Abstract
Background
The DNA-dependent RNA polymerase from T7 bacteriophage (T7 RNAP) has been extensively characterized, and like other phage RNA polymerases it is highly specific for its promoter. A combined in vitro / in vivo selection method has been developed for the evolution of T7 RNA polymerases with altered promoter specificities. Large (103 – 106) polymerase libraries were made and cloned downstream of variant promoters. Those polymerase variants that can recognize variant promoters self-amplify both themselves and their attendent mRNAs in vivo. Following RT / PCR amplification in vitro, the most numerous polymerase genes are preferentially cloned and carried into subsequent rounds of selection.
Results and Conclusions
A T7 RNA polymerase library that was randomized at three positions was cloned adjacent to a T3-like promoter sequence, and a 'specialist' T7 RNA polymerase was identified. A library that was randomized at a different set of positions was cloned adjacent to a promoter library in which four positions had been randomized, and 'generalist' polymerases that could utilize a variety of T7 promoters were identified, including at least one polymerase with an apparently novel promoter specificity. This method may have applications for evolving other polymerase variants with novel phenotypes, such as the ability to incorporate modified nucleotides.
Introduction
The RNA polymerase from bacteriophage T7 has a relatively narrow specificity for a particular promoter sequence, making it an extremely useful tool for molecular biology and biotechnology applications. Most recently, the crystal structure of the polymerase in complex with a DNA promoter has revealed the structural basis for this specificity [1,2]. In addition, a number of researchers have examined the contribution of various nucleotides and functional groups in the promoter and various amino acids in the polymerase to specificity [3-5]. Based upon both structural and mutagenic analyses, it has proven possible to identify polymerase variants with altered specificities for promoters [6,7]. Polymerase variants with altered specificities have also been identified using genetic selections [8]. However, the variant polymerases and variant promoters that have so far been identified are close in sequence to the wild-type. For example, the only known polymerase variant that switches promoters contains a single amino acid change, recognizes a single nucleotide change in the promoter, and closely mimics interactions known to occur for T3 RNA polymerase [9]. The large sequence space that surrounds both polymerases and promoters has so far prevented more sweeping searches for more diverse variants. To address this problem, we have developed a combined in vitro / in vivo selection method based on T7 RNA polymerase autogene construct that allows large (103 – 106) libraries of T7 RNA polymerase variants to be efficiently searched for specificity mutants. The method is novel in that it allows selection of polymerases based on their in vivo enzymatic activity, is generalizable, and may have applications for other polymerase phenotypes, such as intracellular solubility or thermostability.
Results and Discussion
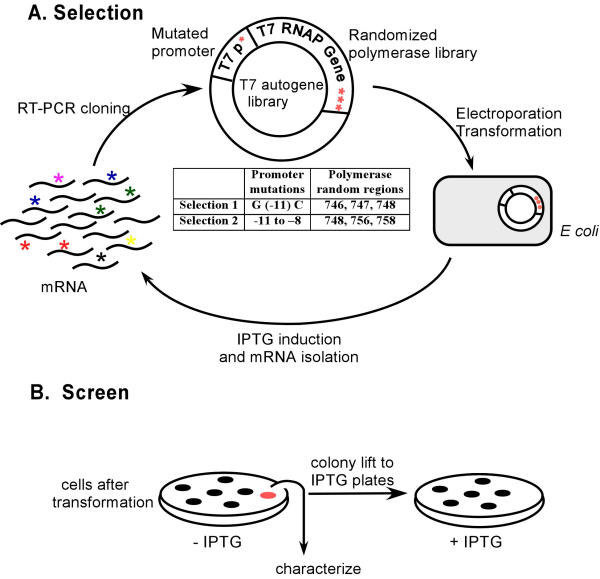
Our combined in vitro / in vivo selection scheme was designed to foster the self-amplification of novel polymerase variants (Figure 1). The T7 RNA polymerase gene was linked to a T7 RNA polymerase promoter, creating a so-called autogene [10], whose activity can be initiated by the basal level expression of polymerase in the cell. Upon transformation into E. coli, the autogene engendered the production of large amounts of T7 RNA polymerase (Figure 2). However, when the polymerase was cloned adjacent to mutant T7 RNA polymerase promoters, little T7 RNA polymerase expression was observed. We reasoned that any polymerase variant that could recognize the mutant promoter would re-establish the feedback loop and concomitantly lead not only to high protein expression levels, but also to high mRNA expression levels. In consequence, mRNA extracted from a population of cells transformed with a polymerase library should represent polymerase variants in rough proportion to their ability to utilize the mutant promoter. These mRNAs could be amplified en masse, re-cloned, and re-transformed into E. coli. Multiple cycles of selection and amplification should ultimately lead to the accumulation of those polymerase variants that were most successful at facilitating their own expression.
Figure 1.
Autogene selection. (A) Scheme. An autogene library containing the polymerase pool and promoter mutations is transformed into cells and induced. Active autogenes overproduce T7 RNA polymerase and mRNAs encoding the polymerase. Total mRNA is extracted, and the gene for T7 RNA polymerase is selectively reverse-transcribed and PCR-amplified. The gene fragments containing sequence variations (shown as *) are re-cloned and re-transformed. Multiple rounds of selection and amplification lead to the accumulation of polymerase variants with altered promoter specificities. (B) Screen for active variants. The autogene library is initially plated on LB agar plates without induction. Colonies are lifted via nitrocellulose filters to a new plate with IPTG and protein production is induced. Colonies that have active autogenes cease to grow due to high polymerase expression levels. These colonies can be identified on the original plate, and subsequently picked and characterized.
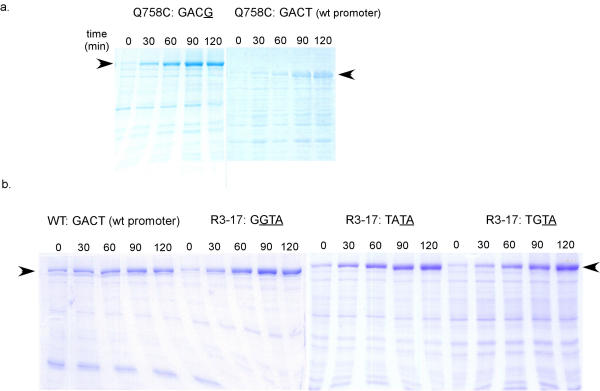
Figure 2.
Analysis of polymerase expression by SDS-PAGE. Cells containing autogene constructs were grown to an O.D600 of 0.4 and induced with IPTG. Aliquots of cells were lysed at time points following induction, and separated on SDS-PAGE. Bands were visualized using Coomassie blue. Arrows indicate the relative position of T7 RNA polymerase. The normalization of samples is apparent from the amount of E. coli proteins present in each lane. (a) Comparison of the activities of the Q758C polymerase on a mutant promoter (GACG at positions -11 to -8) and the wild-type promoter (GACT at positions -11 through -8). (b) Comparison of the activities of the R3-17 polymerase with three different promoters (GGTA, TATA, and TGTA at positions -11 through -8) and the activity of the wild-type autogene with its promoter (GACT at positions -11 through -8).
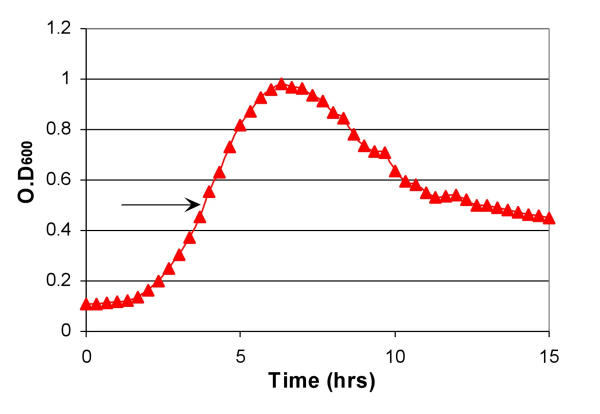
Of course, high level expression of any protein product can potentially be detrimental to cell growth, which would undermine the proposed selection. In order to evaluate the effect of toxicity of autogene on the selection process, the effect of the wild-type autogene construct (pET/T7p/T7) on cell growth was studied before and after induction with IPTG (Figure 3). These initial studies showed that the wild-type autogene did not affect cellular viability for at least 2–3 hours after induction with IPTG. Therefore, we reasoned that isolating the mRNA after an hour of induction should not drastically affect the selection process.
Figure 3.
Growth characteristics of cells containing wild-type autogene. Cells containing the wild-type autogene (pET/T7p/T7) were grown at 37°C and induced with IPTG at an OD600 of 0.5 (indicated by the arrow). The OD600 was monitored every 15 min for 15 hours using an automated Microbiology workstation Bioscreen C (Labsystems Oy, Finland).
We initially searched for polymerase variants that could utilize a promoter variant in which there was a G to C change at position -11. This mutation resembles the bacteriophage T3 promoter. A single asparagine to aspartate substitution at position 748 in T7 RNA polymerase was already known to facilitate the utilization of the T3-like promoter [6]. A library of polymerase variants was constructed in which amino acid residues 746, 747, and 748 were completely randomized. While we expected to obtain sequence changes primarily at position 748, randomization of residues 746 and 747 served as, respectively, positive and negative controls for the experiment. Residue 746 is normally an arginine that interacts with the guanosine [2,5] at position -7 and should remain unchanged following selection. Residue 747 is a leucine residue that does not make direct contacts to the promoter. We therefore expected that the leucine should vary during the course of the selection, or that one of six different leucine codons might be chosen.
The library of polymerase variants was cloned behind the T3-like promoter and three rounds of selection and amplification (as described in Figure 1) were carried out. In each round at least 5,000 individual variants were examined, a number that encompassed a large fraction of the possible variants (8,000). The progress of the selection was monitored in two ways: first, the autogene constructs were under the control of the lac repressor, and we had previously noted that complete depression of the wild-type autogene by IPTG led to cell death. Therefore, the fraction of colonies that were lost on replica plating to IPTG was assumed to be proportional to the accumulation of active autogene variants. Second, the number of PCR cycles that were required to amplify recovered mRNA molecules was assumed to follow the amount of mRNA that was produced in a given round of selection. Both of these variables showed substantial improvement following two rounds of selection, but less improvement following three rounds (for example, the proportion of IPTG-sensitive colonies was 20% after one round of selection, 88% after two rounds, and 96% after three rounds). The selection was therefore deemed to be essentially complete following round three.
Active polymerase variants were identified, cloned, and sequenced following each round of selection. As can be seen in Table 1, the selection not only quickly re-established the wild-type amino acids at positions 746 (arginine) and 747 (leucine), but also converged on the known N748D change, indicating that the combined in vitro / in vivo selection method was working as desired. It is likely that the N748D substitution that arose in the first generation was passed to subsequent generations, since there was no variation in the codons (TTA/G) associated with this gene during the course of the selection.
Table 1.
Summary of selection for "Specialist" and "Generalist" polymerases
| A. Selection of "Specialist" (T3 like polymerase) | |||||
| §Number of variants | Polymerase sequence* | Promoter sequence (-11 to -8) | |||
| 746 | 747 | 748 | |||
| Round – 1 | 4 | R (CGA) | L (TTG) | D (GAT) | CACT |
| 1 | R (CGT) | L (TTA) | Q (CAG) | CACT | |
| 1 | R (CGG) | L (TTA) | S (TCT) | CACT | |
| 1 | R (CGG) | C (TGT) | S (TCT) | CACT | |
| 1 | R (CGC) | P (CCC) | E(GAA) | CACT | |
| 1 | R (AGG) | T (ACA) | N(AAC) | CACT | |
| Round – 2 | 17 | R (CGA) | L (TTA) | D (GAT) | CACT |
| 3 | R (CGA) | L (TTG) | Q (CAG) | CACT | |
| Round – 3 | 16 | R (CGA) | L (TTA) | D (GAT) | CACT |
| * wild type sequence: | R (CGC) | L (TTG) | N (AAC) | GACT | |
| B. Selection of the "Generalist" polymerase | |||||
| §Number of variants | Polymerase sequence* | Promoter sequence (-11 to -8) | |||
| 748 | 756 | 758 | |||
| Round – 3 | |||||
| R3–9 | 3 | N(AAT) | R (CGT) | C (TGC) | GACT(wt) |
| R3–32 | 3 | N(AAT) | R (CGT) | C (TGC) | GACG |
| R3–26 | 1 | N(AAT) | R (CGT) | C (TGC) | TGTA |
| R3–17 | 1 | A (GCC) | M (ATG) | S (AGT) | GGTA |
| R3–5 | 1 | F (TTC) | G (GGC) | I (ATA) | G CTA |
| R3–29 | 1 | T (ACG) | K(AAG) | Q (CAA) | GACT |
| Round – 4 | |||||
| R4–14 | 7 | N(AAT) | R (CGT) | C (TGC) | GACT |
| R4–16 | 3 | N(AAT) | R (CGT) | C (TGC) | GTTA |
| R4–17 | 2 | N(AAT) | R (CGT) | C (TGC) | GTCA |
| ppC6 | 2 | N(AAT) | R (CGT) | C (TGC) | GATA |
| R4–11 | 3 | N(AAT) | R (CGA) | S (AGT) | GACT |
| * wild type sequence | N (AAC) | R (CGC) | Q (CAG) | GACT | |
§number of times the particular variant was isolated.
Interestingly, while the N748D substitution eventually dominates the selection, a N748Q substitution was also prevalent in Round 2. The mutant is known to be able to recognize the T3-like promoter [9] albeit less well than the N748D substitution. Thus, our selection scheme not only identifies functional polymerases, but the selected population seems to represent variants in proportion to their functionality.
To examine a wider range of promoter and polymerase combinations, a promoter library was constructed in which positions -8 through -11 in the promoter were completely randomized; each of these nucleotides had previously been shown to be extremely important for the specificity of interactions with T7 RNA polymerase [6,7,9,11]. Conversely, residues Asn 748, Arg 756, and Gln 758 were known to form sequence specific contacts with this region of the promoter [2]. The promoter library was combined with the gene library in order to create approximately 51,200 (44 × 203) combinations of promoters and polymerases. The joint promoter:polymerase library was transformed into E. coli and variants were again selected according to their transcript representation following RT / PCR amplification. To begin each new cycle, the polymerase variants were re-cloned behind the promoter library.
The success of an individual polymerase variant relied upon either being able to utilize several different promoters in different rounds (a 'generalist' strategy), or upon being very active with the same promoter in different rounds (a 'specialist' strategy). Given the number of promoter variants (256), the number of possible polymerase variants (8,000), and the number of transformants that were assessed in each round (105 – 106), it is likely that there was a population bottleneck during the first round of selection. However, selected polymerase variants should have had access to all possible promoters following the first round.
After three cycles of selection and amplification there was still some diversity in the population (Table 1), but after four cycles a single variant, Q758C, had largely established itself. The Q758C variant was frequently found adjacent to the wild-type T7 RNA polymerase promoter (GACT at positions -11 through -8) but was also found adjacent to a single mutant promoter (GACG), double mutant promoters (GATA, GTCA), a triple mutant promoter (GTTA), and a promoter in which all the residues differed from wild-type (TGTA). Overall, the 'generalist' polymerase appeared to prefer mutant promoters that contained a T at position -9, and G or A at -8 positions. The Q758C mutant has previously been assayed for its activity at position -8 with different bases, and was shown to prefer G or A at -8 compared to the wild type promoter (-8T) [7]. This finding was further borne out in our experiments; the Q758C:GACG polymerase:promoter combination yielded substantially more T7 RNA polymerase as judged by gel electrophoresis, than did Q758C:GACT (wt) (Figure 2a).
Another mutant polymerase, R3–17, selected from round three differed from the wild-type at all the three amino acid positions (N748A; R756M; Q758S). Despite the fact that this variant was extremely novel, it was nonetheless completely functional, and yielded as much or more polymerase expression when paired with its promoter GGTA than did the wild type polymerase paired with the wild type promoter (GACT) (Figure 2b). The ability to use this triple mutant promoter had not previously been observed. To further explore the apparent change in promoter specificity of this polymerase, we carried out an additional screen in which R3–17 was again cloned adjacent to the library of promoter mutations, and replica-plated colonies that grew poorly or not at all upon IPTG induction (indicating polymerase overproduction) were picked and sequenced. In addition to the original promoter (GGTA). two other promoters (TATA and TGTA) were identified. It is interesting that all of the promoters derived from the screen have a cytidine to thymidine substitution at position -9 and a thymidine to adenosine substitution at position -8. This combination of residues has not previously been shown to yield an active T7 RNA polymerase promoter. Consonant with our genetic screen, the R3–17 polymerase paired with each of the three promoters appeared to yield as much or more T7 RNA polymerase as did wild-type autogene paired with the wild-type promoter (Figure 2b). Previous studies of T7 promoter recognition in vitro have indicated that the wild-type T7 RNA polymerase does not tolerate mutations well, particularly at positions -8 to -11 in the T7 promoter [9,11]. In agreement with these studies, we found no mutant promoters that could apparently pair with the wild-type polymerase (see Table 1).
It is becoming increasingly clear that some protein phenotypes can only be accessed by simultaneously generating a large number of mutations [12-15]. However, conventional directed evolution and screening techniques typically examine proteins that contain only one or two amino acid substitutions. For example, the incorporation of single base pair substitutions in the promoter has identified nucleotides critical for polymerase recognition, but did not identify novel promoter specificities [11]. Similarly, a series of polymerases having all possible amino acid substitutions at position 748 were screened along with all possible single base pair substitutions at positions -10 through -12 of the promoter region [9]. While the functional importance of the nucleotides and amino acid residues was confirmed, no new promoter specificities were identified. Our combined in vitro / in vivo selection technique allows the examination of a relatively large number of polymerase sequence variants, and has discovered promoter and enzyme variants that would likely never have been observed by rational mutagenesis or more limited screens.
On the basis of the known crystal structure of T7 RNA polymerase bound to a 17 base-pair, double-stranded promoter sequence, it appears that T7 promoter recognition occurs largely through direct or water-mediated hydrogen bonding [2]. The identities of at least some of the selected mutants can be rationalized on the basis of the known protein:DNA contacts. For example, residue N748 recognizes G-11 in the non-template strand via both direct and indirect hydrogen bonds; the N748D substitution has previously been hypothesized to form an alternate hydrogen-bonding network with the G-11C substitution [2]. Similarly, in the wild-type polymerase, residue Q758 makes a direct hydrogen bond to the adenosine at position -8 in the template strand. The 'generalist' polymerase contains the single Q758C substitution and the shorter cysteine residue may allow (or at the least not interfere with) a larger range of nucleic acid interactions. However, the selected substitutions cannot always be conveniently rationalized: the R3–17 mutant differs from the wild-type T7 RNA polymerase at all the three amino acid positions (N748A; R756M; Q758S), replacing some amino acids with hydrophobic residues that do not likely form specific hydrogen-bonding interactions. Moreover, it is likely that the promoter recognition differs during different stages of transcription initiation [16,17], and it is unclear what particular contacts are made during open and closed complex formation, and during productive versus abortive initiation. Additional characterization of the kinetics and mechanism of the apparently altered promoter specificities will be required before more secure models can be put forth.
Combined in vitro / in vivo selections should generally prove to be useful for altering the properties of many molecular biology enzymes. For example, a compartmentalized self-replication has been reported for the selection of thermostable variants of Taq DNA polymerase. [18]. This method is similar to ours, except that instead of collecting mRNAs from intact cells, the cellular 'test tubes' expressing Taq polymerase are first embedded in water-in-oil emulsions and lysed prior to recovery of functional genes. These methods are nicely complementary: our method potentially allows for in vivo properties to be optimized (e.g., the selection of strong cellular promoters), while cellular emulsion allows for in vitro properties to be optimized (e.g., the utilization of unnatural nucleotides that might not be taken up by cells). Both methods allow similarly large numbers of mutational variants to be simultaneously compared and competed, and both methods can potentially be adapted to a range of molecular biology enzymes that act on their own templates, such as restriction endonucleases or DNA ligases.
Conclusions
In this study, we have developed a facile in vitro / in vivo selection methodology using a T7 RNA polymerase autogene to select polymerase variants with apparently altered promoter specificities. The procedure allows large libraries of promoter variants to be efficiently searched for their promoter recognition ability in vivo. The selection was successfully employed to identify a mutant T7 RNA polymerase 'specialist' which could recognize and transcribe a T3-like promoter, a polymerase 'generalist' which was able to recognize a variety of T7 promoters, and a novel RNA polymerase with an apparently new promoter specificity. The method can potentially be adapted to evolve other polymerases with novel phenotypes.
Materials and Methods
Construction of the wild type autogene
Using plasmid pAR1219, (originally made by Studier et al.[19] and obtained from Dr. David Hoffman, University of Texas at Austin) as a template, a 2.7 kb fragment containing the T7 RNA polymerase gene was amplified; EcoRI and BsmBI restriction sites were added during amplification. The PCR product was cloned into the vector pCR2.1 (TA cloning kit, Invitrogen, Carlsbad, CA) and then into plasmid pET28a+ (Novagen, Madison, WI), which contained the wild-type T7 RNA polymerase promoter. The wild type autogene thus obtained (pET/T7p/T7) was transformed into strain HMS174 pLysS (Novagen, Madison, WI), the same cell line Studier et al. initially used to express the autogene [10].
Construction of autogene libraries
Autogene libraries were constructed by first generating vectors containing promoter mutations, and then ligating polymerase gene libraries into these vectors. A point mutation G(-11)C and the lac operator region were introduced in and adjacent to the T7 RNA polymerase promoter using oligonucleotides ae66.1 and ae66.2. Upon annealing, sticky ends were generated that were suitable for ligation into the pET/T7p/T7 autogene construct cleaved with BglII and XbaI.
ae66.1: GATCTC GAT CCC GCG AAA TTA ATA CCA CTC ACT ATA GGG GAA TTG TGA GCG GAT AAC AAT TCC CCT (BglII site in bold)
ae66.2: CTAGAG GGG AAT TGT TAT CCG CTC ACA ATT CCC CTA TAG TGA GTG GTA TTA ATT TCG CGG GAT CGA (XbaI site in bold)
Similarly, oligonucleotides gcP1.66 and gcP2.66 were used to introduce random residues from positions -8 to -11 in the T7 RNA polymerase promoter.
gcP1 .66: GATCTC GAT CCC GCG AAA TTA ATA CNN NNC ACT ATA GGG GAA TTG TGA GCG GAT AAC AAT TCC CCT (BglII site in bold; N indicates a random sequence region)
gcP2.66: CTAGAG GGG AAT TGT TAT CCG CTC ACA ATT CCC CTA TAG TGN NNN GTA TTA ATT TCG CGG GAT CGA (XbaI site in bold; N indicates a random sequence region)
In the selection for T3-like promoter specificity, T7 RNA polymerase was randomized at amino acid positions 746–748. Two pairs of primers (gcT7a.6 and gcT7lib1) and (gcT7a.9 and gc3'pET and the wild-type (pET/T7p/T7) plasmid were used to generate two gene fragments, which were in turn assembled by overlap PCR. In the selection in which both the promoter and the polymerase pool was used, T7 RNA polymerase was similarly randomized at amino acid positions 748, 756, and 758 using the oligonucleotide gcT7lib2.80.
gcT7a.6: GGACCGATAA CGAAGTAGTT ACCGTGACCG
gcT7lib1.66: CTG cAA cCG GAA CTG cCC GAG GAA CAT CAG NNN NNN NNN CGT CTG AAT gGG CTT CTT GTA TTC CTG (region 2236–2244 is randomized, silent mutations are in lower case)
gcT7lib2.80: C GCT ATC cTT GTT GGT GTT gAT GGT AGG NNN TAA NNN GAA CTG cCC GAG GAA CAT CAG NNN CAA GCG CGT CTG AAT AGG C (regions 2241–2244, 2266–2268, 2272–2274 are randomized, silent mutations are in lower case)
gcT7a.9: CTG ATG TTC CTC GGg CAG TTC CGg TTg CAG (mismatches are in lower case)
gc3'pET: GCT CAG CGG TGG CAG CAG CCA ACT C
Purified, overlap PCR products and autogene vectors containing mutated promoters (e.g., pET/T7p*/T7) were digested with the restriction enzymes AflII and EcoRI. Fragments containing the random region were gel-purified and ligated into the appropriate vectors. In constructing the autogene pool for the second selection, two stop codons were first introduced at amino acid positions 747 and 748 in the wild-type RNA polymerase gene to form pET/T7/T7stop. Oligos containing the promoter pool randomized between -8 and -11 positions were then cloned into pET/T7/T7stop to form the autogene construct with the promoter pool, pET/T7pp/T7stop. This safeguard eliminated the possibility that wild-type RNA polymerases could arise from vector background. Unselected clones from pools were sequenced, and the distribution of random sequence nucleotides was estimated to be 29% G, 21% A, 19% T, and 24% C.
Selection and screening protocols
The autogene pool was transformed into DH5Δlac pLysS cells by electroporation. The culture was incubated at 37°C for 7–10 hrs and T7 RNA polymerase expression was induced by adding IPTG to a final concentration of 0.4 mM. After an hour of induction, RNA was extracted using the Masterpure RNA purification kit (Epicenter Technologies, Madison, WI). The recovered mRNA was reverse-transcribed using AMV-reverse transcriptase (Amersham Pharmacia Biotech Inc., Newark, NJ) and the primer gc3'pET. The cDNA was then PCR-amplified using the primers gcT7a.6 and gc3'pET. The PCR products were gel-purified, digested with AflII and EcoRI, and cloned back into the original autogene vector to form a delimited autogene pool for subsequent rounds of selection.
A colony lift technique was used to monitor the progress of the selection. Cells containing active autogene cease to grow when lifted to LB plates containing IPTG. Colonies from each round of selection were lifted to plates containing IPTG using a butterfly nitrcellulose membrane (Midwest Scientific, Valley Park, MO). The plates were then incubated at 37°C for ~8 hrs. Clones that did not grow upon re-plating were picked from the original plate.
A similar protocol was used for the identification of promoters that could be recognized by the R3–17 polymerase. In this instance, the R3–17 polymerase gene was first isolated via restriction digestion with AflII and EcoRI, cloned downstream of the T7 promoter-pool, and the resultant promoter library was transformed as before. Colonies (ca. 3,000) were lifted to IPTG plates and screened for their inability to thrive.
Acknowledgments
Acknowledgements
We would like to thank the Office of Naval Research and the NIH (grant ID 2R01 AI36083-08) for supporting these efforts.
Contributor Information
Jijumon Chelliserrykattil, Email: jijumon@mail.utexas.edu.
George Cai, Email: cai200x@yahoo.com.
Andrew D Ellington, Email: andy.ellington@mail.utexas.edu.
References
- Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- Diaz GA, Raskin CA, McAllister WT. Hierarchy of base-pair preference in the binding domain of the bacteriophage T7 promoter. J Mol Biol. 1993;229:805–811. doi: 10.1006/jmbi.1993.1086. [DOI] [PubMed] [Google Scholar]
- Schick C, Martin CT. Tests of a model of specific contacts in T7 RNA polymerase-promoter interactions. Biochemistry. 1995;34:666–672. doi: 10.1021/bi00002a034. [DOI] [PubMed] [Google Scholar]
- Li T, Ho HH, Maslak M, Schick C, Martin CT. Major groove recognition elements in the middle of the T7 RNA polymerase promoter. Biochemistry. 1996;35:3722–3727. doi: 10.1021/bi9524373. [DOI] [PubMed] [Google Scholar]
- Raskin CA, Diaz G, Joho K, McAllister WT. Substitution of a single bacteriophage T3 residue in bacteriophage T7 RNA polymerase at position 748 results in a switch in promoter specificity. J Mol Biol. 1992;228:506–515. doi: 10.1016/0022-2836(92)90838-B. [DOI] [PubMed] [Google Scholar]
- Rong M, He B, McAllister WT, Durbin RK. Promoter specificity determinants of T7 RNA polymerase. Proc Natl Acad Sci USA. 1998;95:515–519. doi: 10.1073/pnas.95.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda RA, Chang LL, Warshamana GS. Selection and characterization of a mutant T7 RNA polymerase that recognizes an expanded range of T7 promoter-like sequences. Biochemistry. 1993;32:9115–9124. doi: 10.1021/bi00086a016. [DOI] [PubMed] [Google Scholar]
- Raskin CA, Diaz GA, McAllister WT. RNA polymerase mutants with altered promoter specificities. Proc Natl Acad Sci U S A. 1993;90:T73147–3151. doi: 10.1073/pnas.90.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorff JW, Studier FW. Creation of a T7 autogene. Cloning and expression of the gene for bacteriophage T7 RNA polymerase under control of its cognate promoter. J Mol Biol. 1991;219:61–68. doi: 10.1016/0022-2836(91)90857-3. [DOI] [PubMed] [Google Scholar]
- Imburgio D, Rong M, Ma K, McAllister WT. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39:10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- Zaccolo M, Gherardi E. The effect of high-frequency random mutagenesis on in vitro protein evolution: a study on TEM-1 beta-lactamase. J Mol Biol. 1999;285:775–783. doi: 10.1006/jmbi.1998.2262. [DOI] [PubMed] [Google Scholar]
- Yano T, Oue S, Kagamiyama H. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc Natl Acad Sci USA. 1998;95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness JE, Welch M, Giver L, Bueno M, Cherry JR, Borchert TV, Stemmer WP, Minshull J. DNA shuffling of subgenomic sequences of subtilisin. Nat Biotechnol. 1999;17:893–896. doi: 10.1038/12884. [DOI] [PubMed] [Google Scholar]
- Altamirano MM, Blackburn JM, Aguayo C, Fersht AR. Directed evolution of new catalytic activity using the alpha/beta-barrel scaffold. Nature. 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- Place C, Oddos J, Buc H, McAllister WT, Buckle M. Studies of contacts between T7 RNA polymerase and its promoter reveal features in common with multisubunit RNA polymerases. Biochemistry. 1999;38:4948–4957. doi: 10.1021/bi982689e. [DOI] [PubMed] [Google Scholar]
- Villemain J, Guajardo R, Sousa R. Role of open complex instability in kinetic promoter selection by bacteriophage T7 RNA polymerase. J Mol Biol. 1997;273:958–977. doi: 10.1006/jmbi.1997.1358. [DOI] [PubMed] [Google Scholar]
- Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci USA. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]