Abstract
A new instrument configuration for native ion mobility-mass spectrometry (IM-MS) is described. Macromolecule ions are generated by using a static ESI source coupled to an RF ion funnel, and these ions are then mobility and mass analyzed using a periodic focusing drift tube IM analyzer and an Orbitrap mass spectrometer. The instrument design retains the capabilities for first-principles determination of rotationally averaged ion-neutral collision cross sections and high-resolution measurements in both mobility and mass analysis modes for intact protein complexes. Operation in the IM mode utilizes FT-IMS modes (originally described by Knorr (Knorr, F. J. et al. Anal. Chem. 1985, 57(2), 402–406)), which provides a means to overcome the inherent duty cycle mismatch for drift tube (DT)-IM and Orbitrap mass analysis. The performance of the native ESI-FT-DT-IM-Orbitrap MS instrument was evaluated using the protein complexes Gln K (MW 44 kDa) and streptavidin (MW 53 kDa) bound to small molecules (ADP and biotin, respectively) and transthyretin (MW 56 kDa) bound to thyroxine and zinc.
Graphical Abstract:

Mapping the structural heterogeneity (folded and misfolded states) of proteins and protein complexes1–3 and how post-translational modifications (PTMs)4 and interactions with ligands, i.e., metal ions,5,6 small molecules,7–9 and osmolytes (chemical chaperones),10 influence protein stability as well as the structure–function relationships represents a major challenge to the field of structural biology. Such challenges, previously described as “characterizing the conformationome,”1 are increasingly studied using native electrospray ionization (nESI)-ion mobility (IM)-mass spectrometry (MS). The term “nESI” denotes that the analyte is sprayed from a nondenaturing solvent and conditions (i.e., solvent composition, pH, temperature, etc.) that yield low charge state ions that retain solution-phase conformational preferences and noncovalent interactions.11–13 IM-MS, which provides information on the size of the gas-phase ion, has rapidly gained popularity for structural (2, 3, and 4°) characterization of gas-phase ions.14–17 Combining IM with native MS, which independently measures both size and m/z of the ions, imposes additional constraints; ions formed by “nESI” must not be perturbed during the transition from solution to the gas phase as well as during subsequent analysis of the gas-phase ions.18,19 Although nESI-IM-MS does not provide the same level of structural detail as does spectroscopic techniques such as circular dichroism (CD), Förster resonance energy transfer (FRET), X-ray crystallography, and NMR, which measure the signals averaged in a populational manner, IM-MS is the only biophysical structural characterization technique capable of determining how protein structure(s) responds to specific changes in the local environment at the populational level.20–22 Moreover, temperature-dependent MS binding studies are the only method to elucidate thermochemistry of individual binding events, particularly for systems that bind multiple ligands.23,24 However, the resolution of current IM-MS instrumentation is often inadequate for a number of biophysical studies of protein–ligand interactions because of poor resolving power. New instrumentation is needed to address how ligand binding may alter conformational preferences of the target protein and whether these binding events alter binding of additional ligands. Addressing these types of questions is essential for a better understanding of allostery and cooperativity, fundamental properties of macromolecules. Great strides have been made in the development of IM-MS instrumentation for structural characterization of biomolecules, but the instruments were largely developed for proteomics research. Moreover, these instruments are not optimized for studies of large proteins and their complexes. Realizing the full potential of nESI-IM-MS for studies of large proteins and their complexes necessitates major advances in instrumentation, most notably enhanced mobility (RIM) and mass (Rm/z) resolution, while retaining capabilities for preserving noncovalent interactions and accurate determinations of ion-neutral collision cross sections (CCS).
Coupling DT-IM to high-performance MS, such as in hybrid IM-q-ToF instruments, is relatively straightforward, because IM separation is slow relative to the acquisition of the ToF mass spectrum. However, when using ion trapping MS (e.g., Orbitraps), an inherent duty cycle mismatch exists as both the DT IM separation and the mass scans occur on approximately the same time scale. Consequently, the MS cannot efficiently capture mobility information across the full arrival time distribution (ATD); such limitations have severely hampered the development of IM-Orbitrap instruments.25,26
There exists little doubt that biomolecule IM-MS was greatly accelerated by the introduction of traveling-wave (TW) IM-q-ToF instruments,27 and remarkable progress has been realized, in spite of the limitations of TWIMS-MS, viz. low-resolution ion mobility (td/Δtd) measurements, the requirement for calibration methods for determining rotationally averaged ion-neutral collision cross sections (CCS), and limited mass resolving power (Rp = M/ΔM, where ΔM = (M2 – M1), the difference in mass of two ions having different masses) of the ToF instrument. Here, we describe a novel nESI-FT-DT-IM-Orbitrap MS (Figure 1) specifically designed for structural studies of large proteins, protein complexes, and their interactions with small molecules and other proteins. Although other IM-Orbitraps have been developed,25,26 these instruments were not designed for preserving noncovalent interactions and are unable to be used for native MS because of their configuration and design. Conversely, this new instrument, which we denote as “next-generation”, incorporates all the essential components developed over the past decade that define “native MS”. Specifically, (i) ion formation conditions are optimized for retention of solution-phase structure(s), viz. static-spray ESI emitters ranging in size from micrometer to submicrometer that are compatible with the use of “native-like” solvents for forming low charge states that have low internal energy;11,12,28 (ii) a periodic focusing (PF) drift tube (DT) IM that operates under low electric field strengths, which minimizes collisional heating of the ions, allows for first-principles determinations of the ion’s rotationally averaged CCS, and provides ion radial focusing for increased ion transmission;29,30 (iii) IM data acquisition is performed using a Fourier transform (FT) IM-MS method first described by Hill and more recently by Clowers;31,32 and (iv) mobility separated ions are then mass analyzed using the high-Rp Orbitrap MS.33–35
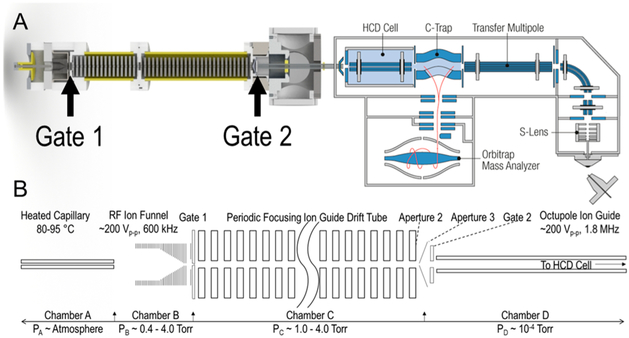
Figure 1.
(A) SolidWorks rendering of the home-built nESI-FT-DT-IM coupled to the HCD cell of an Orbitrap MS. (B) A detailed schematic representation of the home-built platform. From left to right depicts a heated capillary for ion introduction via nESI (not pictured) and an RF ion funnel to focus ions into the gating region. Ions are injected via gate 1 into a 58 cm PF DT and selectively transmitted through gate 2 into an RF-only octupole ion guide. Mobility modulated ions are then loaded and trapped in the HCD cell of the Orbitrap, where they are subsequently transferred to the C-Trap and then injected into the Orbitrap for mass analysis.
We have previously shown the importance of improved Rp in native MS using an Orbitrap over IM-ToF MS to characterize heterogeneous lipid binding events to the trimeric ammonia transport channel (AmtB), an integral membrane protein.35 More specifically, IM-ToF MS simply does not possess sufficient Rp to separate the individual lipids bound, whereas the Orbitrap MS successfully separated 46 different combinations of lipids bound to AmtB. Notably, Rm/z of native mass spectra are markedly lower than those of small molecules (viz. metabolites, carbohydrates, lipids, and peptides) because of the size and heterogeneity of large proteins and their complexes; therefore, direct Rm/z comparisons between native MS and small molecule MS should not be made.
The major impetus for development of new IM-MS technologies focused on structural biology is the need to study protein complexes and their interactions with small molecules (e.g., drugs), metal ions, peptides/proteins, and nucleic acids. As a first step, the instrument performance was characterized using a number of well-studied model monomeric soluble proteins, i.e., cytochrome C, ubiquitin, and lysozyme. We then demonstrate the novel capabilities of an nESI-FT-DT-IM-Orbitrap MS instrument by investigating protein complexes (streptavidin, Gln K, and transthyretin [TTR]) and their interactions with small molecule(s) (streptavidin·biotin, Gln K·ADP, TTR·Zn(II), and TTR·thyroxine [T4]).
EXPERIMENTAL SECTION
Instrumentation.
For these experiments, the nano-ESI source previously described was mounted onto a PF DT.35 The nano-ESI uses pulled borosilicate glass capillaries, prepared in-house, that are either gold coated or contain a platinum wire (300 μm) inserted into the capillary. ESI potentials of 1.50–2.00 kV were used for all studies. Ions formed by nano-ESI enter a heated metal capillary, where the final stages of ion dehydration occur, and then focused using a radiofrequency (RF) ion funnel (200–250 Vp-p, 600 kHz, Ardara Technologies, Ardara, PA) maintained at a gas pressure between 2.0 and 2.5 Torr. Ions exiting the RF ion funnel are focused through gate 1 to introduce a packet of ions into the DT. A 58 cm PF DT, maintained at a constant helium flow, is used for IM separation. PF DTs rely on ion optic geometries (8 mm I.D., 6.35 mm width, 6.35 mm spacing) to produce a distant-dependent effective potential mimicking RF focusing as ions traverse each electrode of the DT.36–38 A voltage gradient of 10 V/cm at ~2.0 Torr was used for all experiments, which corresponds to being expressed as 5 V/(cm·Torr) or Townsends (Td). Mobility separated ions exit the DT by modulating gate 2 and are subsequently introduced to an RF-only octupole ion guide (200–250 Vp-p, 2.5 MHz), which is used to focus the ions into the HCD cell of an Exactive Plus with extended mass range Orbitrap MS (Thermo Fisher Scientific, San Jose, CA). This interface region and Orbitrap operating parameters have been described in detail previously.35 Briefly, the Orbitrap mass spectrometer was tuned using typical operating parameters: collision energy (CE) in the HCD cell was set to 10 V to minimize postmobility fragmentation, the maximum injection time was set to 200 ms, the trapping gas pressure was set between 4 and 7 au, and an Orbitrap resolution of 17 500–35 000 was selected, as it yielded higher quality mass spectra. IMS and Orbitrap electronics were synchronized externally using an Arduino Leonardo to trigger FT-IMS pulsing and contact closure, respectively.
Operation of a Dual-Gate Fourier Transform IMS.
Many conventional IM-MS instruments use single-gate instrument configurations to introduce discrete ion packets into a drift cell followed by detection using comparatively fast mass analyzers to acquire nested IM-MS spectra. This so-called “pulse-and-wait” sampling mode has a duty cycle of less than 1%, i.e., greater than 99% of the total ion population is not sampled. Using a dual-gate platform, ion entry into the DT is controlled by gate 1. Gate 2 is positioned at the rear of the DT to select a specific ion arrival time that is then transmitted for mass analysis.39–42 The time delay between pulsing gates 1 and 2 defines the drift time of the detected ions; the experiment is repeated with different time delays to acquire an entire ATD. An added benefit to the dual-gate platform is the potential to eliminate the need for multifield calibrations to determine the time ions spend outside of the DT by placing gates directly before and after the DT. While this approach is effective, only 0.01% of ions are analyzed, reducing instrument sensitivity and significantly slowing data acquisition, and results are entirely dependent upon highly stable ionization sources.
To overcome the low duty cycle of a basic dual-gate pulsing platform, a variety of multiplexed acquisition modes have been developed for IM-MS,43–45 and very recently, Clowers and coworkers have reintroduced FT-IMS originally introduced by Hill.31,32 Here, we implemented FT-IMS, which was first described to improve the duty cycle of time dispersive IMS platforms by modulating the dual gates of the DT. This provides 25% ion transmission with improved spectral quality and dramatically improved acquisition times. Operating in FT-IM, the DT is used as a frequency-dependent filter (or 1/td filter) by synchronously modulating gates 1 and 2 with square waves that are linearly swept from low (5 Hz) to high (7 kHz) frequencies over multiple minutes. The frequency encoding of ion mobility information is possible, because ions are transmitted only when their drift time (td) is correlated with the frequency (v = 1/td) of the gating; therefore, by sampling across a range of frequencies, a signal (S) will be obtained with a frequency dependence of
Where I0 is the ion intensity without any pulsing. The resultant oscillating signal, for an isolated ion, can be Fourier transformed to determine the frequency of transmission. This frequency is directly related to ATD (td = v−1) and can therefore be correlated by dividing the frequency axis by the sweep rate, resulting in the ATD of an ion. The FT-IMS workflow is summarized in Figure 2.
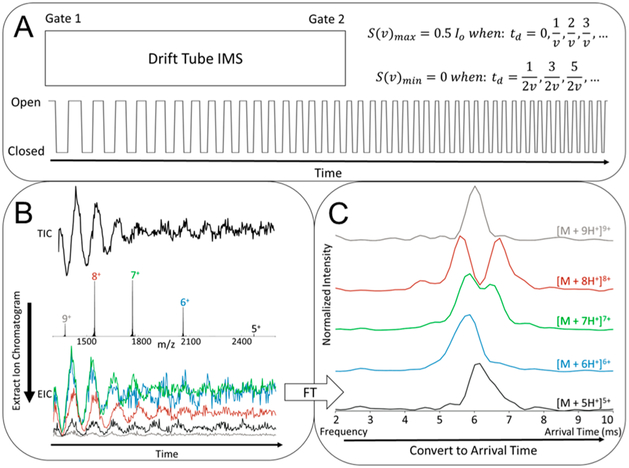
Figure 2.
(A) Dual gates at the entrance and exit of the DT are synchronously pulsed with a square waveform with a linear chirp frequency from 5 to 7000 Hz. Applying this waveform turns the DT into a frequency-dependent filter with transmission characteristics aligning with the stated equations. (B) Mass spectral data is acquired with an associated total ion chromatogram (TIC), from which an extracted ion chromatogram (EIC) can be determined by isolating a single m/z abundance over time. A TIC, MS, and EIC are shown for the model protein cytochrome C. (C) EICs can be Fourier transformed to determine the frequency of transmission, which is correlated to arrival time by a direct relationship. Exemplary ATDs were extracted from the cytochrome C data in panel B.
For this study, FT-IMS was implemented using custom linear sweep waveforms (5 to 7000 Hz over 8 min) generated via a Python script and uploaded to a National Instruments PXI-5421 waveform generator used to trigger gating events. DEI PVX-4140 pulse generators were used to apply square waves to gates 1 and 2.
Benchmarking the Instrument.
To benchmark the new instrument, we first analyzed cytochrome C (Figure 2B,C), ubiquitin (Figure S2), and lysozyme (Figure S3). Figure 2B,C shows the MS and extracted ATDs of the five observed charge states of cytochrome C analyzed in water with 1% acetic acid and align well with ATDs reported by a number of studies.46–49 The lower charge states of cytochrome C, [M + 5H+]5+, and [M + 6H+]6+, exhibit compact, native-like conformers. A charge-dependent unfolding is then observed where [M + 7H+]7+ populates the partially unfolded intermediate conformer, [M + 8H+]8+ populates both the intermediate and fully unfolded extended conformer, and [M + 9H+]9+ populates only the extended conformer. Moreover, ATDs for both ubiquitin and lysozyme are also in agreement with previous studies (Figures S2 and S3).18,46,49,50
Data Processing.
Mass spectral data were acquired using the Exactive software to generate RAW format data. RAW data was converted using a Python script making use of Multiplierz.51 Extracted ion chromatograms were obtained from RAW MS data using custom Python scripts written in-house, and the extracted data were processed and subjected to Fourier transformation using custom Python scripts.
Chemicals and Materials.
Gln K and TTR were expressed and purified in-house as described previously.23,52 Streptavidin, cytochrome C, bovine ubiquitin, and lysozyme were purchased from Sigma-Aldrich and used without further purification. All complexes were buffer exchanged using a centrifugal buffer exchange device (Micro Bio-Spin 6, Bio-Rad) into 200 mM ammonium acetate before analysis. ADP (ammonium salt), biotin, and T4 were purchased from Sigma-Aldrich. Biotin and T4 were first diluted in DMSO before diluting to 15 μM in 200 mM ammonium acetate. ADP was dissolved and diluted to 15 μM in 200 mM ammonium acetate.
RESULTS AND DISCUSSION
A major challenge of uniform field IM for the analysis of protein complexes is poor transmission of these ions due to radial diffusion as they traverse the DT. To overcome this issue, the IM-Orbitrap MS takes advantage of a PF DT, which exhibits high ion transmission by minimizing radial diffusion. The PF DT device also removes the necessity for an ion funnel at the end of the DT as employed in other IM devices.29,36 PF-DT IM improves radial focusing through the unique geometry of electrodes utilizing thicker electrodes and a smaller I.D. to generate effective RF (~kHz frequency) potentials between each electrode to periodically focus ions radially, thereby improving ion transmission.37,38 Although previous descriptions of PF-IM were limited to studies of peptides and small proteins, more recent SIMION 8.1 trajectory simulations clearly show that the increased numbers of charges and masses of larger proteins provide even higher radial focusing.53
Protein Complexes: Streptavidin, Gln K, and Transthyretin.
Improvements over traditional IM-ToF instruments, where Rp is most often a limitation, are illustrated by analyzing the 3° and 4° structures of protein complexes and their interactions with ligands. Here, we explored streptavidin·biotin, Gln K·ADP, TTR·T4, and TTR·Zn(II) complexes. Monitoring such ligand binding events are difficult or nearly impossible to resolve using ToF mass analyzers.7,35,54,55 These observations lend insight into the effects of small molecule and ligand binding on the protein structure–function relationships at unprecedented detail.
Streptavidin, a homotetrameric 53 kDa protein complex, was analyzed using the nESI-FT-DT-IM-Orbitrap MS. Each streptavidin monomer can individually bind one biotin molecule, wherein each of the four binding sites of the tetramer are thermodynamically equivalent.56 The streptavidin–biotin interaction, one of the most stable in nature (Ka of ~2.5 × 1013 M−1),57 has been well-characterized using a variety of techniques.58 The streptavidin–biotin interaction has been observed in previous IM-MS studies; however, these studies utilized non-natural fluorescein tagged derivatives (biotin-4-fluorescein) to increase mass separation between the unbound and ligated streptavidin complexes to overcome limited Rp.7 Here, we analyzed the streptavidin–biotin complex using natural biotin. Mass spectra of the apo-streptavidin and holo-streptavidin complexes are shown in Figure 3A, where a shift in m/z denotes the addition of four biotin molecules to the complex. The additional peaks (*) observed represent streptavidin with additional methionine residues on the N-terminus, a result of the protein expression process. While these modified proteins were observed mass spectrally, extracted ATDs did not include these modifications. Figure 3B shows the extracted ATDs of apo-streptavidin and holo-streptavidin. Apo-streptavidin exhibits a slightly lower drift time compared with holo-streptavidin, an expected shift with the addition of four small molecules. The ATD for holo-streptavidin is narrower than that of the apo-streptavidin. This compaction is associated with an increased structural stability and homogeneity, an observation aligned with previous findings.58,59 To our knowledge, this represents the first example of IM-MS resolving the streptavidin and natural biotin interaction.
Figure 3.
(A) Mass spectrum and (B) extracted ATD of [M + 14H+]14+ apo-streptavidin and the holo-streptavidin complexes. A small shift in drift time was observed upon binding four biotin molecules as well as a compaction of peak width. *Denotes additional N-terminal methionine residues on streptavidin and were not included in the extracted ATDs.
The PII transduction protein Gln K (MW 44 kDa) is a homotrimeric protein complex that negatively regulates the ammonium ion transport of AmtB by plugging the channel of AmtB,60 and this process plays a key role in nitrogen regulation of cells.61 Crystallography data suggest that two conformations of Gln K exist where the T-loop of a monomer can form an α-helix (CCSTJM = 2746 Å2) or extended β-hairpin (CCSTJM = 3427 Å2);62 the two conformers are potentially important in regulating its interactions with AmtB.63 Additionally, each monomer of Gln K is able to bind a single ADP; however, the effects of individual ADP binding on protein structure and function is not fully understood.64 Gln K was analyzed via the nESI-FT-DT-IM-Orbitrap MS to explore its structure in complex with ADP as seen in Figure 4. Mass spectral data for Gln K after expression and purification showed the presence of the apo-Gln K complex and up to two ADPs bound (Figure 4A). A 3-fold addition of ADP shifts the equilibrium to the holo-Gln K complex (three ADP molecules bound), and additional, nonspecific ADP binding is observed with four and five ADP bound (Figure 4B).
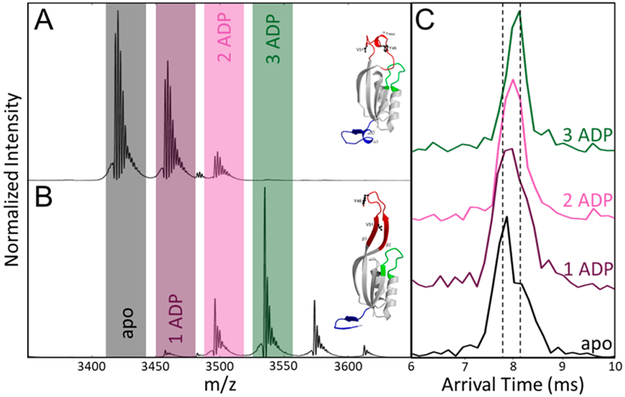
Figure 4.
Mass spectra of (A) Gln K and (B) Gln K with a 3-fold addition of ADP. The observed adducts on the Gln K were sodium adducts and were not included in the ATD extractions. (C) The extracted ATDs of the [M + 11H+]11+ Gln K·ADP complexes. PDB ID: (top) 1HWU and (bottom) 1QY7.63
Extracted ATD of apo-Gln K (Figure 4C) confirms the presence of two distinct protein conformers with the appearance of two peaks potentially representing the T-loop adopting an α-helix or β-hairpin. The more compact, α-helical T-loop conformer “closed state” appears as the dominant peak, and the right shoulder shows the presence of the larger β-hairpin T-loop “open state”. The stepwise addition of ADP to Gln K promotes the open state, where Gln K·ADP1 shows a slightly more abundant open state, Gln K·ADP2 populates both the open and closed states with a slightly higher abundance of open state, and holo-Gln K shows the open state with little closed state present. Sakai et al. previously suggested that holo-Gln K exhibits the disordered T-loop; however, it remains unclear if one or two ADP molecules bound to Gln K is sufficient to promote a regulatory interaction with AmtB.63
TTR is a 56 kDa, homotetrameric protein complex involved in degenerative diseases such as amyloidosis, where partially unfolded monomers from tetramer dissociation can result in amyloid fibril formation.65 TTR participates in the transport of the natural hormone T4, and the association of TTR and T4 is effective in inhibiting amyloid fibril formation.66 Here, TTR was studied in the presence of its transport partner T4 to explore the effect of binding on protein structure. Figure 5A shows the mass spectrum obtained of TTR in the presence of T4 and up to two T4 molecules bound to TTR. In addition to T4 binding, each protein–ligand complex surprisingly exhibits additional binding to Zn(II) as discussed in greater detail below. ATDs in Figure 5B exhibit slightly longer drift times of TTR with each successive T4 bound. The peak width of the holo-TTR·T4 structure was reduced with respect to apo-TTR, indicative of greater homogeneity in protein structure as well as greater protein stability.
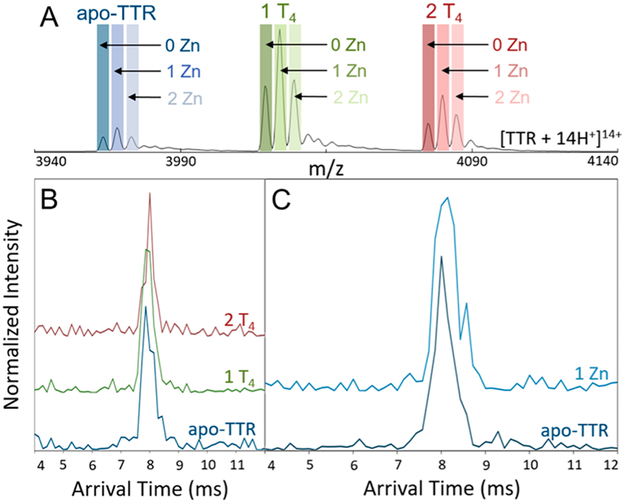
Figure 5.
(A) Representative mass spectrum of the [M + 14H+]14+ TTR complex with binding of up to two T4 and Zn(II). A mass resolving power of 840 is required to separate the apo-Zn containing ions with the Orbitrap. (B) The extracted ATDs of TTR binding to one and two T4’s. (C) The extracted ATDs of TTR bound to Zn(II).
In addition to its transport activity, TTR is reported to be a metallopeptidase when complexed with Zn(II).67 While Zn(II) binding is important for proteolytic activity, Palmieri et al. reported Zn(II) binding noticeably increased the rate of TTR aggregation.68 The high-Rp mass analysis fully resolves TTR·Zn(II) complexes, and the overall sensitivity of the instrument is sufficient for IM-MS analysis as shown in Figure 5C. The addition of Zn(II) extends the drift time of TTR, which is consistent with the formation of an extended conformation with partial unfolding of the complex. Owing to the limited Rp of IM-ToF, previous IM-MS studies for the TTR·T4 complex may have contained signals relating to TTR·Zn(II) complexes masked by poor resolving power. It should be noted that the abundances of Zn(II) and T4 are independent of one another, indicating that there is no cooperative or competitive binding between the two.
Observations of protein–ligand complexes such as the streptavidin·biotin, Gln K·ADP, TTR·Zn(II), and TTR·T4 highlight the importance of high-resolution IM-MS measurements for biophysical studies. Lower Rp often masks the intricate details that underlie protein–ligand interactions, as it simply cannot adequately resolve such species. Moreover, protein heterogeneity can lead to inaccurate structural measurements of protein complexes, rendering protein–ligand species unresolvable. The presented data show the possibility that these ligand-bound species have distinct structures and play a role in the functionality of the protein.
CONCLUSION
For the first time, IMS was coupled to the HCD cell of an Orbitrap MS for high-resolving power IM-MS measurements, and these results clearly illustrate increased performance of ion mobility-mass spectrometry necessary for the next generation of biophysical studies of intact protein complexes. In its current configuration, a RIM of ~40 was achieved for intact protein complexes and, to the best of our knowledge, represents the highest resolution IM measurements made on such systems. The union of PF-DT and FT-IMS provides not only improved ion transmission that aids data acquisition but also allows for higher throughput sample processing over that obtained using duty cycle mismatched instruments. For example, using the nESI-FT-DT-IM-Orbitrap MS instrument, the full IM and MS spectrum can be acquired in 8 min or less. The range of protein complexes analyzed illustrates the importance of understanding the role that metal ions and small molecules play in protein conformational preferences, which would otherwise not be observed. The results reported herein make more detailed studies on other protein complexes possible, including exploring the effects of PTMs, protein–ligand interactions, and protein misfolding using high-resolution IM-MS.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank Will Seward of the TAMU Chemistry Department Machine Shop for the fabrication of all custom instrumentation, Greg Matthijetz of the TAMU Laboratory for Biological Mass Spectrometry for his electronics expertise, and Alexander Makarov of Thermo Fisher Scientific for numerous helpful discussions relating to interfacing ion mobility to the Orbitrap. Funding for this work was provided by the National Science Foundation (CHE-1707675 (DHR) and CHE-1506672 (BHC)), National Institutes of Health (DP2GM123486 (AL), R01GM121751-01A1 (DHR and AL), P41GM128577-01 (DHR)), and endowment funds from the MDS Sciex Professorship (DHR).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b02463.
Additional mass spectra, arrival time distributions, and mass resolutions for proteins and protein complexes (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Clemmer DE; Russell DH; Williams ER Acc. Chem. Res 2017, 50 (3), 556–560. [DOI] [PubMed] [Google Scholar]
- (2).Sharon M J. Am. Soc. Mass Spectrom 2010, 21 (4), 487–500. [DOI] [PubMed] [Google Scholar]
- (3).Sharon M; Robinson CV Annu. Rev. Biochem 2007, 76 (1), 167–193. [DOI] [PubMed] [Google Scholar]
- (4).Aebersold R; Agar JN; Amster IJ; Baker MS; Bertozzi CR; Boja ES; Costello CE; Cravatt BF; Fenselau C; Garcia B; Ge Y; Gunawardena J; Hendrickson RC; Hergenrother PJ; Huber CG; Ivanov AR; Jensen ON; Jewett MC; Kelleher NL; Kiessling LL; Krogan NJ; Larsen MR; Loo JA; Ogorzalek Loo RR; Lundberg E; MacCoss MJ; Mallick P; Mootha VK; Mrksich M; Muir TW; Patrie SM; Pesavento JJ; Pitteri SJ; Rodriguez H; Saghatelian A; Sandoval W; Schlüter H; Sechi S; Slavoff SA; Smith LM; Snyder MP; Thomas PM; Uhlén M; Van Eyk JE; Vidal M; Walt DR; White FM; Williams ER; Wohlschlager T; Wysocki VH; Yates NA; Young NL; Zhang A Nat. Chem. Biol 2018, 14, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chen L; Gao YQ; Russell DH J. Phys. Chem. A 2012, 116 (1), 689–696. [DOI] [PubMed] [Google Scholar]
- (6).Chen S-H; Russell DH Biochemistry 2015, 54 (39), 6021–6028. [DOI] [PubMed] [Google Scholar]
- (7).Allison TM; Reading E; Liko I; Baldwin AJ; Laganowsky A; Robinson CV Nat. Commun 2015, 6, 8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Laganowsky A; Reading E; Allison TM; Ulmschneider MA Degiacomi MT; Baldwin AJ; Robinson CV Nature 2014, 510 (7503), 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kitova EN; El-Hawiet A; Schnier PD; Klassen JS J. Am. Soc. Mass Spectrom 2012, 23 (3), 431–441. [DOI] [PubMed] [Google Scholar]
- (10).O'Brien EP; Ziv G; Haran G; Brooks BR; Thirumalai D Proc. Natl. Acad. Sci. U. S. A 2008, 105 (36), 13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Heck AJR Nat. Methods 2008, 5, 927. [DOI] [PubMed] [Google Scholar]
- (12).Leney AC; Heck AJR J. Am. Soc. Mass Spectrom 2017, 28 (1), 5–13. [DOI] [PubMed] [Google Scholar]
- (13).Heuvel R. H. H. v. d. ; Heck AJR Curr. Opin. Chem. Biol 2004, 8 (5), 519–526. [DOI] [PubMed] [Google Scholar]
- (14).Lanucara F; Holman SW; Gray CJ; Eyers CE Nat. Chem 2014, 6 (4), 281–294. [DOI] [PubMed] [Google Scholar]
- (15).von Helden G; Wyttenbach T; Bowers MT Int. J. Mass Spectrom. Ion Processes 1995, 146–147, 349–364. [Google Scholar]
- (16).Wittmer D; Chen YH; Luckenbill BK; Hill HH Anal. Chem 1994, 66 (14), 2348–2355. [Google Scholar]
- (17).Jurneczko E; Barran PE Analyst 2011, 136 (1), 20–28. [DOI] [PubMed] [Google Scholar]
- (18).Chen S-H; Russell DH J. Am. Soc. Mass Spectrom 2015, 26 (9), 1433–1443. [DOI] [PubMed] [Google Scholar]
- (19).Breuker K; McLafferty FW Proc. Natl. Acad. Sci. U. S. A 2008, 105 (47), 18145–18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).El-baba TJ; Woodall DW; Raab SA; Fuller DR; Laganowsky A; Russell DH; Clemmer DE J. Am. Chem. Soc 2017, 139, 6306. [DOI] [PubMed] [Google Scholar]
- (21).Shi L; Holliday AE; Shi H; Zhu F; Ewing MA; Russell DH; Clemmer DE J. Am. Chem. Soc 2014, 136 (36), 12702–12711. [DOI] [PubMed] [Google Scholar]
- (22).Servage KA; Silveira JA; Fort KL; Clemmer DE; Russell DH J. Phys. Chem. Lett 2015, 6 (24), 4947–4951. [DOI] [PubMed] [Google Scholar]
- (23).Cong X; Liu Y; Liu W; Liang X; Russell DH; Laganowsky A J. Am. Chem. Soc 2016, 138 (13), 4346–4349. [DOI] [PubMed] [Google Scholar]
- (24).Patrick JW; Boone CD; Liu W; Conover GM; Liu Y; Cong X; Laganowsky A Proc. Natl. Acad. Sci. U. S. A 2018, 115, 2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ibrahim YM; Garimella SVB; Prost SA; Wojcik R; Norheim RV; Baker ES; Rusyn I; Smith RD Anal. Chem 2016, 88 (24), 12152–12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Keelor JD; Zambrzycki S; Li A; Clowers BH; Ferrández FM Anal. Chem 2017, 89 (21), 11301–11309. [DOI] [PubMed] [Google Scholar]
- (27).Giles K; Pringle SD; Worthington KR; Little D; Wildgoose JL; Bateman RH Rapid Commun. Mass Spectrom 2004, 18 (20), 2401–2414. [DOI] [PubMed] [Google Scholar]
- (28).Susa AC; Xia Z; Williams ER Anal. Chem 2017, 89 (5), 3116–3122. [DOI] [PubMed] [Google Scholar]
- (29).Gillig KJ; Ruotolo BT; Stone EG; Russell DH Int. J. Mass Spectrom 2004, 239 (1), 43–49. [Google Scholar]
- (30).Silveira JA; Jeon J; Gamage CM; Pai P-J; Fort KL; Russell DH Anal. Chem 2012, 84 (6), 2818–2824. [DOI] [PubMed] [Google Scholar]
- (31).Knorr FJ; Eatherton RL; Siems WF; Hill HH Anal. Chem 1985, 57 (2), 402–406. [DOI] [PubMed] [Google Scholar]
- (32).Morrison KA; Siems WF; Clowers BH Anal. Chem 2016. 88 (6), 3121–3129. [DOI] [PubMed] [Google Scholar]
- (33).Hu Q; Noll RJ; Li H; Makarov A; Hardman M; Graham Cooks R J. Mass Spectrom 2005, 40 (4), 430–443. [DOI] [PubMed] [Google Scholar]
- (34).Makarov AA Mass spectrometer. US5886346A, 1995. [Google Scholar]
- (35).Poltash ML; McCabe JW; Patrick JW; Laganowsky A; Russell DH Development and Evaluation of a Reverse-Entry Ion Source Orbitrap Mass Spectrometer. J. Am. Soc. Mass Spectrom 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Blase RC; Silveira JA; Gillig KJ; Gamage CM; Russell DH Int. J. Mass Spectrom 2011, 301 (1–3), 166–173. [Google Scholar]
- (37).Gamage CM; Silveira JA; Blase RC; Russell DH Int. J. Mass Spectrom 2011, 303 (2–3), 154–163. [Google Scholar]
- (38).Silveira JA; Gamage CM; Blase RC; Russell DH Int. J. Mass Spectrom 2010, 296 (1–3), 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Karasek FW; Kilpatrick WD; Cohen MJ Anal. Chem 1971, 43 (11), 1441–1447. [Google Scholar]
- (40).Karasek FW; Denney DW; Dedecker EH Anal. Chem 1974, 46 (8), 970–973. [DOI] [PubMed] [Google Scholar]
- (41).Karasek FW; Tatone OS Anal. Chem 1972, 44 (11), 1758–1763. [DOI] [PubMed] [Google Scholar]
- (42).Clowers BH; Hill HH Anal. Chem 2005, 77 (18), 5877–5885. [DOI] [PubMed] [Google Scholar]
- (43).Davis AL; Liu W; Siems WF; Clowers BH Analyst 2016. 142 (2), 292–301. [DOI] [PubMed] [Google Scholar]
- (44).Tummalacherla M; Garimella SVB; Prost SA; Ibrahim YM Analyst 2017, 142 (10), 1735–1745. [DOI] [PubMed] [Google Scholar]
- (45).Szumlas AW; Ray SJ; Hieftje GM Anal. Chem 2006, 78 (13), 4474–4481. [DOI] [PubMed] [Google Scholar]
- (46).Pringle SD; Giles K; Wildgoose JL; Williams JP; Slade SE; Thalassinos K; Bateman RH; Bowers MT; Scrivens JH Int. J. Mass Spectrom 2007, 261 (1), 1–12. [Google Scholar]
- (47).May JC; Jurneczko E; Stow SM; Kratochvil I; Kalkhof S; McLean JA Int. J. Mass Spectrom 2018, 427, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Clemmer DE; Hudgins RR; Jarrold MF J. Am. Chem. Soc 1995, 117 (40), 10141–10142. [Google Scholar]
- (49).Clemmer DE; Jarrold MF J. Mass Spectrom 1997, 32 (6), 577–592. [Google Scholar]
- (50).Wyttenbach T; Bowers MT J. Phys. Chem. B 2011, 115 (42), 12266–12275. [DOI] [PubMed] [Google Scholar]
- (51).Alexander WM; Ficarro SB; Adelmant G; Marto JA Proteomics 2017, 17 (15–16), 1700091. [DOI] [PubMed] [Google Scholar]
- (52).Lashuel HA; Wurth C; Woo L; Kelly JW Biochemistry 1999, 38 (41), 13560–13573. [DOI] [PubMed] [Google Scholar]
- (53).Manura D; Dahl D SIMION (R) 8.0 User Manual; Scientific Instrument Services, Inc: Ringoes, NJ, USA, https://simion.com/manual/, 2008. [Google Scholar]
- (54).Gault J; Donlan JAC; Liko I; Hopper JTS; Gupta K; Housden NG; Struwe WB; Marty MT; Mize T; Bechara C; Zhu Y; Wu B; Kleanthous C; Belov M; Damoc E; Makarov A; Robinson CV Nat. Methods 2016, 13 (4), 333–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Hyung S-J; Robinson CV; Ruotolo BT Chem. Biol 2009, 16 (4), 382–390. [DOI] [PubMed] [Google Scholar]
- (56).Jones ML; Kurzban GP Biochemistry 1995, 34 (37), 11750–11756. [DOI] [PubMed] [Google Scholar]
- (57).Chilkoti A; Stayton PS J. Am. Chem. Soc 1995, 117 (43), 10622–10628. [Google Scholar]
- (58).Gonzalez M; Bagatolli LA; Echabe I; Arrondo JL; Argarana CE; Cantor CR; Fidelio GD J. Biol. Chem 1997, 272 (17), 11288–94. [DOI] [PubMed] [Google Scholar]
- (59).Deng L; Kitova EN; Klassen JS J. Am. Soc. Mass Spectrom 2013, 24 (1), 49–56. [DOI] [PubMed] [Google Scholar]
- (60).Conroy MJ; Durand A; Lupo D; Li X-D; Bullough PA; Winkler FK; Merrick M Proc. Natl. Acad. Sci. U. S. A 2007, 104 (4), 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Arcondéguy T; Jack R; Merrick M Microbiol. Mol. Biol. Rev 2001, 65 (1), 80–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Marklund EG; Degiacomi MT; Robinson CV; Baldwin AJ; Benesch JLP Structure 2015, 23 (4), 791–799. [DOI] [PubMed] [Google Scholar]
- (63).Sakai H; Wang H; Takemoto-Hori C; Kaminishi T; Yamaguchi H; Kamewari Y; Terada T; Kuramitsu S; Shirouzu M; Yokoyama S J. Struct. Biol 2005, 149 (1), 99–110. [DOI] [PubMed] [Google Scholar]
- (64).Radchenko MV; Thornton J; Merrick M J. Biol. Chem 2010, 285 (40), 31037–31045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Hammarström P; Jiang X; Hurshman AR; Powers ET; Kelly JW Proc. Natl. Acad. Sci. U. S. A 2002, 99 (Suppl 4), 16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Hammarström P; Wiseman RL; Powers ET; Kelly JW Science 2003, 299 (5607), 713. [DOI] [PubMed] [Google Scholar]
- (67).Liz MA; Leite SC; Juliano L; Saraiva MJ; Damas AM; Bur D; Sousa MM Biochem. J 2012, 443 (3), 769. [DOI] [PubMed] [Google Scholar]
- (68).de C. Palmieri L ; Lima LMTR; Freire JBB; Bleicher L; Polikarpov I; Almeida FCL; Foguel D J. Biol. Chem 2010, 285 (41), 31731–31741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.