Abstract
Hyperemesis Gravidarum (HG), severe nausea/vomiting in pregnancy (NVP), can cause poor maternal/fetal outcomes. Genetic predisposition suggests the genetic component is essential in discovering an etiology. We performed whole-exome sequencing of 5 families followed by analysis of variants in 584 cases/431 controls. Variants in RYR2 segregated with disease in 2 families. The novel variant L3277R was not found in any case/control. The rare variant, G1886S was more common in cases (p = 0.046) and extreme cases (p = 0.023). Replication of G1886S using Norwegian/Australian data was supportive. Common variants rs790899 and rs1891246 were significantly associated with HG and weight loss. Copy-number analysis revealed a deletion in a patient. RYR2 encodes an intracellular calcium release channel involved in vomiting, cyclic-vomiting syndrome, and is a thyroid hormone target gene. Additionally, RYR2 is a downstream drug target of Inderal, used to treat HG and CVS. Thus, herein we provide genetic evidence for a pathway and therapy for HG.
Keywords: Hyperemesis gravidarum, Nausea, Vomiting, Pregnancy, RYR2
1. Introduction
Nausea and vomiting of pregnancy is a common symptom affecting 70% of pregnant women (Goodwin, 1998). Clinical intervention is necessary in the severest form, Hyperemesis Gravidarum (HG), which affects up to 2% of pregnancies (Christodoulou-Smith et al., 2011). HG leads to significant weight loss, dehydration, electrolyte imbalance, and ketonuria (Fairweather, 1968; Goodwin et al., 1992a,b; Goodwin, 1998). Although maternal mortality is rare, 6 deaths due to HG have been reported recently (MacGibbon et al., 2015), as well as morbidity including Wernicke’s encephalopathy (Chiossi et al., 2006), acute renal failure (Hill et al., 2002), liver function abnormalities (Ahmed et al., 2013), splenic avulsion (Nguyen et al., 1995), esophageal rupture (Woolford et al., 1993), pneumothorax (Schwartz and Rosoff, 1994), and post-traumatic stress symptoms (Christodoulou-Smith et al., 2011). HG is also associated with poor fetal/child outcomes including a 4-fold increased risk of preterm birth and a 3-fold increased risk of neurodevelopmental delay in children (Fejzo et al., 2013, 2015).
A variety of potential causative factors have been investigated, but the etiology remains unknown. Evidence for a genetic predisposition is provided by classic twin studies of Norwegian, Spanish, and Finnish cohorts (Colodro-Conde et al., 2016a; Corey et al., 1992). Family based studies provide evidence that female relatives of patients with HG are more likely to be affected, with a 17-fold increased risk if a sister has HG (Gadsby et al., 1993; Vellacott et al., 1998; Vikanes et al., 2010; Zhang et al., 2011). Recently, mutations in the thyrotropin receptor gene have been linked to hyperemesis gravidarum accompanied by gestational thyrotoxicosis. This suggests a genetic etiology has already been identified in, at minimum, a subgroup of cases (Coulon et al., 2016). Thus, understanding the genetic component is essential in discovering the causal pathway(s).
The objective of this study was to perform whole-exome sequencing on HG families to identify rare variants conferring susceptibility to HG and to validate these findings in a large cohort of affected and unaffected individuals from the United States, followed by replication in cohorts from Australia and Norway.
2. Materials and methods
2.1. Study summary
The size and minimum HG CASE and CONTROL criteria for the 3 populations (US, Norway, Australia) used in this study are summarized in Fig. 1A. The genetic analysis methods used on each population are summarized in Fig. 1B.
Fig. 1. Summary of A) Populations and B) Methods used in this study.
The size and minimum HG CASE and CONTROL criteria for the 3 populations (US, Norway, Australia) used in this study are summarized in Fig. 1A. The genetic analysis methods used on each population are summarized in Fig. 1B. *MILD NVP was defined as participants who answered: ‘had some NVP for more than 7 days, but did not see a doctor or nurse and did not disrupt daily routine very much’ **MODERATE NVP was defined as ‘disrupted daily routine but did not affect weight and did not need medication’.
2.2. US population
2.2.1. Eligibility criteria
The source population for HG CASES in the US included patients primarily recruited through advertising on the HER Foundation website (www.helpher.org). The stringent study criteria were designed to exclude all cases and controls that would increase phenotypic uncertainty. Briefly, the inclusion criteria for affected individuals were a diagnosis of HG and treatment with intravenous (IV) fluids or total parenteral nutrition/nasogastric feeding tube. Each participant was asked to recruit a non-blood related acquaintance with at least 2 pregnancies that went beyond 27 weeks. Controls were eligible if they experienced normal or no nausea/vomiting in their pregnancy, no weight loss due to nausea/vomiting and no medical attention in their pregnancy due to nausea/vomiting. Participants were enrolled in the family study if an HG CASE had 2 or more additional family members with HG. Additional affected family members were eligible if they reported severe NVP accompanied by > 5% weight loss, and medication or hospitalization for HG. Control family members had the same eligibility requirements as Controls.
2.2.2. Description of US families analyzed by whole-exome sequencing and sanger sequencing
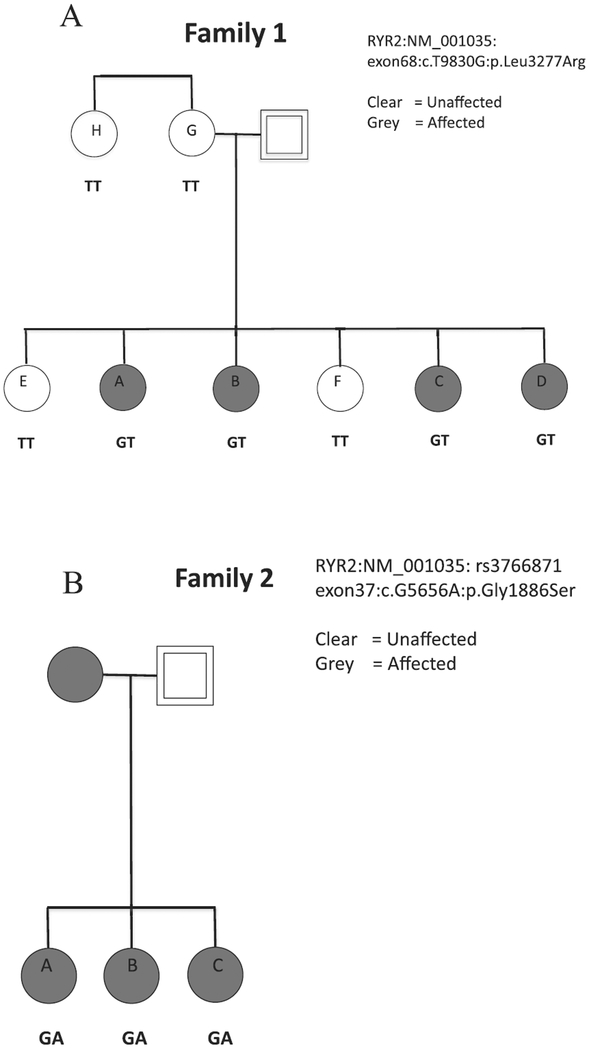
The whole-exome sequencing study included 15 affected individuals and 3 unaffected individuals. These 18 individuals came from 5 families (Fig. 1A). Follow-up analysis to confirm segregation in Family 1 included additional family members −3 unaffected and 1 affected individual from Family 1. Each family submitted saliva samples for a minimum of three affected individuals. We chose to analyze a total of 18 individuals: 3 affected individuals from each of 5 families with HG in addition to 3 unaffected controls from 3 of the five families to further limit potential causal variants by dismissing those variants identified in unaffected family members. Pedigrees of two families of Caucasian/European descent analyzed in this whole-exome sequencing study, and whose variants are described herein, are shown in Fig. 2. Family 1 is of mixed Finnish, Swedish, English, and German descent. We collected DNA from 4 CASES (4 sisters) and 4 CONTROLS (2 sisters, mother, and maternal aunt) from Family 1. This family consists of 9 sisters, 5 affected and 4 unaffected, but only those siblings who participated are shown in Fig. 2A. Family 2 is of mixed Scottish, German, Swiss, English, and Italian descent and we collected DNA from 3 affected sisters (Fig. 2B).
Fig. 2. Pedigrees and Genotypes of A) Family 1 and B) Family 2.
In Family 1, A, B, C, and D all were affected with HG. Participant A reported pic line, medication and weight loss to treat HG, B reported a 10 pound weight loss and medication to treat symptoms until birth, C reported intravenous fluids (IV) and a 23 pound weight loss, and D reported IV fluids, hospitalization, weight loss, and medication to treat her HG. Among the unaffected family members, participant E reported 2 pregnancies with mild nausea, no weight loss, and no medication; F reported 5 pregnancies with no nausea and vomiting, no weight loss, and no medication in any pregnancy, G reported 13 pregnancies with normal nausea and vomiting, no weight loss, no medication, and H reported 3 pregnancies with normal nausea and vomiting, no weight loss, and no medication. For Family 2, all 3 sisters were affected and all 3 sisters required medication and IV fluids to treat their HG. Sisters A and C were both hospitalized for HG. Their mother was also affected but did not participate.
2.2.3. Description of US CASE/CONTROL population analyzed by genotyping and copy-number analysis
The follow-up CASE/CONTROL population from the United States included 584 HG CASES and 431 unaffected CONTROLS that were all genotyped. A subgroup analysis was performed comparing the most severe HG CASES requiring total parenteral nutrition/nasogastric feeding tube to Controls who reported no NVP in any pregnancy. The subgroup was also used for a copy-number analysis. All participants gave informed consent. This study was approved by the UCLA Institutional Review Board.
2.3. Norwegian population
2.3.1. Eligibility criteria
Eligibility was determined using data obtained from self-reported questionnaires (Nilsen et al., 2009). HG CASES were eligible if they were admitted to hospital for prolonged nausea and vomiting in pregnancy. Controls were eligible if they were NOT admitted to hospital for prolonged nausea and vomiting of pregnancy.
2.3.2. Description of Norwegian population analyzed by GWAS and correlation with weight loss
The samples in this HG study included 385 HG CASES and 2280 unaffected Controls. The samples included were singleton pregnancies of Norwegian ancestry. Summary statistics for RYR2 were analyzed for independent replication in the Norwegian Mother and Child Cohort Study (MoBa), a prospective population-based pregnancy cohort conducted by the Norwegian Institute of Public Health recruited from Norway during 1999–2008 (http://www.fhi.no/moba-en; Magnus et al., 2016). In addition, data was collected for each participant on maternal pre-pregnancy weight and weight change until week 18 of gestation and used as a continuous variable in a regression analysis to study genetic associations with weight loss in early pregnancy. Ethical approval for the MoBa study has been approved by the Regional Committee for Medical Research Ethics and all women provided informed written consent.
2.4. Australian population
2.4.1. Eligibility criteria
Eligibility was determined using data collected from health and wellbeing surveys. Women reported their experience in their pregnancy with the most severe NVP using a five-point questionnaire adapted from Zhang et al. (2011).
HG CASES (1) were defined as NVP that disrupted daily routine accompanied by weight loss and medication and/or IV fluids and/or feeding tube. A second less stringent CASE criteria included HG CASES (1) in addition to HG CASES (2) that reported NVP that disrupted their daily routine and medication treatment, but did not lose weight. CONTROLS were defined as experiencing no NVP.
Women in between the extreme ends who reported MILD NVP (NVP for more than 7 days, but did not see a doctor or nurse/did not disrupt daily routine very much) and MODERATE NVP (disrupted daily routine but it did not affect my weight and did not need medication, were not included in the CASE: CONTROL study, but were included in a continuous analysis of the GWAS data.
2.4.2. Description of Australian population GWAS analyzed using CASE:CONTROL and continous phenotype
The Australian sample is composed of genotyped women unselected for HG who are part of the Australian Endogene Study and the QIMR Mothers of Twins Study, which are two of the cohorts participating in the NVP Genetics Consortium (Colodro-Conde et al., 2016b).
As part of health and wellbeing surveys, a total of 1440 women reported on NVP severity. We conducted a CASE;CONTROL analysis using 946 women who were in the extremes of the severity scale. Women reporting no NVP (n = 677) were used as controls and women reporting severe NVP (n = 269) with disruption of their daily routine and medication prescription, including those losing weight and put on a drip or feeding tube, were used as CASES. We also conducted a more stringent CASE/CONTROL analysis that excluded the 139 HG CASES with no weight loss, thus limiting the study to 130 HG CASES and 677 CONTROLS. Finally, we analyzed NVP as a continuous phenotype for all 1440 study participants, which included an additional 163 CASES of MILD NVP and 331 CASES of MODERATE NVP.
3. Genetic methods
3.1. Genetic methods for US population
3.1.1. Whole-exome sequencing of families
Each study participant was asked to submit a saliva sample for DNA analysis. A saliva collection kit (Oragene, Ottawa, Canada) was self-administered for submitting 2 mL of saliva. DNA was extracted from 75% of the saliva sample according to the manufacturer’s instructions (Oragene, Ottawa Canada).
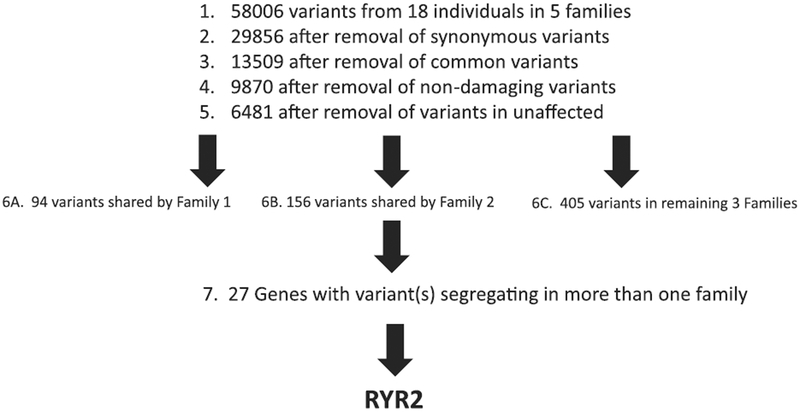
We sequenced the entire exomes (~50 Mb) of 15 affected individuals and 3 unaffected individuals from 5 HG families. Paired end reads 100 nucleotides (2 × 100 nucleotides) were generated on an Illumina HiSeq 2000. Each sample was sequenced on 3 different lanes to avoid lane bias. Qseq files were converted into Sanger-formatted FASTQ files and reads were mapped to the reference human genome build hg19 using the Burrows Wheeler Alignment algorithm (BWA) (Li and Durbin, 2009). Duplicated reads were marked by Picard. The Genome Analysis Toolkit (GATK) was used for local realignment around indel sites followed by a base quality recalibration (McKenna et al., 2010). For reliable SNP calling we used genotype quality ≥ 10; read QUAL ≥ 30 and a minimum read depth of 4. The combined total variants from all 18 individuals were filtered as shown in Fig. 3. Synonymous variants, which are unlikely to be causal, were discarded. The identified variants were further filtered against variants present in the HapMap, 1000 Genomes Project and dbSNP132 databases, selecting for novel variants and known variants with minor allele frequency <5% (McKenna et al., 2010; International HapMap 3 Consortium et al., 2010). These variants were further filtered by selecting variants predicted to affect protein function using PolyPhen and SIFT (Ramensky et al., 2002; Ng and Henikoff, 2003). Variants were further filtered by deleting variants present in the 3 unaffected family controls. All variants were discarded that were not shared by all 3 whole-exome sequenced affected family members with each family. Finally, we identified a subgroup of genes involved in more than one family, and screened these genes for a functional effect, which included genes 1) functionally relevant to reproduction (ie hormones), 2) nausea and vomiting (ie gastric tract, vomiting center of the brain), and 3) genes expressed in relevant tissues (ie ovary, placenta, vomiting center of the brain).
Fig. 3.
Whole-exome sequencing filtering steps identifies RYR2 variants.
3.1.2. Sanger Sequencing to analyze segregation in family 1
Sanger Sequencing of the novel variant in Family 1 (RYR2 exon68:c.T9830G:p.Leu3277Arg) was performed to confirm whole-exome sequencing results and to confirm or deny segregation with the disease in the remaining family members who were not included in exome sequencing (1 affected sister and 1 unaffected sister, the unaffected mother, and an unaffected maternal aunt). PCR primer pairs GGAAGTCATACTGCCCATGC and GGGGTACAATGTCTTCTTCCA were designed from genomic DNA to amplify and sequence the variant. PCR amplification and sequencing were carried out using standard methods.
3.1.3. Protein prediction tools used to predict functional effect of L3277R
The SIFT protein prediction tool was used to determine that the novel SNP encoding L3277R resulted in a damaging protein product, and the Provean Prediction tool was used to determine that it was deleterious. (Choi et al., 2012; Ng and Henikoff, 2003).
3.1.4. Genotyping
Taqman primers were designed for both the novel variant L3277R in Family 1 and the rare variant G1886S identified in Family 2, and used to screen individuals from ≥573 HG CASES and ≥426 controls using Applied Biosystems PRISM 7900HT Sequence Detection System (TaqMan) for large-scale screening. The call rate was >96%.
3.1.5. Statistical analysis
Statistical significance of association of genotype with HG was determined by calculating the p-values using a 1-tailed Fisher’s exact test (http://graphpad.com/quickcalcs/contingency1/) and odds ratios were calculated using the odds ratio calculator (https://www.medcalc.org/calc/odds_ratio.php). A p-value <0.05 was considered statistically significant.
3.1.6. Copy-number analysis of RYR2
Quantitative real-time PCR analysis of RYR2 was performed in triplicate on 10 ng from 240 DNA samples (101 extreme CASES requiring tube feeding and 139 extreme controls with no nausea/vomiting in ≥2 pregnancies) on 384-microwell optical plates using the predesigned Taqman Copy Number Assay covering 90 base pairs within a likely pathogenic duplicated region in autism (Soueid et al., 2016) (Assay ID: Hs00137466_cn FAM labeled, MGB probe, Thermofisher Scientific, Waltham, MA) and the RNaseP Copy Number Reference Assay (VIC labeled, TAMRA probe). Melt-curve analysis was applied and all results were normalized to RNaseP levels and calculated using the ΔΔCT method. One sample with a deletion originally identified in the above triplicate assay along with 5 normal control samples, were assayed a second time in duplicate (re-diluted to 10 ng from the original DNA sample) to verify the deletion.
3.1.7. Genetic methods for Norwegian population
Maternal genome-wide data were obtained using Illumina HumanCoreExome genotyping BeadChip v1.1. Imputation was performed with reference panel HapMap phase 3 build 36 using IMPUTE2 (Howie et al., 2009). Standard association analyses were performed in PLINK 1.7 (Purcell et al., 2007). Genotypes were analyzed with allelic and genotypic approach. Regression analysis was performed using a z-score transformed gestational weight gain (GWG) based on maternal pre-pregnancy weight and weight change until week 18 of gestation.
3.1.8. Genetic methods for Australian population
The samples were genotyped using Illumina arrays and genotype imputation was completed using 1000 Genome Phase 3 version 5 as reference data. For validation of the rare imputed SNP G1886S, we also conducted CASE/CONTROL analysis in a more stringent subset by removing the participants who did not report weight loss from the CASES, thus limiting to a more stringent CASE phenotype (n = 130).
4. Results
4.1. Whole-exome sequencing identifies RYR2 variants linked to HG in 2 of 5 families
We sequenced the entire exomes (~50 Mb) of 15 affected individuals and 3 unaffected individuals from 5 families with HG. The mean coverage was 54 fold. Reads were mapped to the human genome reference build UCSC hg19 using BWA (Li and Durbin, 2009). On average, 3223 single-nucleotide variants were detected in each individual and a total of 58,006 variants were detected in all 5 families combined (Fig. 3). The synonymous variants were subsequently discarded resulting in 29,856 variants. The identified variants were further filtered against variants present in the Hap-Map, 1000 Genomes Project, and dbSNP132 databases, resulting in 13,509 novel variants and known variants with minor allele frequency <5% (McKenna et al., 2010; International HapMap 3 Consortium et al., 2010). These variants were further filtered by selecting variants predicted to affect protein function using Poly-Phen and SIFT (Ramensky et al., 2002; Ng and Henikoff, 2003). Filtering for missense and stop gain or stop loss variants that were shared by any of the 3 whole-exome sequenced unaffected family members resulted in 6481 variants.
As we did not find any single variant that was shared by all the affected members across all of the families, we focused on variants within each family shared by all 3 whole-exome sequenced affected subjects. For example, 94 variants were shared by all 3 affected individuals in Family 1, and 227 variants were shared by all 3 affected individuals in Family 2. We searched for variants and/or genes that were shared by more than one family. 27 genes were identified that carried rare variants in the affected family members in more than one family. These variants were evaluated based on a functional effect, which included variants located in genes functionally relevant to reproduction (ie hormones), nausea and vomiting (ie gastric tract, vomiting center of the brain), and genes expressed in relevant tissues (ie ovary, placenta). This resulted in identification of the gene RYR2 involved in 2 of 5 families as a strong candidate based on its functional potential: RYR2 encodes an intracellular calcium release channel that is part of a signaling pathway for emesis expressed in the vomiting center of the brain (Giannini et al., 1995; Zhong et al., 2014). It is the only gene identified with variants significantly linked to cyclic vomiting syndrome, is a thyroid hormone target gene, and is differentially expressed in cumulus cells of the pre-ovulatory follicle (Grøndahl et al., 2012; Jiang et al., 2000; Lee et al., 2015).
Synonymous and common variants in RYR2 that were originally removed in the filtering steps were re-investigated. None of the 6 variants identified segregated with disease. In addition, variants in RYR2 in the remaining families (Family 3,4,5) were not identified in this study. Potentially causal variants in other genes in the remaining families are currently under investigation.
4.2. Genotyping the novel and rare RYR2 variants in the US cohort provides confirmation
In the largest HG family (Family 1), the novel heterozygous variant in the RYR2 gene (RYR2:NM_001035:exon68:c.T9830G:p.Leu3277Arg) was confirmed by Sanger Sequencing to be shared by four affected sisters and was not shared by either of 2 unaffected sisters, the unaffected mother, nor the unaffected maternal aunt (Fig. 2A). The phenotype and genotype results suggest L3277R is of paternal rather than maternal origin in this family. However, the DNA from the father who is presumed to be a carrier, nor his sister who reportedly did not have HG, was unfortunately not available. We do not have any additional information about phenotype on the father’s side (ie father’s mother). Genotyping via Taqman showed the RYR2 variant to be unique in the sample to Family 1, as it was not identified in 584 HG CASES and 431 unaffected CONTROLS (Table 1). The nucleotide at the location of L3277R is 100% conserved across vertebrates and invertebrates. The mutation changes a hydrophobic amino acid to an electrically charged amino acid, and is predicted to be damaging and deleterious (SIFT Prediction Score = 0; Provean Prediction Score = −5.38).
Table 1.
RYR2 variant table.
| RYR2 variant | Source | EXON/INTRON | Method | Screened | OR | P-value |
|---|---|---|---|---|---|---|
| L3277R* | FAMILY 1, USA | 68:c.T9830G | Genotyping | 584 HG, 431 C | NA | NA |
| G1886S | FAMILY 2, USA | 37:c.G5656A | Genotyping | 584 HG, 431 C | 1.29 | 0.046 |
| G1886S | FAMILY 2, USA2 | 37:c.G5656A | Genotyping | 106 HG, 141 C | 4.27 | 0.023 |
| G1886S | NORWEGIAN | 37:c.G5656A | GWAS | 318 HG, 1823 C | 1.32 | 0.661 |
| G1886S | AUSTRALIAN | 37:c.G5656A | GWAS | 269 HG, 677 C | 0.89 | 0.693 |
| G1886S | AUSTRALIAN2 | 37:c.G5656A | GWAS | 130 HG, 677 C | 1.17 | 0.665 |
| rs790899 | NORWEGIAN | Intron 95 | GWAS | 385 HG, 2280 C | 1.19 | 0.033 |
| rs790899 | AUSTRALIAN | Intron 95 | GWAS | 269 HG, 677 C | 1.33 | 0.013 |
| rs1891246 | NORWEGIAN | Intron 100 | GWAS | 385 HG, 2280 C | 1.23 | 0.009 |
| rs1891246 | AUSTRALIAN | Intron 100 | GWAS | 269 HG, 677 C | 1.3 | 0.014 |
| NOVEL DEL* | USA | 16:237619976 | Copy Number | 101 HG, 139C | NA | NA |
HG=Hyperemesis Gravidarum, C=Unaffected Control, TPN = Severe HG requiring tube feeding.
L3277R* = novel deleterious SNP (c.9830T > G, p.Leu3277Arg).
DEL = deletion in exon 16 of unknown size.
rs3766871 G1886S (NM_001035.2:c.5656G > A, NP_001026.2:p.Gly1886Ser).
rs790899 (NM_001035.2:c.13913 + 381G > A; XM_006711804.2:c.13943 + 381G > A).
rs1891246 (NM_001035.2:c.14434–490T > G; XM_005273224.1:c.14491–490T > G).
USA2 and AUSTRALIAN2 are datasets with more stringent criteria.
USA2 (HG = requiring iv feeding, C = no NVP).
AUSTRALIAN2 (HG = weight loss, C = no NVP).
In Family 2, the heterozygous variant G1886S RYR2:NM_001035:exon37:c.G5656A:p.Gly1886Ser rs3766871 was shared by all 3 affected sisters (Fig. 2B). Genotyping via Taqman identified the heterozygous variant G1886S (Family 2) to be twice as common (p = 0.046) in CASES than CONTROLS (in 38 out of 580 additional CASES and 17 out of 431 CONTROLS) and four times more common (p = 0.023) when comparing the extreme ends of the clinical spectrum, 9 out of 106 CASES requiring tube feeding compared to 3 out of 141 controls who reported no nausea/vomiting in pregnancy) (Table 1). The SNP G1886S is already known to have a biological effect in the homozygous state. Homozygous substitution of serine for glycine causes a significant increase in cellular calcium oscillation activity compared to wild-type RYR2 in HEK293 cells (Koop et al., 2008). Interestingly, calcium oscillations are completely abolished by homozygous substitution of a neighboring SNP in the double mutant G1885E/G1886S. The estimated frequency of G1886S in the European_American population (ESP6500) is 0.031 and the estimated frequency of G1885E is 0.023 (The 1000 Genomes Project Consortium, 2015). Therefore the estimated frequency of carrying both mutations is very rare (<0.001), but may be selected for in the extreme control population where the phenol-type (no NVP in at least 2 pregnancies) is also rare. The SNP G1886S has also been associated with ventricular arrhythmias and is an independent predictor of sudden cardiac death, while a neighboring SNP rs790896 (G > A) was linked to a decreased risk of sudden cardiac death (Ran et al., 2010). The frequency of the protective A allele rs790896 is predicted to be 0.415 in the European population (The 1000 Genomes Project Consortium, 2015), so the estimated frequency of carrying both G1886S and rs790896 is 0.013. In this study we identified 3/141 extreme controls carrying G1886S and 2 cases of G1886S/rs790896 are predicted in 141 extreme controls. Therefore, it will be interesting to investigate if additional variants in controls carrying RYR2 G1886S (such as G1885E and rs790896) explain why this variant is also present in a subset of controls with no NVP.
4.3. Summary statistics were supportive but not statistically significant for variant G1886S in RYR2 in both a Norwegian and an Australian GWAS
Genotype data for G1886S were imputed in both Norwegian and Australian datasets. Although statistical significance was not achieved probably due to the rarity of G1886S and the small number of affected individuals, there is a supportive trend in both cohorts. In the Norwegian cohort there is a 1.3-fold OR for this SNP (reference allele A), and in the Australian cohort, after removal of the CASES with no weight loss to better reflect the severe end of the clinical spectrum of NVP, there was a 1.2-fold OR for G1886S (Table 1).
Common variants in RYR2 (rs790899 and rs1891246) are significantly linked to HG and are highly significant with respect to weight loss in early pregnancy.
In addition to the rare variants, common RYR2 SNPs (rs790899 and rs1891246) were significantly linked to HG in both the Norwegian and Australian GWAS using the CASE/CONTROL phenotypes (Table 1). No other common variants were identified that reached statistical significance in both the Norwegian and Australian datasets.
In the Norwegian dataset, adding the zscore for weight change until gestational week 18 as a covariate increased the odds ratio and significance for the common RYR2 variants, and suggested a strong association with weight loss (p = 2.12E-31 for rs790899 and p = 1.19E-31 for rs1891246, Table 2A). In the smaller Australian dataset, using the continuous severity measure, neither rs790899 nor rs1891246 reached statistical significance.
Table 2.
Adding zscore weight change until gestational week 18 as covariate shows rs790899 and rs1891246 associated with weight.
| CHR | SNP | BP | A1 | Test | NMISS | OR | Stat | P-VALUE |
|---|---|---|---|---|---|---|---|---|
| 1 | rs790S99 | 237957678 | C | ADD | 2499 | 1.267 | 2.649 | 0.00808 |
| 1 | rs790S99 | 237957678 | C | COV1 | 2499 | 0.2447 | −11.66 | 2.12E-31 |
| 1 | rs1891246 | 237981846 | G | ADD | 2499 | 1.292 | 2.968 | 0.002993 |
| 1 | rs1891246 | 237981846 | G | COV1 | 2499 | 0.2424 | −11.71 | 1.19E-31 |
rs790899 (NM_001035.2:c.13913 + 381G > A; XM_006711804.2:c.13943 + 381G > A).
rs1891246 (NM_001035.2:c.14434–490T > G; XM_005273224.1:c.14491–490T > G).
4.4. Copy-number analysis identifies a deletion in RYR2 in an extreme HG CASE requiring intravenous feeding (TPN)
We also performed copy-number analysis to search for pathogenic duplications and/or deletions in RYR2. A deletion in exon 16 was identified in RYR2 in DNA from one woman with HG requiring total parenteral nutrition (TPN) among DNA isolated from 101 CASES requiring TPN for severe HG (Table 1). The deletion was not observed in any of the remaining samples including 139 extreme controls reporting no nausea and no vomiting in any of their pregnancies.
5. Discussion
his is the first whole-exome association study of HG and suggests RYR2 may play a role in the biology of HG. We have successfully identified two rare variants in RYR2 that are linked to HG using a whole-exome sequencing approach followed by genotyping a large validation cohort from the US. Independent replication in GWAS studies from Norway and Australia are suggestive of a role for RYR2 and revealed 2 common variants very significantly associated with weight loss in early pregnancy. Copy-number screening identified a deletion in RYR2 in an extreme HG CASE requiring intravenous feeding.
RYR2 encodes an intracellular calcium release channel which localizes to the intracellular Ca2+ stores (ER/SR), and that, in excitable cells, including cardiac muscles and neurons, controls contraction and activity, respectively (Santulli and Marks, 2015). RYR2 is expressed in several other cell types in many tissues, including the thyroid gland, ovaries, and pancreas (Craps et al., 2015; Grøndahl et al., 2012; Santulli et al., 2015). An animal model of emesis shows that intracellular Ca2+ release through ryanodine receptors in the brainstem initiate Ca2+-dependent activation of CaMKIIa and ERK1/2, leading to emesis, which can be blocked by dantrolene, an inhibitor of emesis (Zhong et al., 2014). Thus, we speculate that the variants and deletions described here may result in abnormalities in the emesis-signaling pathway via a hyper-functioning Ca2+ channel. Indeed, variants in RYR2, including G1886S identified in this study (albeit in a heterozygous state), have been shown to cause increased RYR2 channel activity in a homozygous state (Koop et al., 2008). Functional evidence for the other variants identified in this study remains to be determined, but there are some clues. The novel variant L3277R is predicted to be deleterious and map between a putative phosphorylation site (AA2947) and a CALM interacting site (AA3581). And exon 16, the location of the deletion, contains, an RIH Domain (CDD:250561) which may form a binding site for IP3. The common variants identified in this study (rs790899 and rs1891246) both map in introns toward the end of the gene in sites with no predicted regulatory significance (Kent et al., 2002), and therefore, are not likely to have a phenotype on their own, but may be linked to functional mutations/deletions not identified in this study.
The role RYR2 variants play in HG etiology is unknown, but there are several intriguing avenues to explore further. Firstly, RYR2 encodes an intracellular calcium release channel that is the only ryanodine receptor expressed in the vomiting center of the brain (Giannini et al., 1995) and has been implicated in a signaling pathway underlying emesis in an animal model (Zhong et al., 2014). Secondly, the thyroid hormone has been shown to induce RYR2 overexpression (Jiang et al., 2000), while the drug Inderal (Propranolol, used to treat hyperthyroidism) blocks RYR2 phosphorylation and lowers its expression (Yoshida et al., 1992). Hyperthyroidism accompanies HG in as many as 60% of pregnancies (Goodwin et al., 1992a,b) and mutations in the thyrotropin receptor have been linked to HG (Coulon et al., 2016), providing additional genetic evidence that this pathway may be causal in some cases. However, because thyrotoxicosis is not normally associated with nausea and vomiting, it is likely that another factor is involved, such as the additional requirement of an emetic stimulus (ie pregnancy hormones). There could be a viscious cycle of deterioration from NVP to HG as a result of hormone-induced nausea/vomiting in conjunction with aberrant RYR2 Ca2+ signaling caused by progressive thyroid dysfunction and/or mutant RYR2. Of note, two of 6 recent maternal deaths secondary to HG were accompanied by severe thyrotoxicosis/thyroid storm (Knight et al., 2014; MacGibbon et al., 2015). Thirdly, in a NextGen sequencing study of Cyclic Vomiting Syndrome (CVS), RYR2 was the only gene among over 1000 genes screened, with variants significantly linked to the disease (Lee et al., 2015), and Inderal has been used to effectively treat 92% of children with CVS (Haghighat et al., 2007). Likewise, in 1980 a patient presenting with severe thyrotoxicosis and hyperemesis gravidarum reportedly responded dramatically to Inderal treatment (Valentine et al., 1980). As her thyroid function improved and the Inderal was discontinued, she again returned to the hospital with severe vomiting. Upon restarting medication, her vomiting ceased and she continued treatment until term. Our findings provide evidence for a biological pathway, diagnostic marker, and potential targeted therapy for the etiology and treatment of HG.
Lastly, limited evidence suggests RYR2 may play a role in fertility. It is expressed more than 50-fold in cumulus cells compared to mural granulosa cells of human pre-ovulatory follicle, and its expression correlates with amphiregulin, a key mediator of the effect of LH/hCG and a marker for oocyte competence (Grøndahl et al., 2012). Ryanodine receptor variants are significantly associated with pig litter size (Omelka et al., 2004), and women with HG produce an abnormally high number of mature oocytes when undergoing follicle stimulation (Fejzo et al., 2010). A genetic link that explains both the symptoms of HG and a potential increase in fertility, would provide a rationale for why severe nausea in pregnancy has not been selected out in nature despite its link to adverse outcomes.
The limitation of this study stems from the fact that full sequencing and copy number analysis of RYR2 in all cases and controls, is cost-prohibitive. We were only able to study the 2 mutations involved in the 2 HG families in this study, not the complete gene sequence, in the validation cohorts from the United States. Also, while the US cohort used intravenous fluid treatment as its clinical criteria for HG, and the Norwegian cohort used hospitalization, the Australian cohort used a less severe phenotype, which may have led to a reduced effect for that dataset. The small sizes for GWAS may also contribute to an underestimate of the effect for the validation cohorts. Alternatively, the multiple analyses in different relatively small cohorts can lead to over-interpretation of the results. Finally, the copy-number analysis that identified a deletion only surveyed 90 bp of exon 16 in 240 individuals. Mutation and copy-number analysis of the full RYR2 gene in cases and controls is now warranted to determine their frequencies in affected individuals and to understand the role of RYR2 in HG pathogenesis.
In conclusion, this study uses an innovative approach to identifying the etiology of HG. This disease has thus far eluded both scientists and clinicians, resulting in largely ineffective treatments, significant maternal morbidity, and an increased risk in adverse fetal outcome (Fejzo et al., 2013). Mutations in genes in the ryanodine receptor-signaling pathway may account for a substantial amount of the attributable risk of HG, although just how much must be deferred to a follow-up study as causality has not been definitively established. Additional studies are required, such as functional analysis of the deleterious RYR2 variant L3277R, complete deletion analysis of RYR2, and a larger GWAS. However, this novel discovery may provide the first step in understanding the etiology of HG. The identification of genes linking HG to RYR2 provides an intriguing new avenue for diagnosis, research, and therapy.
Acknowledgements
We thank the participants for volunteering their time to be part of this study.
Funding
The research was supported, in part, by the Hyperemesis Education and Research Foundation (MSF), the Paul and Janis Plotkin Family Foundation, the UCLA Clinical and Translational Science Institute (CTSI) UL1TR000124 (MSF), and NIH Grant GM053275 (JSS). NICHD Grant R01HD058008. The Norwegian Mother and Child Cohort Study was also supported by the Norwegian Ministry of Health and the Ministry of Education and Research, NIH/NIEHS (contract no N01-ES-75558), NIH/NINDS (grant no.1 UO1 NS 047537–01 and grant no.2 UO1 NS 047537–06A1). Australian Research Council (ARC) Grants A79600334, A79906588, A79801419, DP0212016, DP0343921 and the National Health and Medical Research Council Project Grant (NHMRC) Grants 241944, 389875, 552485, 552471, 1031119, 1049894, 1084325. This work was also supported by Fundación Seneca-Regional Agency for Science and Technology, Murcia, Spain [19151/PD/13 to L.C.C.]. SEM is supported by NHMRC fellowship APP1103623.
Details of ethics approval
This study was approved by the UCLA Institutional Review Board on 5/20/2011 as IRB#11–001681 and the Queensland Institute of Medical Research Human Research Ethics Committee.
Footnotes
Conflict of interest
The authors report no competing financial interests or conflicts of interest.
Disclosure of interests
The authors declare no competing financial interests.
References
- Ahmed KT, Almashhrawi AA, Rahman RN, Hammoud GM, Ibdah JA, 2013. Liver diseases in pregnancy: diseases unique to pregnancy. World J. Gastroenterol 19 (43), 7639–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiossi G, Neri I, Cavazutti M, Basso G, Fucchinetti F, 2006. Hyperemesis gravidarum complicated by Wernicke’s encephalopathy: background, case report and review of the literature. Obstet. Gynecol. Surv 61, 255–268. [DOI] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP, 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou-Smith J, Gold JI, Romero R, Goodwin TM, Macgibbon KW, Mullin PM, Fejzo MS, 2011. Posttraumatic stress symptoms following pregnancy complicated by hyperemesis gravidarum. J. Matern. Fetal Neonatal Med 24 (11), 1307–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colodro-Conde L, Jern P, Johansson A, Sánchez-Romera JF, Lind PA, Painter JA, Ordoñana JR, Medland SE, 2016a. Nausea and vomiting during pregnancy is highly heritable. Behav. Genet 46 (4), 481–491. [DOI] [PubMed] [Google Scholar]
- Colodro-Conde L, Cross SM, Lind PA, Painter JN, Gunst A, Jern P, Johansson A, Maegbaek ML, Munk-Olsen T, Nyholt DR, Ordonana JR, Paternoster L, Sanchez Romera JF, Wright MJ, Medland SE, 26 February 2016b. Cohort profile: nausea and vomiting during pregnancy genetics Consortium (NVP genetics Consortium). Int. J. Epidemiol pii: dyv360, PMID: 2692160; doi: 10.1093/ije/dyv360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey LA, Berg K, Solaas MH, Nance WE, 1992. The epidemiology of pregnancy complications and outcome in a Norwegian twin population. Obstet. Gynecol 80 (6), 989–994. [PubMed] [Google Scholar]
- Coulon AL, Savagner F, Briet C, Vernin M, Munier M, Chabre O, Rodien P, 2016. Prolonged and severe gestational thyrotoxicosis due to enhanced hCG sensitivity of a mutant thyrotropin receptor. J. Clin. Endocrinol. Metab 101 (1), 10–11. [DOI] [PubMed] [Google Scholar]
- Craps J, Wilvers C, Joris V, De Jongh B, Vanderstraeten J, Lobysheva I, Balligand JL, Sonveaux P, Gilon P, Many MC, Gérard AC, Colin IM, 2015. Involvement of nitric oxide in iodine deficiency-induced microvascular remodeling in the thyroid gland: role of nitric oxide synthase 3 and ryanodine receptors. Endocrinology 156 (2), 707–720. [DOI] [PubMed] [Google Scholar]
- Fairweather DVI, 1968. Nausea and vomiting in pregnancy. Am. J. Obstet. Gynecol 102 (1), 135–175. [DOI] [PubMed] [Google Scholar]
- Fejzo MS, Magtira A, Schoenberg FP, MacGibbon K, Mullin P, Romero R, Tabsh K, 2013. Antihistamines and other prognostic factors for adverse outcome in hyperemesis gravidarum. Eur. J. Obstet. Gynecol. Reprod. Biol 170 (1), 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejzo MS, Magtira A, Schoenberg FP, Macgibbon K, Mullin PM, 2015. Neuro-developmental delay in children exposed in utero to hyperemesis gravidarum. Eur. J. Obstet. Gynecol. Reprod. Biol 189, 79–84. [DOI] [PubMed] [Google Scholar]
- Fejzo MS, Romero R, Goodwin TM, 2010. Patients with a history of hyperemesis gravidarum have similar symptoms during egg stimulation and develop ovarian hyperstimulation syndrome: case series. Fertil. Steril 93 (1), 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby R, Barnie-Adshead AM, Jagger C, 1993. A prospective study of nausea and vomiting during pregnancy. Br. J. Gen. Pract 43, 245–248. [PMC free article] [PubMed] [Google Scholar]
- Giannini G, Conti A, Mammarella S, Scrobogna M, Sorrentino V, 1995. The ryanodine receptor/calcium channel genes are widely and differentially expressed in murine brain and peripheral tissues. J. Cell. Biol 128 (5), 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TM, Montoro M, Mestman JH, 1992a. Transient hyperthyroidism and hyperemesis gravidarum: clinical aspects. Am. J. Obstet. Gynecol 167, 648–652. [DOI] [PubMed] [Google Scholar]
- Goodwin TM, Montoro M, Mestman JH, Pekary AE, Hershman JM, 1992b. The role of chorionic gonadotropin in transient hyperthyroidism of hyperemesis gravidarum. J. Clin. Endocrinol. Metab 75 (5), 1333–1337. [DOI] [PubMed] [Google Scholar]
- Goodwin TM, 1998. Hyperemesis gravidarum. Clin. Obstet. Gynecol 41 (3), 597–603. [DOI] [PubMed] [Google Scholar]
- Grøndahl ML, Andersen CY, Bogstad J, Borgbo T, Boujida VH, Borup R, 2012. Specific genes are selectively expressed between cumulus and granulosa cells from individual human pre-ovulatory follicles. Mol. Hum. Reprod 18 (12), 572–584. [DOI] [PubMed] [Google Scholar]
- Haghighat M, Rafie SM, Dehghani SM, Fallahi GH, Nejabat M, 2007. Cyclic vomiting syndrome in children: experience with 181 cases from southern Iran. World. J. Gastroenterol 13 (12), 1833–1836, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JB, Yost NP, Wendel GD Jr., 2002. Acute renal failure in association with severe hyperemesis gravidarum. Obstet. Gynecol 100 (5 Pt 2), 1119–1121. [DOI] [PubMed] [Google Scholar]
- h, http://graphpad.com/quickcalcs/contingency1/, http://graphpad.com/quickcalcs/contingency1/(accessed 18.8.16).
- ht, http://www.fhi.no/moba-en, http://www.fhi.no/moba-en(accessed 18.8.16).
- htt, https://www.medcalc.org/calc/odds_ratio.php,https://www.medcalc.org/calc/odds_ratio.php (accessed 18.8.16).
- Howie BN, Donnelly P, Marchini J, 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap 3 Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. , 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Xu A, Tokmakejian S, Narayanan N, 2000. Thyroid hormone-induced overexpression of functional ryanodine receptors in the rabbit heart. Am. J. Physiol. Heart Circ. Physiol 278 (5), H1429–H1438. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, 2002. The human genome browser at UCSC. Genome Res. 12 (6), 996–1006 (accessed 18.8.16). https://genome.ucsc.edu/cgi-bin/hgGatewayGRCh38/hg38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- on behalf of MBRRACE- UK, 2014. In: Knight M, Kenyon S, Brocklehurst P, Neilson J, Shakespeare J, Kurinczuk JJ (Eds.), Saving Lives, Improving Mothers’ Care - Lessons Learned to Inform Future Maternity Care from the UK and Ireland Confidential Enquiries into Maternal Deaths and Morbidity 2009–12. National Perinatal Epidemiology Unit, University of Oxford, Oxford. [Google Scholar]
- Koop A, Goldmann P, Chen SR, Thieleczek R, Varséanyi M, 2008. ARVC-related mutations in divergent region 3 alter functional properties of the cardiac rya-nodinereceptor. Biophys. J 94 (12), 4668–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Wong SA, Li BU, Boles RG, 2015. NextGen nuclear DNA sequencing in cyclic vomiting syndrome reveals a significant association with the stress-induced calcium channel (RYR2). Neurogastroenterol. Motil 27 (7), 990–996. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R, 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGibbon KW, Fejzo MS, Mullin PM, 2015. Mortality secondary to hyper-emesis gravidarum: a case report. Women’s Health Gynecol 1 (2), 1–7. [Google Scholar]
- Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, Handal M, Haugen M, Høiseth G, Knudsen GP, Paltiel L, Schreuder P, Tambs K, Vold L, Stoltenberg C, 2016. Cohort profile update: the norwegian mother and child cohort study (MoBa). Int. J. Epidemiol 45 (2), 382–388. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome. Res 20,1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S, 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N, Deitel M, Lacy E, 1995. Splenic avulsion in a pregnant patient with vomiting. J. C. C 38, 464–465. [PubMed] [Google Scholar]
- Nilsen RM, Vollset SE, Gjessing HK, et al. , 2009. Self-selection and bias in a large prospective pregnancy cohort in Norway. Paediatr. Perinat. Epidemiol 23 (6), 597–608. [DOI] [PubMed] [Google Scholar]
- Omelka R, Vasícek D, Martiniakova M, Bulla J, Bauerové M, 2004. Simultaneous detection of malignant hyperthermia and genetic predisposition for improved litter size in pigs by multiplex PCR-RFLP. Folia Biol. (Krakow) 52 (1–2), 113–115. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC, 2007. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet 81 (accessed 18.8.16). http://pngu.mgh.harvard.edu/purcell/plink/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S, 2002. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y, Chen J, Li N, Zhang W, Feng L, Wang R, Hui R, Zhang S, Pu J, 2010. Common RyR2 variants associate with ventricular arrhythmias and sudden cardiac death in chronic heart failure. Clin. Sci. (Lond) 119 (5), 215–223, 4. [DOI] [PubMed] [Google Scholar]
- Santulli G, Marks AR, 2015. Essential roles of intracellular calcium release channels in muscle, brain, metabolism, and aging. Curr. Mol. Pharmacol 8 (2), 206–222. Review. [DOI] [PubMed] [Google Scholar]
- Santulli G, Pagano G, Sardu C, Xie W, Reiken S, D’Ascia SL, Cannone M, Marziliano N, Trimarco B, Guise TA, Lacampagne A, Marks AR, 2015. Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J. Clin. Invest 125 (5), 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Rosoff L, 1994. Pneumomediastinum and bilateral pneumothoraces in a patient with hyperemesis gravidarum. Chest 106, 1904–1906. [DOI] [PubMed] [Google Scholar]
- Soueid J, Kourtian S, Makhoul NJ, Makoukji J, Haddad S, Ghanem SS, Kobeissy F, Boustany RM, 2016. RYR2, PTDSS1 and AREG genes are implicated in a Lebanese population-based study of copy number variation in autism. Sci. Rep 6, 19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium, 2015. A global reference for human genetic variation, the 1000 Genomes Project Consortium. Nature 526, 68–74 (accessed 18.8.16). http://browser.1000genomes.org/index.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine BH, Jones C, Tyack AJ, 1980. Hyperemesis gravidarum due to thyrotoxicosis. Postgrad. Med. J 56 (660), 746–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellacott ID, Cooke EJA, James CE, 1988. Nausea and vomiting in early pregnancy. Int. J. Gynecol. Obstet 27, 57–62. [DOI] [PubMed] [Google Scholar]
- Vikanes A, Skjaerven R, Grjibovski AM, Gunnes N, Vangen S, Magnus P, 2010. Recurrence of hyperemesis gravidarum across generations: population based cohort study. B. M. J 29, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford TJ, Birzgalis AR, Lundell C, Farrington WT, 1993. Vomiting in pregnancy resulting in oesophageal perforation in a 15-year-old. J. Laryngol. Otol 107,1059–1060. [DOI] [PubMed] [Google Scholar]
- HYPERLINK “http://www.HelpHer.org“\o“http://www.HelpHer.org“www.HelpHer.org (accessed 18.8.16).
- Yoshida A, Takahashi M, Imagawa T, Shigekawa M, Takisawa H, Nakamura T, 1992. Phosphorylation of ryanodine receptors in rat myocytes during beta-adrenergic stimulation. J. Biochem 111 (2), 186–190. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cantor R, MacGibbon K, Romero R, Goodwin TM, Mullin P, et al. , 2011. Familial aggregation of hyperemesis gravidarum. A. J. O. G 204 (3), 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Hutchinson TE, Chebolu S, Darmani NA, 2014. Serotonin 5-HT3 receptor-mediated vomiting occurs via the activation of Ca2+/CaMKII-dependent ERK1/2 signaling in the least shrew (Cryptotis parva). PLoS One 9 (8), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]