAs are released into the circulation during severe polymicrobial sepsis and that both tissue RNA and miRNAs are capable of stimulating complement factor production and AP activation through specific innate immune signaling.
MATERIALS AND METHODS
Reagents
Antibodies against phospho-p38, phospho-ERK1/2, phospho-JNK, IκB-α, and GAPDH were purchased from Cell Signaling Technology (Beverly, MA). Total p38, ERK1/2, and JNK antibodies were from Santa Cruz Biotechnology (Dallas, Texas). Bovine pancreas RNase A, lipofectamine 3000, TRIzol LS, SYTO RNASelect Green fluorescent cell stain, and Quant-iT™ RNA assay kit were from Invitrogen Life Technology (Carlsbad, CA). The TLR ligands, poly(I:C), Pam3Cys and CpG, were from Enzo Life (Plymouth Meeting, PA). DNase was from Thermo Scientific Inc. (Waltham, MA). Imiquimod (R837, TLR7 ligand) and CL075 (TLR7/8 ligand) were from Invivogen (San Diego, CA). Human cfB antibody and cobra venom factor (CVF) were purchased from Complement Technology (Texas). TRIzol reagent used to extract RNA from cell or tissues and HRP-conjugated donkey anti-goat IgG are from Sigma-Aldrich (St. Louis, MO). Protease and phosphatase inhibitors were from Roche Diagnostics (Indianapolis, IN). miRNA mimics were synthesize by Integrated DNA Technologies (Coralville, IA) with sequences listed in Table 1. miScript II RT kit, miScript SYBR green PCR kit and miRNA primers for qRT-PCR were purchased from Qiagen (Valencia, CA). Criterion™ XT Bis-Tris Precast gels were purchased from Bio-Rad (Hercules, CA). Luminata Forte Western HRP substrate was from Millipore Corporation (Billerica, MA).
Table 1.
Sequence of miRNA mimics
| miRNA | Sequence |
|---|---|
| mmu-miR-146a-5p | 5′-UGAGAACUGAAUUCCAUGGGUU-3′ |
| mmu-miR-145-5p | 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′ |
| mmu-miR-122-5p | 5′-UGGAGUGUGACAAUGGUGUUUG-3′ |
| mmu-miR-34a-5p | 5′-UGGCAGUGUCUUAGCUGGUUGU-3′ |
| mmu-miR-210-3p | 5′-CUGUGCGUGUGACAGCGGCUGA-3′ |
| mmu-miR-192-5p | 5′-CUGACCUAUGAAUUGACAGCC-3′ |
Animals
Eight to 16 week-old gender and age-matched mice were used. C57BL/6J wild-type (WT), TLR7−/−, and TLR3−/− mice were purchased from the Jackson Laboratories and housed in a Massachusetts General Hospital (MGH) animal facility for at least one week before experiments. cfB−/− mice were kindly provided by Xiaobo Wu at Washington University in St. Louis School of Medicine. MyD88−/− mice were provided water supplemented with sulfamethoxazole (4 mg/ml) and trimethoprim (0.8 mg/ml). Antibiotics were stopped for at least two weeks prior to experiments. All animals were housed in pathogen-free, temperature-controlled, and air-conditioned facilities with 12 h/12 h light/dark cycles and fed with the same bacteria-free diet. All animal care and procedures were performed according to the protocols approved by the Subcommittee on Research Animal Care of MGH and are in compliance with the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health.
Mouse model of polymicrobial sepsis
A clinically relevant rodent model of sepsis was created by cecal ligation and puncture (CLP) as described previously (3, 24, 25). In brief, mice were anesthetized and the cecum was ligated 1.2 cm from the tip and punctured with an 18-gauge needle. A small amount of fecal materials was squeezed out gently. The sham-operated mice underwent laparotomy but without CLP. After surgery, pre-warmed normal saline (0.05 ml/gram body weight) was administered subcutaneously. Postoperative pain control was managed with subcutaneous injection of bupivacaine (3 mg/kg) and buprenorphine (0.1 mg/kg). Postoperatively, mice were monitored for rectal temperature and sepsis severity score (0 = bright, alert, and responsive; 1 = slightly lethargic; 2 = lethargic and hunched; 3 = very lethargic, hunched, and shaky; 4 = dead) as described previously (26).
MicroRNA array
Plasma was prepared from EDTA anticoagulant blood of both sham and septic mice at 24 h after surgery. The profile of 68 miRNAs related to an immunology panel was analyzed using a novel technology provided by Firefly BioWorks (Boston, MA). The technology employed a unique post-hybridization ligation-based scheme to fluorescently label bound microRNA targets. Since RNA extraction was no longer needed with this technology, we were able to detect miRNA array in a small volume of samples (300 μl) without risk of losing RNA.
RNA extraction
Tissue and cell RNA extraction
Heart, spleen, or macrophages were disrupted and placed in TRIzol reagent. RNA was extracted according to the manufacturer’s protocol. RNA in DEPC-treated water was quantified and stored at −80°C for future experiments. Plasma RNA extraction: Plasma was prepared from EDTA anti-coagulated blood. Two hundred μl of plasma was mixed with 50 μl of DEPC-treated water and 750 μl of TRIzol LS. Caneorhabditis elegans miR-39 (Cel-miR-39) was added as the spike-in control. Plasma RNA was extracted followed the manufacturer’s protocol, quantified using a Quant-iT™ RNA assay kit, and stored at −80°C for future experiments.
Cell treatments
Rat neonatal cardiomyocytes or bone marrow-derived macrophages were incubated in serum-free culture medium containing 0.05% BSA for 1 h prior to treatment with splenic RNA (10 μg/ml), miRNA mimics (50 nM), poly (I:C) (10 μg/ml), R837 (1 μg/ml), CL075 (1 μg/ml) or pam3cys (1 μg/ml) for the indicated time period. The miRNA mimic sequences are listed in Table 1. Media or cells were collected for analysis. RNA, miRNA mimics and poly (I:C) were complexed with the transfection agent lipofectamine 3000 and incubated for 5 min at room temperature prior to the treatment. For nuclease treatment in Fig. 2E, 4 μg RNA was incubated with RNase (10 μg), DNase (1 U) at room temperature for 30 min before complexed with lipofectamine 3000.
Fig. 2. Role of RNA in cfB production in the heart and isolated cardiomyocytes.
A. RNA digestion by serum RNase - Testing the efficacy of systemic administration of RNase. As detailed in the Methods, sera were prepared from mice injected with saline or RNase A, and incubated with 3 μg of purified RNA at 37°C for 2 hours. Lane 1: Untreated RNA; Lane 3–8: RNA treated with serum from the mice injected with saline; Lane 9–14: RNA treated with serum from the mice injected with RNase A. B–D. Effect of systemic RNase administration on cardiac gene expression of cfB, C3 and C5. RNase was administrated before and 12h after sham or CLP surgery. cfB/C3/C5 gene expression was detected in the heart 24h after surgery using qRT-PCR. n=4 in each group. # P < 0.05, δ P < 0.001 vs. Sham-Saline; ** P < 0.01. E. Effect of RNase on RNA-induced cfB protein production. Cultured rat neonatal cardiomyocytes were treated lipofectamine alone (Lipo), or with cardiac RNA (10 μg/ml) for 24 hours. As indicated, in some treatment groups, RNA was first pretreated with RNase or DNase before applied to cardiomyocyte cultures. Medium cfB was detected by Western blot as described in the Methods. F. Effect of TLR ligands on cfB protein production in rat neonatal cardiomyocytes. Cardiomyocytes were treated with poly (I:C) (10 μg/ml), R837 (1 μg/ml), or CL075 (1 μg/ml) for 24 hours. Medium cfB was detected with Western blot.
qRT-PCR
Gene expression in cells and tissues
Cell and tissue RNA was extracted with TRIzol reagent and reversely transcribed to cDNA by reverse-transcription (RT) reaction. cDNA was quantified using qPCR as described previously (25). The PCR primer sequences for host cellular RNA (3) and bacterial RNA (27) are listed in Table 2. Transcript expression was calculated using the comparative Ct method normalized to GAPDH (2−ΔΔCt) and expressed as fold change in CLP or treatment group over sham or control group. MiRNA expression in plasma: Plasma RNA was extracted with TRIzol LS as described above. Reverse transcription was performed using miScript II RT kit. miRNAs expression was analyzed using miScript SYBR green PCR kit following the manufacture’s protocol. Relative miRNA expression was calculated using the comparative Ct method normalized to spike-in Cel-miR-39 (2−ΔΔCt) and expressed as fold change in CLP group over sham group.
Table 2.
Primer sequence for qRT-PCR
| Gene | Primer Sequence |
|---|---|
| Mouse GAPDH | Forward 5′-AACTTTGGCATTGTGGAAGG-3′ Reverse 5′-GGATGCAGGGATGATGTTCT-3′ |
| Mouse C3 | Forward 5′-GGCAAGACAGTCGTCATCCT-3′ Reverse 5′-CCAAGACAAAGGCAAGATGC-3′ |
| Mouse C5 | Forward 5′-TACCACAGAACCCAGGAGGA-3′ Reverse 5′-GCCATCCGCAGGTATGTTAG-3′ |
| Mouse cfB | Forward 5′-GAAACCCTGTCACTGTCATTC-3′ Reverse 5′-CCCCAAACACATACACATCC-3′ |
| Bacterial 16S rRNA (27) | Forward 5′-ATTAGATACCCTGGTAGTCCACGCC-3′ Reverse 5′-CGTCATCCCCACCTTCCTCC-3′ |
In vivo RNase administration and efficacy test
In vivo RNase administration and activity was tested as described previously with modifications (15). In brief, bovine pancreatic RNase A was dialyzed and filtered through 30 kDa cut-off filter column and stored at 4 °C. To test RNase activity, 3mg RNase or the same volume of normal saline was administrated s.c.. Blood was collected at 3 h, 6 h and 9 h after the injection. The serum was prepared and filtered through a 30 kDa filter column to remove large proteins. Filtered serum was incubated with 3 μg of purified kidney RNA at 37 °C for 2 hour followed by SYTO RNASelect green fluorescence staining and RNA electrophoresis. To test the impact of RNase administration on cardiac cfB expression in sepsis, RNase or normal saline was administrated as follows: RNase 0.5 mg, i.p., 1 h before surgery, 3 mg s.c. right after surgery, and 3 mg s.c. 12 h after surgery. At 24 h, the hearts were harvested and stored at −80°C for future experiment.
Isolation and culture of cardiomyocytes and macrophages
Rat neonatal cardiomyocytes and mouse bone marrow-derived macrophages were isolated and cultured as described previously (3).
Immunoblotting of cfB, MAPKs, and IκBα
Medium proteins were separated in 4–12% gradient SDS-PAGE under reducing condition, transferred to PVDF membrane, and blotted with goat anti-human cfB antibody. cfB protein bands were visualized using Luminata Forte Western HRP substrate. For MAP kinase and IκBα detection, macrophages were washed with cold DPBS and lysed in NP-40 buffer containing various protease and phosphatase inhibitors following various treatments. Protein was quantified with Bradford method and equal amount of protein was subjected to SDS-PAGE and immunoblotting.
Complement alternative pathway activity assay
The alternative pathway (AP) activity was assayed as described previously (28) with modifications. Specifically in this study, AP activation was achieved in the medium containing 11μg/ml of cobra venom factor (CVF, a functional C3b analog), cfB-deficient serum containing factor D, and 5 mM MgCl2. In the presence of Mg2+, CVF binds to cfB, which is cleaved by factor D into Ba that is released, and Bb that remains bound to CVF. The CVF.Bb complex is a C3 convertase. The mixture was then incubated at 37°C water bath for 1 hour. The assay solutions were mixed with SDS sample buffer and subjected to SDS-PAGE. Total cfB or Ba fragments were immunoblotted with a specific anti-cfB antibody.
In vivo administration of RNA and miRNA mimics
In vivo RNA and miRNA mimics administration was performed as previously described (18). In Brief, after fur removed and the abdominal skin disinfected, mice were administered with 1) lipofectamine or lipofectamine-complexed RNA (50μg/mouse), 2) lipofectamine-complexed miR-146a (20μg/mouse) or miR-146a mutant via intra-peritoneal (i.p.) injection with a 31 gauge needle insulin syringe. The injection site was immediately covered with an adhesive 3M Tegaderm film to prevent any potential contamination and infection. Twenty hours later, two ml saline was injected into the peritoneal space and mixed thoroughly. Peritoneal lavage was collected and centrifuged. The cell-free supernatant was stored at −80°C for further cfB protein analysis.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6 software (GraphPad Software Inc., La Jolla, CA). The distributions of the continuous variables were expressed as the mean ± SE. Two-way ANOVA with Bonferroni adjusted P-value for post-hoc analysis was applied to test the differences in cardiac cfB expression between Saline and RNase group (Fig. 2B), cfB expression in the macrophages among WT, TLR7−/− and TLR3−/− groups (Fig 3B, 5E), MAPKs/IκBα expression between WT and TLR7−/− groups (Fig. 7B, C, D, E), the role of MAPK inhibitors on cfB expression (Fig. 7) and lavage cfB protein expression in WT, MyD88−/− and TLR7−/− (Fig. 9). Pearson correlation analysis was used to assess the relationship between the plasma RNA concentration to rectal temperature, sepsis severity, and splenic cfB mRNA level The relationship of plasma miRNA-146a, 122, 145, 210 level to splenic cfB mRNA was also analyzed by Pearson correlation. Student’s t test was used for statistical analysis between the groups of all other data. The null hypothesis was rejected for P < 0.05 with the two-tailed test.
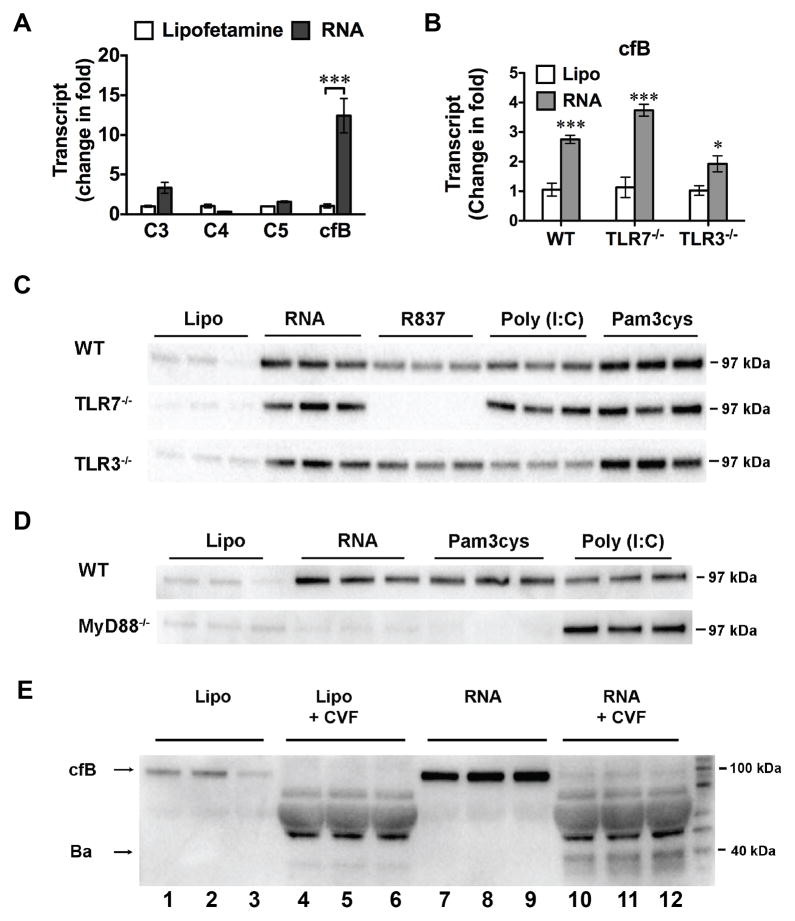
Fig. 3. Splenic RNA-induced cfB production was mediated via MyD88 signaling in macrophages.
A. Complement gene expression in macrophages treated with splenic RNA. Mouse macrophages were treated with lipofectamine alone or splenic RNA (10 μg/ml) complexed with lipofectamine. Six hours later, C3, C4, C5, cfB mRNA was analyzed by qRT-PCR. *** P < 0.001, n=3 in each group. The experiments were repeated three times. B. cfB gene expression in macrophages isolated from WT, TLR7−/−, or TLR3−/− mice. Cells were treated with lipofectamine alone or RNA (10 μg/ml) for 6 hours. * P < 0.05, *** P < 0.001 vs. the corresponding lipofectamine control (Lipo). n=3 in each group. C. Splenic RNA- and TLR ligand-induced cfB protein production in WT, TLR7−/− or TLR3−/− macrophages. Macrophages were treated with lipofectamine, RNA (10 μg/ml), R837 (0.25 μg/ml), poly (I:C) (10 μg/ml) or pam3cys (1 μg/ml). Twenty-four hours later, medium cfB was analyzed. The experiment was repeated 3 times. D. Impact of MyD88 on RNA- and TLR ligand-induced cfB production in the medium. The experiment was repeated twice. E. AP activity assay. Macrophages were incubated in serum-free (complement-free) medium and treated with lipofectamine (lane 1–6) or 10 μg/ml RNA (lane 7–12) for 24 hours as indicated. At the end of treatment, media were collected and incubated in the presence of 2.5% cfB−/− mouse serum, 11 μg/ml of cobra venom factor (CVF) and 5 mM of MgCl2 at 37°C for 1 hour, and analyzed for cfB and Ba fragment by Western blot. The experiment was repeated twice. Lipo=lipofectamine.
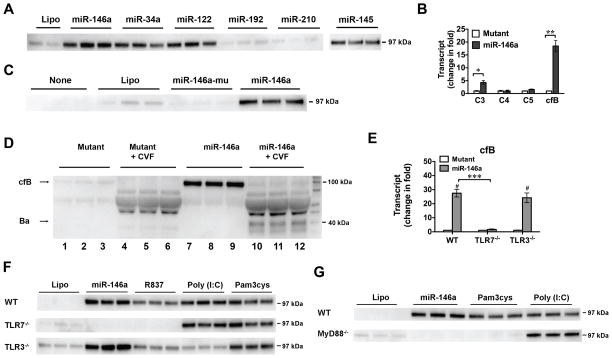
Fig. 5. miRNA-induced cfB production is via TLR7-MyD88 signaling in the macrophage.
A. miRNA mimics induce cfB production. Macrophages were treated with 50 nM of miRNAs for 18 hours and media were analyzed for cfB expression. B. Effect of miR-146a on complement gene expression in macrophages. miR-146a mimic or miR-146a mutant (50 nM) was incubated with WT macrophages. Six hours later, C3, C4, C5 and cfB mRNAs were measured using qRT-PCR. * P < 0.05, ** P < 0.01. n=3 in each group. The experiment was repeated twice. C. Effect of miR-146a and its U→A mutant on cfB protein production in the medium. D. AP activity assay. Macrophages were incubated in serum-free (complement-free) medium and treated with 50 nM of miR-146a mutant (lane 1–6) or miR-146a (lane 7–12) for 24 hours. At the end of treatment, media were collected and incubated in the absence (lane 1–3, 7–9) or presence (lane 4–6, 10–12) of 2.5% cfB−/− mouse serum, 11 μg/ml cobra venom factor (CVF) and 5 mM MgCl2 at 37°C for 1 hour, and analyzed for cfB and Ba fragment by Western blot. The experiment was repeated twice. E. Impact of TLR7 and TLR3 deletion on miR-146a-induced cfB gene expression. WT, TLR3−/− and TLR7−/− macrophages were treated with miR-146a (50 nM) or its mutant for 6 hours. cfB mRNA was quantified by qRT-PCR. # P < 0.001 vs. the corresponding mutant control in each strain. *** P < 0.001, n=3 in each group. The experiment was repeated twice. F. Impact of TLR7 and TLR3 deletion on miR-146a-induced cfB protein production. Macrophages from WT, TLR3−/−, TLR7−/− mice were treated with lipofectamine, miR-146a (50 nM), R837 (0.25 μg/ml), poly (I:C) (10 μg/ml), or pam3cys (1 μg/ml) for 18 hours. Media were collected for cfB analysis. The experiment was repeated twice. G. Effect of MyD88 deletion on miR-146a-induced cfB protein production. Cells were treated with lipofectamine, miR-146a, pam3cys, or poly (I:C) for 18 hours and the culture media were tested for cfB expression. The experiment was repeated twice. Lipo=lipofectamine.
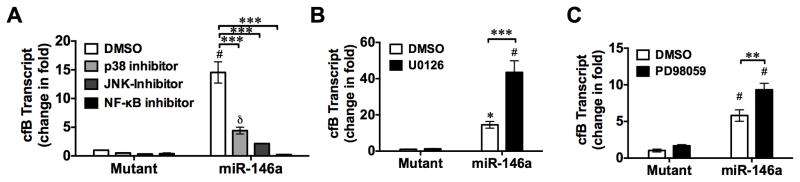
Fig. 7. Inhibition of JNK, p38, and NF-κB attenuates miR-146a-induced cfB gene expression.
WT macrophages were pre-incubated with p38 inhibitor (SB203580, 20 μM), JNK inhibitor II (SP600125, 20 μM), ERK inhibitors (U0126, 10 μM; PD 98095, 20 μM), or NF-κB signaling inhibitor (Bay 11-7082,10 μM) for 1 hour prior to miR-146a (50 nM) or its U→A mutant treatment. cfB mRNA level was quantified by qRT-PCR 6 hours after treatment. * P < 0.05, δ P < 0.01, # P < 0.001, vs. corresponding mutant control. ** P < 0.01, *** P < 0.001. n=3 in each group.
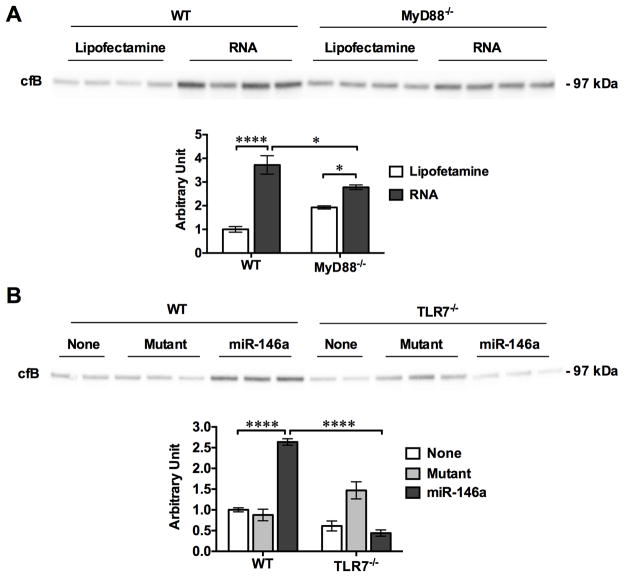
Fig. 9. Cellular RNA and miR-146a mimics stimulate cfB production in vivo via TLRs signaling.
RNA (50 μg/mouse) or miR-146a (20 μg/mouse) complexed with lipofectamine was injected i.p. into WT, MyD88−/− or TLR7−/− mice with lipofectamine or mutant as the respective control. Twenty hours later, the peritoneal lavage was harvested and cfB protein expression was analyzed by Western blot. The cfB expression levels were quantitated by a NIH ImageJ software and expressed as fold change over Lipofectamine control (A) or no injection group (None, B) in WT mice. A. cfB expression in the peritoneal lavage after RNA injection. n=4 per group, * P < 0.05, **** P < 0.0001. B. cfB expression in the peritoneal lavage after miRNA-146a injection. n=2–3 per group, **** P < 0.0001. None, no injection; Mutant, miR-146a-mutant.
RESULTS
Polymicrobial infection leads to an increase in circulating RNA
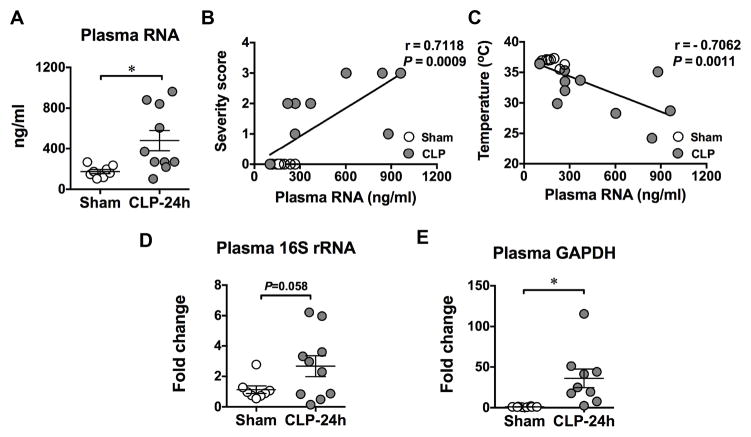
To determine whether or not cellular RNA was released into the circulation during bacterial infection, we created a CLP model of polymicrobial infection and tested the circulating RNA in both sham and CLP mice. As shown in Fig. 1A, while there was a large variation among different infected mice, the mean RNA concentration in the plasma of CLP mice was significantly higher than that of sham mice (478.1 ± 100 vs. 174.6 ± 20 ng/ml). Interestingly, the amount of circulating RNA was correlated with the level of sepsis severity as well as with body temperature of mice including both sham and CLP groups (Fig. 1B–C). Mice with more severe sepsis and lower body core temperature tended to have higher circulating RNA levels. Moreover, we found that a significant amount of cell-free16S rRNA, common to many species of bacteria, rose in the plasma of septic mice compared to the sham-operated mice (Fig. 1D). Similarly, the circulatory cell-free mRNA of GAPDH, which is stably and constitutively expressed at high levels in most tissues and cells, was also found significantly increased in septic mice (Fig. 1E). These data suggest that during polymicrobial infection, both host cells and invading bacteria release their cellular RNA into the host circulation and that the circulating RNA level is associated with the overall sepsis severity.
Fig. 1. Polymicrobial sepsis leads to host and bacterial RNA release into the blood circulation.
Twenty-four hours after sham or CLP surgery, mice were euthanized and blood was collected via inferior vena cava. RNA was extracted from plasma and quantified as described in Methods. A. Plasma total RNA concentrations in sham and CLP mice. * P < 0.05. n=8 in sham group, n=10 in CLP group. B-C. Association of plasma RNA concentration and sepsis severity score or core temperature, respectively. Sepsis severity score was monitored as detailed in the Methods. Rectal temperature was recorded as the core temperature. n=18 pair samples. D. Bacterial 16S rRNA in the plasma as measured by qRT-PCR. E. Host GAPDH mRNA in the plasma as measured by qRT-PCR, * P < 0.05. n=8 in sham group, n=10 in CLP group.
Eliminating extracellular RNA reduces cardiac cfB gene expression in septic mice
Our previous study has established an important role of cfB and the complement AP activation in septic cardiac dysfunction and mortality (3). In the study, we show that cardiac cfB is markedly and specifically upregulated in response to polymicrobial infection. Lack of systemic cfB improves cardiac function as well as survival of septic mice. To determine if extracellular RNA contributes to cardiac cfB upregulation during bacterial sepsis, we treated mice with RNase A before and after CLP surgery aiming to eliminate extracellular RNA. To determine the efficacy of RNase A administration, we tested the serum RNase activity. As shown in Fig. 2A, when sera were incubated with purified cellular RNA, the sera derived from the saline-injected mice possessed endogenous RNase activity resulting in partial degradation of 28S/18S rRNA and consequently increase in RNA of small molecular weight around 5S (Fig. 2A, lane 3–8). In comparison, systemic administration of exogenous RNase led to higher serum RNase activity between 3 and 9 hours after injection as demonstrated by almost complete digestion of cellular RNA including 28S/18S rRNA and 5S small RNA (Fig. 2A, lane 9–14). As shown in Fig. 2B–C, 24 hours after CLP, there was a significant increase in cfB and a slightly increase in C3 gene expression (cfB: 6.6 ± 0.7; C3: 1.5 ± 0.1, fold change), but not that of C5 (Fig. 2D) in the heart as compared with the sham control. Importantly, there was 46% reduction in cardiac cfB gene expression in septic mice treated with RNase before and after CLP surgery compared to septic mice treated with saline alone (Fig. 2B). These data suggest that circulating RNA may contribute in part to cardiac cfB production during polymicrobial infection.
RNA and TLR ligands induce cfB production in cardiomyocytes
To determine the effect of cellular RNA on cardiac cfB production, we isolated rat neonatal cardiomyocytes and treated the cultured cells with RNA that was isolated from mouse heart and complexed with lipofectamine. As shown in Fig.2E, 24 hours after RNA treatment, there was a significant amount of cfB detected in the culture media, whereas there was minimal cfB detected in lipofectamine-treated cells. The effect of the cardiac RNA was almost abolished by pretreatment with RNase, but not DNase. Moreover, media collected from cardiomyocyte cultures treated with TLR3 ligand (poly (I:C)), TLR7 ligand (R837), or TLR7/8 ligand (CL075) all displayed elevated cfB levels compared to the lipofectamine control (Fig. 2F).
Splenic RNA induces cfB production via MyD88 signaling
We next tested the effect of splenic RNA on complement gene expression in bone marrow-derived macrophages. As shown in Fig. 3A, consistent with what we found in cardiomyocytes, RNA induced a marked cfB gene expression, whereas it had a minimal effect on C3, C4 or C5 gene expression. Loss of function study indicated that neither TLR7 nor TLR3 gene deletion had significant impact on the RNA-induced cfB expression (Fig. 3B). This was confirmed by Western blot. Neither TLR7- or TLR3-deficiency had any effect on the RNA-induced cfB protein production (Fig. 3C). Similar to cfB protein production in cardiomyocytes, R837 (TLR7 agonist) or poly (I:C) (TLR3 agonist) induced a significant increase in cfB protein production and their effect was completely or near completely blocked in the cells lacking TLR7 or TLR3, respectively (Fig. 3C). The reduction of cfB in these TLR-deficient macrophage was not due to poor cell condition, as both TLR7−/− and TLR3−/− macrophages had a similar robust response to Pam3cys, a TLR2 ligand compared to WT macrophages. On the other hand, MyD88, an adaptor for all TLRs with the exception of TLR3, completely blocked RNA- and TLR2-induced cfB production, but had no impact on TLR3-mediated cfB protein production (Fig. 3D). Finally, to determine the biological activity of the de novo synthesized cfB in macrophages treated with splenic RNA, we tested the ability of medium cfB in promoting the AP activity. As illustrated in Fig. 3E, compared with lipofectamine (Lipo) (lane 1–3), splenic RNA induced a marked increase in the cfB level of serum-free medium (lane 7–9). In the presence of CVF (a functional C3b analog), there was an increased AP activity as demonstrated by an increased cfB cleavage and consequent Ba generation (lane 10–12). In contrast, treatment with CVF alone without the newly synthesized medium cfB only exhibited a minimal level of AP activity (lane 4–6).
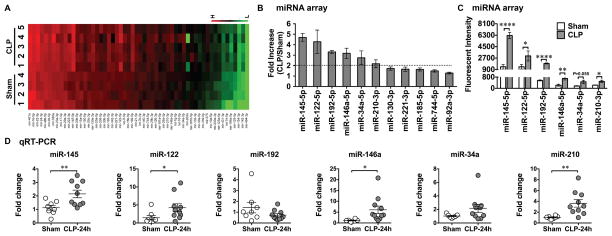
Plasma microRNA array in septic mice
To identify host miRNA released into the circulation during polymicrobial infection, we subjected mice to sham or CLP surgery. Twenty-four hours later, we collected the plasma from both groups of mice and tested them for miRNA expression using miRNA array (Fig. 4A). The complete set of the original miRNA array data (accession no: GSE74952) can be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74952. Among the 68 miRNAs reportedly related to immunologic functions, six miRNAs had more than 2-fold increase in the CLP mice compared to the sham control mice (CLP/Sham ratio > 2) with fluorescence counts > 100 (Fig. 4B). These were miR-145-5p, miR-122-5p, miR-192-5p, miR-146a-5p, miR-34a-5p, and miR-210-3p. Fig. 4C illustrates the mean fluorescent intensity of these six miRNAs in both groups as measured by the miRNA array. The differences in the fluorescent intensities of these six miRNAs between the sham and CLP groups were statistically significant (Fig. 4C). To validate these miRNA array results, we tested these six miRNAs using qRT-PCR in the plasma collected from a separate set of animals that underwent either sham or CLP procedures. As shown in Fig. 4D, out of the six miRNAs tested, four were confirmed with significant increase in septic mice compared to sham mice, namely miR-145-5p, miR-122-5p, miR-146a-5p, and miR-210-3p. To determine whether or not these miRNAs were capable of inducing cfB production, we treated macrophages with the miRNA mimics complexed with lipofectamine or lipofectamine alone for 24 hours. As shown in Fig. 5A, miR-146a, miR-34a, miR-122, or miR-145, all at 50 nM, induced a marked increase in cfB protein production in macrophages. In contrast, miR-192 and miR-210 at the same concentration failed to induce cfB production. In the case of miR-146a, the cfB-inducing effect was sequence-specific as its U→A mutant failed to induce cfB production at both gene and protein levels (Fig. 5B–C). The effect of miR-146a also appeared to be specific for cfB as it only up-regulated the gene expression of cfB, mild increase in C3, but not C4, or C5, in macrophages (Fig. 5B). Moreover, similar to splenic RNA-induced cfB response, miR-146a-induced cfB synthesis in macrophages promoted the AP activation. As illustrated in Fig. 5D, the de novo synthesized cfB was released from macrophages stimulated by miR-146a (lane 7–12) and in the presence of the functional C3b analog CVF and cfB −/− serum containing factor D and Mg2+, led to the AP activation as evidenced by increased cfB cleavage and Ba formation (lane 10–12). In contrast, miR-146a mutant failed to induce cfB production or AP activation (lane 1–6).
Fig. 4. Plasma miRNA profiling in septic mice.
A. Heat map of miRNA array. Mice were subjected to Sham or CLP procedure. At 24 hours, plasma was collected and miRNAs were analyzed using Firefly microRNA array provided by Firefly BioWorks. Sham (n=4) and CLP (n=5) mice. The fluorescence intensity of 56 miRNAs was expressed from low (green) to high (red). B. Fold change in the plasma miRNAs in the CLP compared to that of Sham mice (CLP/Sham) as measured by miRNA array. C. Mean fluorescent intensity of plasma miRNAs as measured by miRNA array in Sham and CLP mice. Six miRNAs (miR-145, miR-122, miR-192, miR-146a, miR-34a, miR-210) were significantly increased for more than 2-fold in the septic mice compared to the sham control. .* P < 0.05, ** P < 0.01, **** P < 0.001. n=4 in sham group, n=5 in CLP group. D. qRT-PCR validation of miRNAs. The 6 target miRNAs were tested using qRT-PCR in a separate set of plasma. * P < 0.05, ** P < 0.01. n=8 in sham group, n=10 in CLP group.
miR-146a induces cfB production via TLR7-MyD88 signaling in macrophages
To determine the mechanisms responsible for miRNA-induced cfB production, we tested miR-146a for its signaling pathway in mediating cfB production. As shown in Fig. 5E–F, treatment with miR-146a led to a marked increase in cfB gene and protein expression. The effect was completely blocked in TLR7−/−, but unaffected in TLR3−/− macrophages. Moreover, R837 and poly (I:C) induced a robust cfB production and their effects were specifically blocked in TLR7−/− or TLR3−/− macrophages, respectively. In contrast, Pam3cys, a TLR2 ligand, maintained its ability to induce cfB synthesis in both WT and TLR3- and TLR7-deficient macrophages. Finally, the effect of miR-146a seemed completely dependent on MyD88 signaling as it failed to induce cfB synthesis in MyD88−/− macrophages (Fig. 5G). As anticipated, Pam3cys (TLR2), but not poly (I:C) (TLR3), induced cfB production via MyD88-dependent mechanism.
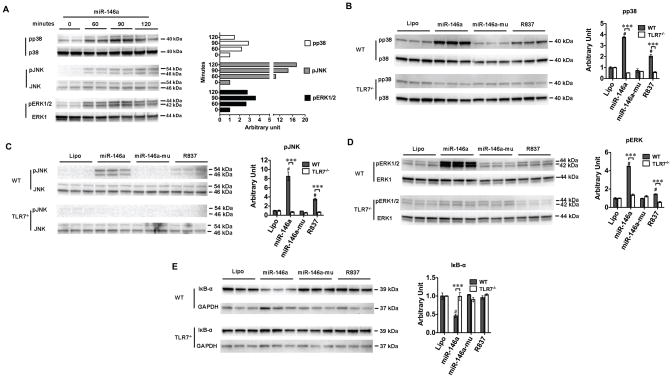
miR-146a activates MAPKs and NF-κB via TLR7 signaling
To investigate the downstream signaling of miR-146a, we tested the intracellular MAP kinase and NF-κB pathways. WT macrophages were stimulated with 50 nM of miR-146a for a period of 60 min, 90 min, or 120 minutes. Immunoblotting of the cell lysates revealed a strong phosphorylation of p38, JNK and ERK1/2 with the peak effect seen between 90 – 120 minutes (Fig. 6A). R837, a TLR7 ligand, but not the miR-146a mutant, induced a similar effect as miR-146a (Fig. 6B–D). The MAPK activation appeared to be mediated via TLR7 as TLR7−/− macrophages failed to respond to miR-146a or R837 (Fig. 6B–D). Activation of NF-κB involves the phosphorylation and proteolysis of the IκB-α protein and subsequently nuclear translocation of the NF-κB factors. As shown in Fig. 6E, stimulation with miR-146a (not its mutant) of macrophages led to the degradation of IκB-α, suggesting an activation of NF-κB signaling in the miR-146a-treated cells. In contrast, the IκB-α protein level was maintained at the baseline level in TLR7−/− macrophages despite of miR-146a treatment.
Fig. 6. miR-146a activates MAPK and NF-κB signaling via TLR7.
A. Immunoblotting of p38, JNK and ERK1/2. Macrophages were treated with 50 nM of miR-146a. At time 0, 60, 90, 120 min, cells were lysed and tested for phosphorylated and total p38, JNK, and ERK1/2. The difference was expressed as the ratio to time 0 as quantified by Image J. B–D. TLR7 deletion completely blocked miR-146a- or R837-induced phosphorylation of p38, JNK, and ERK1/2, respectively. WT and TLR7−/− macrophages were treated with miR-146a (50 nM) or R837 (0.25 μg/ml) for 90 minutes. Cell lysate extracted and analyzed for phosphorylation of p38, JNK and ERK1/2. Total MAP kinase expression was served as the internal control. # P < 0.001 vs. lipofectamine control (Lipo). *** P < 0.001, WT vs. TLR7−/− group. n=3 in each group. The difference was expressed as the ratio to lipofectamine control (Lipo) in each strain as quantified by Image J. E. TLR7 deletion blocked miR-146a-induced IκB-α degradation. GAPDH served as the protein loading control. # P < 0.001 vs. lipofectamine control (Lipo). *** P < 0.001, WT vs. TLR7−/− group. n=3 in each group. The difference was expressed as the ratio to lipofectamine control (Lipo) in each strain as quantified by ImageJ.
Inhibition of MAP kinases and NF-κB signaling attenuates miR-146a-induced cfB production
To determine whether or not MAP kinases and NF-κB activation are involved in cfB de novo synthesis in response to miR-146a treatment, we treated cultured macrophages with specific inhibitors of p38, JNK, ERK1/2, or NF-κB. As shown in Fig. 7A, miR-146a induced a robust repsonse in cfB gene expression compared to its U→A mutant. Pretreatment of macrophages with the inhibitor of p38 or JNK markedly reduced the miR-146a-induced cfB gene expression. Bay 11-7082, an inhibitor of IκB-α kinase, completely blocked the cfB gene expression induced by miR-146a. In contrast, blocking of ERK signaling by MEK1/2 inhibitor PD98059 or U0126, significantly enhanced the miR-146a-mediated cfB gene expression (Fig. 7B–C).
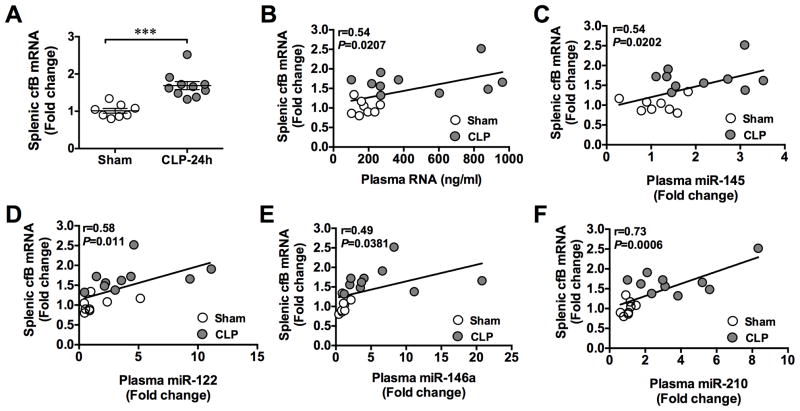
Splenic cfB expression is correlated with plasma RNA and miRNAs during polymicrobial infection
As shown in Fig. 8A, 24 h after CLP procedure, there was a significant increase in cfB gene expression in the spleen as compared to that of the sham-operated mice. To determine the relationshiop between the splenic cfB expression and the plasma RNA/miRNA levels, we plotted the two sets of data (splenic cfB vs. plasma RNA or miRNAs) from both sham and CLP groups. As shown in Fig. 8B–F, the splenic cfB expression was significantly correlated with the plasma RNA and miRNAs (miR-145, miR-122, miR-146a, miR-210) levels. The r values ranged between 0.49 – 0.73.
Fig. 8. Splenic cfB gene expression is correlated with plasma RNA and miRNAs during polymicrobial sepsis.
Mice were subjected to sham or CLP procedure. Twenty hours later, the plasma and spleens were collected and extracted for total RNA. The plasma RNA concentrations were measured and analyzed for miRNAs as indicated using qRT-PCR. Splenic RNA was measured for cfB mRNA. A. Splenic cfB mRNA expression. *** P < 0.001. n=8 in sham group, n=10 in CLP group. B. Association of the plasma RNA concentrations and splenic cfB mRNA expression. n=18 pair samples. C–F. Association of the plasma miR-145, miR-122, miR-146a, miR-210 levels and the splenic cfB mRNA expression. n=18 pair samples.
RNA and miR-146a activate cfB production in vivo
To test whether or not in vivo administraton of RNA or miRNA mimics was able to induce cfB production, we injected tissue RNA or miR-146a into the peritoneal cavity. Twenty hours later, the peritioneal lavage was collected and tested for cfB protein level. As shown in Fig. 9, i.p. Injection of splenic RNA or miR-146a in WT mice induced a rise in cfB expression in the peritoneal cavity compared to the controls (lipofectamine and miR-146 mutant, respectively). Somewhat similar to cfB production in RNA-treated macrophages in vitro, the RNA-induced cfB production in vivo was in part dependent on MyD88 as MyD88−/− mice had significantly lower peritoneal cfB expression as compared with WT mice (Fig. 9A). On the other hand, systemic deletion of TLR7 completely prevented the miR-146a-induced cfB production in vivo.
DISCUSSION
We have previously documented a critical role for cfB and complement AP activation in an animal model of polymicrobial sepsis (3). Peritoneal bacterial infection induces a marked systemic as well as local tissue expression of cfB and AP activation. Systemic cfB deficiency results in improved survival and organ functions in animals subjected to severe sepsis. However, the cellular and molecular mechanisms responsible for cfB up-regulation and AP activation in the context of bacterial sepsis remain largely unexplored. In the current study, we tested the hypothesis that severe sepsis leads to release of bacterial and host cellular RNA into the circulation and that cellular RNA such as miRNAs are capable of inducing cfB production and AP activation. We found a significant increase in bacterial as well as host RNA in the plasma of septic animals. The plasma RNA concentration was closely associated with sepsis severity. Also, the plasma RNA and selective miRNA levels were correlated with splenic cfB expression during polymicrobial sepsis. miRNA array identified and qRT-PCR confirmed multi-fold increase in the plasma level of four host miRNAs, i.e., miR-146a, miR-145, mi-122, and miR-210, in septic mice. Moreover, treatment of cardiomyocytes and macrophages with splenic RNAs or the synthetic miRNA mimics (miR-146a, -145, -122, -34a) induced a robust cfB production and the AP activation. In vivo, i.p. injection of RNA or miR-146a led to a significant rise in the peritoneal cfB expression. Loss-of-function experiments demonstrated that signaling via MyD88 was important for RNA-induced cfB production in macrophages and in intact animals, and that TLR7 signaling proved to be critical for the miR-146a-induced cfB production in vitro and in vivo. Finally, we showed that miR-146a mimic activated intracellular MAPKs and NF-κB signaling via TLR7 signaling and that inhibition of p38, JNK and NF-κB singling, but not ERK, led to a significant reduction in miRNA-146a-mediated cfB gene expression in macrophages.
Cell apoptosis and necrosis occur in severe sepsis and is associated with the mortality of septic animals (29, 30). The injured cells release danger-associated molecules such as nucleic acids, HMGB1, heat shock proteins, mitochondrial components, which trigger innate immune responses such as cytokine production, immune cell activation and recruitment, and free radical species production (31). In an effort to search for an endogenous mediator(s) for cfB production during polymicrobial sepsis, we hypothesize that cellular RNA released as result of tissue injury during sepsis is capable of inducing cfB de novo synthesis. Our observations that systemic RNase administration attenuates myocardial cfB gene expression and that plasma RNA concentration is correlated with splenic cfB expression during bacterial infection are consistent with the notion that extracellular RNA is in part responsible for the cfB expression in the septic organs and provides us a clear rationale to test the ability of cellular RNA in activating cfB production. Indeed, our in vitro experiments demonstrate that tissue-derived RNA is capable of inducing a robust cfB production in and release from both cardiomyocyte and macrophage cultures. This effect is clearly RNA-mediated as RNase, not DNase, near completely inhibits the RNA-induced cfB production. To assess the biological activity of the de novo synthesized cfB, we tested the AP activation in the media derived from the macrophage cultures treated with splenic RNA in the presence of CVF (a C3b analog) and cfB-deficient serum containing complement factor D and supplemented with Mg2+. The increase in medium Ba fragment in the RNA-treated macrophage cultures demonstrates that the RNA-induced cfB can bind to CVF and actively participates in the complement AP activation. Of note, while splenic RNA was used in most of the experiments in the study, the ability of cellular RNA to trigger cfB production appears not organ/tissue-specific as cellular RNA isolated from mouse heart exhibits a similar effect. Moreover, the ability of cellular RNA to activate cfB production was further demonstrated in vivo following i.p. injection of cardiac RNA. Finally, in a separate study, we have observed that cardiac cellular RNA from human, rat, and mouse, and extracellular RNA released from injured cardiomyocytes all exhibit an ability to induce various cytokine responses in cardiomyocytes and macrophages (18). Taken together, these data demonstrate that significant amount of cellular RNA is released during bacterial sepsis and that host cellular RNA is capable of activating cfB production and promoting the AP activation.
TLR3 and TLR7 are two important innate immune receptors reportedly sensing RNA of viral origins (32–34). TLR3 senses double-stranded (ds) RNA and signals primarily through Trif, whereas TLR7 senses single-stranded (ss) RNA and signals via MyD88 (35). The finding that several TLR ligands including pam3cys, poly (I:C), R837, and CL075 induce cfB production in cardiomyocytes and macrophages suggests that activation of TLR2, TLR3, TLR7, and TLR8 is probably sufficient to initiate cfB synthesis. However, when we tested the cfB production in macrophages of several KO mice and compared it with that in WT mice, we found that neither TLR3- nor TLR7-deficiency had any impact on the cellular RNA-induced complement synthesis. This was somewhat surprising since several previous studies have suggested the role of TLR3 in sensing endogenous RNA. Kariok, et al, report that in vitro transcribed mRNA elicits cytokine production via a TLR3-dependent mechanism in human dendritic cells (36). A recent study by Bernard, et al, shows that RNA from UVB-irradiated keratinocytes induces cytokine production in normal human epidermal keratinocytes via TLR3 (37). Moreover, in a stable-transfected human HEK 293 cell line, in vitro transcribed mRNA can induce cytokine production via TLR7- and TLR8-dependent manner (38). Moreover, we show that cardiac RNA elicits cytokine production in part via TLR7-dependent signaling (18). Nevertheless, the current study identified that MyD88 deficiency abolished the effect of RNA in vitro and partially in vivo, suggesting the important role for MyD88 signaling to mediate the cellular RNA-induced cfB response. Several possibilities may explain these data. TLR8, which like TLR7, recognizes ssRNA, could mediate the effect of splenic RNA via MyD88-dependent signaling. In human, TLR8, not TLR7, is responsible for ssRNA-induced cytokine response. HEK 293 cells expressing human TLR8, but not TLR3 or TLR7, respond to viral RNA40 (39) or human mitochondrial RNA (38). However, while TLR7 and TLR8 are closely phylogenetically related, mouse TLR8 was thought to be nonfunctional (34). Alternatively, splenic RNA may signal through all three RNA sensors, namely TLRs 3/7/8, to induce the complement response since both single- and double-stranded RNA may exist in the spleen. Lacking one of them may prove insufficient to impact on the final outcome (i.e., cfB production and AP activation) because of the shared and somewhat redundant signaling pathways among the three RNA sensors. Finally, it is also possible, although probably less likely, that different cell response to RNA may depend on different RNA sensors and that RNA from different tissues, e.g., splenic vs. cardiac RNA, may use different RNA sensors.
We employed miRNA array to identify the different expression of plasma miRNAs between the sham and CLP mice. Validated by qRT-PCR, we identified four miRNAs (miR-146a, miR-145, miR-122, and miR-210) that were significantly elevated in the septic mice compared to the sham control. The miRNA array was quite limited and contained only 68 miRNAs. It is likely that the actual number of miRNAs being up-regulated during polymicrobial sepsis is higher. An excellent study by Wu, et al, used a similar approach and identified that out of more than 1000 miRNAs tested, 10 miRNAs were up-regulated in the whole blood and serum following CLP procedure (14). Interestingly, while it is well documented that TLR signaling (e.g., TLR2, TLR4, and TLR5) promotes miRNA biogenesis and miRNAs regulate TLR signaling (40–42), this study found that systemic deficiency of TLR2, TLR4, or NF-κB had no significant impact on the serum miRNA up-regulation induced by CLP (14).
Expanding the miRNA array assays, we tested the biological activities of the identified miRNAs. We found that miR146a, miR-145, miR-122 and miR-34a induced a significant cfB production in macrophages. miRNA146a is an immediate early-response gene induced by various microbial components and pro-inflammatory mediators. It reportedly inhibits TLR and cytokine signaling by targeting IRAK-1 and TRAF-6 translation (40) and is widely known for its role in negative regulation of inflammation and endotoxin response (42–46). Different from these studies, the present study identified a novel role of miR-146a – promoting the innate immune complement response by stimulating cfB de novo synthesis and AP activation. Moreover, we found that the effect of miR-146a mimic was entirely dependent on TLR7→MyD88 signaling as either TLR7 or MyD88 deficiency completely blocked the miR146-induced cfB production in macrophages and in the peritoneum following i.p. injection.
Consistent with these findings, several previous studies have demonstrated the role of extracellular miRNA as a signal molecule via TLR7. Fabbri, et al. (47) report that miR-21 and -29a secreted by tumor cells bind to TLR7 and activate the receptors in immune cells, leading to TLR-mediated NF-κB activation and secretion of prometastatic inflammatory cytokines. Another secreted extracellular miRNA, let-7, also reportedly activates TLR7 signaling and mediates pain (48) or caused neurodegeneration (49). These data suggest that miRNA, similar to ssRNA from virus (39), can be recognized by TLR7 and induce inflammatory response. Of note, we could not identify any consensus pattern of oligonucleotide sequences among the miRNAs that induce cfB production. However, all four miRNAs that exhibit the strong ability to activate cfB synthesis are rich in U nucleotides (7–9 Us), accounting for 30–40% of their entire sequences. This is in consistent with the report by Heil, et al. that U-rich or U/G-rich oligonucleotides are essential for TLR7 recognition of ssRNA (39). Indeed, the U→A mutants of miR-146a abolished its ability to induce cfB synthesis in macrophages.
One of the major signaling families that participate in the intracellular transmission of extracellular signals is the group of mitogen-activated protein kinases (MAPKs). This group of MAPKs is composed of serine/threonine kinases that phosphorylate and activate each other (50, 51). ERK1/2, p38, JNK1/2 have been well studied in the context of innate immunity (52). Together with NF-κB signaling, these MAPKs participate in the regulation of cellular inflammatory response. The current finding that miRNA-146a, but not its U→A mutant, elicits phosphorylation of MAPKs and enhanced degradation of IκB-α suggests that miR-146a signals through these MAP kinases and activates NF-κB pathways. The data derived from WT and TLR7−/− macrophages demonstrate that miR146a induces these intracellular events exclusively via TLR7 sensing. Importantly, employing specific inhibitors, our data further demonstrate that miR-146a activates cfB production via p38-, JNK-, and NF-κB-dependent mechanism. Interestingly, the ERK inhibitor enhanced the cfB-inducing effect of miR-146a suggesting that ERK may negatively regulate the cfB synthesis upon miR-146 stimulation.
In summary, the current study showed that polymicrobial sepsis induced a significant increase in the plasma RNA level, which included bacterial and host cellular RNA including several miRNA, e.g., miR-146a, miR-145, miR-122, and miR-210. Treatment with endogenous tissue RNA or the synthetic miRNA mimics such as miR-146a induces de novo cfB synthesis in and release from cultured cardiomyocytes and macrophages in vitro as well as in the peritoneal space when injected i.p. in vivo. Both RNA and miR-146a elicited the cfB production via similar and specific innate immune and intracellular MAPK signaling mechanisms. Thus, our study has identified a novel role of splenic/cardiac RNA and selective miRNAs in promoting the complement alternative pathway via specific innate immune signaling.
Footnotes
The authors declare no competing interests.
Disclosure:
This work was supported in part by the NIH (Bethesda, Maryland) grant, GM097259 (WC), and a mentored research award from International Anesthesia Research Society (San Francisco, California) (LZ).
References
- 1.Deutschman CS, Tracey KJ. Sepsis: current dogma and new perspectives. Immunity. 2014;40:463–475. doi: 10.1016/j.immuni.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Walport MJ. Complement. First of two parts. New Eng J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 3.Zou L, Feng Y, Li Y, Zhang M, Chen C, Cai J, Gong Y, Wang L, Thurman JM, Wu X, Atkinson JP, Chao W. Complement factor B is the downstream effector of TLRs and plays an important role in a mouse model of severe sepsis. J Immunol. 2013;191:5625–5635. doi: 10.4049/jimmunol.1301903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mollnes TE, Song WC, Lambris JD. Complement in inflammatory tissue damage and disease. Trends Immunol. 2002;23:61–64. doi: 10.1016/s1471-4906(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 5.Niederbichler AD, Hoesel LM, Westfall MV, Gao H, Ipaktchi KR, Sun L, Zetoune FS, Su GL, Arbabi S, Sarma JV, Wang SC, Hemmila MR, Ward PA. An essential role for complement C5a in the pathogenesis of septic cardiac dysfunction. J Exp Med. 2006;203:53–61. doi: 10.1084/jem.20051207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Kimura Y, Fang C, Zhou L, Sfyroera G, Lambris JD, Wetsel RA, Miwa T, Song WC. Regulation of Toll-like receptor-mediated inflammatory response by complement in vivo. Blood. 2007;110:228–236. doi: 10.1182/blood-2006-12-063636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harboe M, Mollnes TE. The alternative complement pathway revisited. J Cell Mol Med. 2008;12:1074–1084. doi: 10.1111/j.1582-4934.2008.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pratt JR, Basheer SA, Sacks SH. Local synthesis of complement component C3 regulates acute renal transplant rejection. Nat Med. 2002;8:582–587. doi: 10.1038/nm0602-582. [DOI] [PubMed] [Google Scholar]
- 9.Farrar CA, Zhou W, Lin T, Sacks SH. Local extravascular pool of C3 is a determinant of postischemic acute renal failure. FASEB J. 2006;20:217–226. doi: 10.1096/fj.05-4747com. [DOI] [PubMed] [Google Scholar]
- 10.Daha MR, van Kooten C. Is there a role for locally produced complement in renal disease? Nephrol Dial Transplant. 2000;15:1506–1509. doi: 10.1093/ndt/15.10.1506. [DOI] [PubMed] [Google Scholar]
- 11.Morgan BP, Gasque P. Extrahepatic complement biosynthesis: where, when and why? Clin Exp Immunol. 1997;107:1–7. doi: 10.1046/j.1365-2249.1997.d01-890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczorowski DJ, Afrazi A, Scott MJ, Kwak JH, Gill R, Edmonds RD, Liu Y, Fan J, Billiar TR. Pivotal advance: The pattern recognition receptor ligands lipopolysaccharide and polyinosine-polycytidylic acid stimulate factor B synthesis by the macrophage through distinct but overlapping mechanisms. J Leukoc Biol. 2010;88:609–618. doi: 10.1189/jlb.0809588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. The role of immunostimulatory nucleic acids in septic shock. Int J Clin Exp Med. 2012;5:1–23. [PMC free article] [PubMed] [Google Scholar]
- 14.Wu SC, Yang JC, Rau CS, Chen YC, Lu TH, Lin MW, Tzeng SL, Wu YC, Wu CJ, Hsieh CH. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS One. 2013;8:e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Feng Y, Zou L, Wang L, Chen HH, Cai JY, Xu JM, Sosnovik DE, Chao W. Role of Extracellular RNA and TLR3-Trif Signaling in Myocardial Ischemia-Reperfusion Injury. J Am Heart Assoc. 2014;3:e000683. doi: 10.1161/JAHA.113.000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabrera-Fuentes HA, Ruiz-Meana M, Simsekyilmaz S, Kostin S, Inserte J, Saffarzadeh M, Galuska SP, Vijayan V, Barba I, Barreto G, Fischer S, Lochnit G, Ilinskaya ON, Baumgart-Vogt E, Boning A, Lecour S, Hausenloy DJ, Liehn EA, Garcia-Dorado D, Schluter KD, Preissner KT. RNase1 prevents the damaging interplay between extracellular RNA and tumour necrosis factor-alpha in cardiac ischaemia/reperfusion injury. Thromb Haemost. 2014;112:1110–1119. doi: 10.1160/TH14-08-0703. [DOI] [PubMed] [Google Scholar]
- 17.Fischer S, Grantzow T, Pagel JI, Tschernatsch M, Sperandio M, Preissner KT, Deindl E. Extracellular RNA promotes leukocyte recruitment in the vascular system by mobilising proinflammatory cytokines. Thromb Haemost. 2012;108:730–741. doi: 10.1160/TH12-03-0186. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Chen H, Cai J, Zou L, Yan D, Xu G, Li D, Chao W. Cardiac RNA induces inflammatory responses in cardiomyocytes and immune cells via Toll-like receptor 7 signaling. J Biol Chem. 2015;290:26688–26698. doi: 10.1074/jbc.M115.661835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rooij E. Introduction to the series on microRNAs in the cardiovascular system. Circ Res. 2012;110:481–482. doi: 10.1161/CIRCRESAHA.111.257311. [DOI] [PubMed] [Google Scholar]
- 21.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 22.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Zou L, Feng Y, Chen YJ, Si R, Shen S, Zhou Q, Ichinose F, Scherrer-Crosbie M, Chao W. Toll-like receptor 2 plays a critical role in cardiac dysfunction during polymicrobial sepsis. Crit Care Med. 2010;38:1335–1342. doi: 10.1097/CCM.0b013e3181d99e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou L, Feng Y, Zhang M, Li Y, Chao W. Nonhematopoietic toll-like receptor 2 contributes to neutrophil and cardiac function impairment during polymicrobial sepsis. Shock. 2011;36:370–380. doi: 10.1097/SHK.0b013e3182279868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker WE, Bozzi AT, Goldstein DR. IRF3 contributes to sepsis pathogenesis in the mouse cecal ligation and puncture model. J Leukoc Biol. 2012;92:1261–1268. doi: 10.1189/jlb.0312138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergin PF, Doppelt JD, Hamilton WG, Mirick GE, Jones AE, Sritulanondha S, Helm JM, Tuan RS. Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction. J Bone Joint Surg Am. 2010;92:654–663. doi: 10.2106/JBJS.I.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pangburn MK, Muller-Eberhard HJ. The alternative pathway of complement. Springer Semin Immunopathol. 1984;7:163–192. doi: 10.1007/BF01893019. [DOI] [PubMed] [Google Scholar]
- 29.Hotchkiss RS, Tinsley KW, Swanson PE, Chang KC, Cobb JP, Buchman TG, Korsmeyer SJ, Karl IE. Prevention of lymphocyte cell death in sepsis improves survival in mice. Proc Natl Acad Sci U S A. 1999;96:14541–14546. doi: 10.1073/pnas.96.25.14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotchkiss RS, Tinsley KW, Karl IE. Role of apoptotic cell death in sepsis. Scand J Infect Dis. 2003;35:585–592. doi: 10.1080/00365540310015692. [DOI] [PubMed] [Google Scholar]
- 31.Angus DC, van der Poll T. Severe sepsis and septic shock. New Eng J Med. 2013;369:2063. doi: 10.1056/NEJMc1312359. [DOI] [PubMed] [Google Scholar]
- 32.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 33.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crozat K, Beutler B. TLR7: A new sensor of viral infection. Proc Natl Acad Sci U S A. 2004;101:6835–6836. doi: 10.1073/pnas.0401347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.{Akira ON, LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 36.Kariko K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 37.Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, Martinez L, Greidinger EL, Yu BD, Gallo RL. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med. 2012;18:1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 40.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 42.Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. MicroRNA-146a and RBM4 form a negative feed-forward loop that disrupts cytokine mRNA translation following TLR4 responses in human THP-1 monocytes. Immunol Cell Biol. 2013;91:532–540. doi: 10.1038/icb.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Gazzar M, Church A, Liu T, McCall CE. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-alpha during TLR4-induced gene reprogramming. J Leukoc Biol. 2011;90:509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI, Fish JE. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med. 2013;5:949–966. doi: 10.1002/emmm.201202318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao M, Wang X, Zhang X, Ha T, Ma H, Liu L, Kalbfleisch JH, Gao X, Kao RL, Williams DL, Li C. Attenuation of Cardiac Dysfunction in Polymicrobial Sepsis by MicroRNA-146a Is Mediated via Targeting of IRAK1 and TRAF6 Expression. J Immunol. 2015;195:672–682. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park CK, Xu ZZ, Berta T, Han Q, Chen G, Liu XJ, Ji RR. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron. 2014;82:47–54. doi: 10.1016/j.neuron.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kalin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL, Golenbock D, Schott E, Ploegh HL, Wulczyn FG, Lehnardt S. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 50.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. MAP kinases. Chem Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 52.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]