The Pharmacogene Variation (PharmVar) Consortium is the successor of the Human Cytochrome P450 (CYP) Allele Nomenclature website that served the pharmacogenetics community by designating CYP star (*) alleles. The aim of PharmVar is to continue the mission of serving as an official allele designation authority for the global pharmacogenetics community.1 Herein, we describe the introduction of the first non‐CYP gene to PharmVar. Pharmacogenetic variation of NUDT15 plays a significant role in thiopurine response variability and toxicity.
The NUDT15 Gene
NUDT15 is a member of a large phosphatase protein family that shares a common NUDIX catalytic domain and metabolizes a wide range of nucleotide substrates (Table 1 ). Originally characterized as a pyrophosphatase, NUDT15 converts oxidized GTP to its monophosphate form, preventing the integration of the damaged purine nucleotides into DNA and subsequent mismatch repair. However, 8‐oxo‐GTP is a weak substrate for NUDT15 compared with its main metabolizer NUDT1; thus, the physiological functions of NUDT15 remain unclear. NUDT15 has been linked to xenobiotic drug metabolism in genome‐wide association studies of drug toxicity, and subsequent mechanistic studies have demonstrated that it plays a key role in the conversion of the active thiopurine metabolite thioguanosine triphosphate to thioguanosine monophosphate.2 NUDT15 variants encoding no or severely decreased function predispose patients to excessive thiopurine activation and hematopoietic toxicities when receiving this class of drugs for either benign (e.g., inflammatory bowel diseases) or malignant (e.g., acute lymphoblastic leukemia) conditions. Given the compelling underlying biology and clinical relevance of this pharmacogenetic association, there is a growing interest in preemptive NUDT15‐guided thiopurine dosing to avoid severe adverse events. The importance of including NUDT15 genetic information in dosing recommendations for preventing thiopurine toxicity is further evidenced by the updated Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline on TPMT‐guided thiopurine dosing recommendation that includes NUDT15 in addition to TPMT.3
Table 1.
Gene summary
| Gene data | Description |
|---|---|
| Alias |
NUDT15D (nudix nucleoside diphosphate linked moiety X–type motif 15), MTH2, MutT homolog 2 |
| Gene IDs |
HGNC: 23063 Entrez gene: 55270 Ensembl: ENSG00000136159 OMIM: 615792 UniProtKB: Q9NV35 PharmGKB accession ID: PA134963132 |
| Genomic RefSeq | NG_047021.1 |
| Transcript RefSeq | NM_018283.3 |
| Protein RefSeq | NP_060753.1 |
| LRG | NA |
NA, not applicable; LRG, Locus Reference Genomic record
The initial genome‐wide studies associated only a single nonsynonymous single‐nucleotide polymorphism in exon 3 of NUDT15 with thiopurine toxicity.4 There is, however, growing evidence that additional functional variants exist. A total of 16 coding region variants have been reported to date (Figure 1 ), most of which affect function and/or thermal stability of the NUDT15 protein. On the basis of NUDT15 pyrophosphatase activity with thioguanosine triphosphate as the substrate, haplotypes containing p.R139C (c.414C>T, rs116855232) or the p.G17_V18del (c.55_56insGAGTCG, rs869320766) variant showed a severe decrease in activity2 and were thus classified as “no function” alleles by CPIC.3
Figure 1.
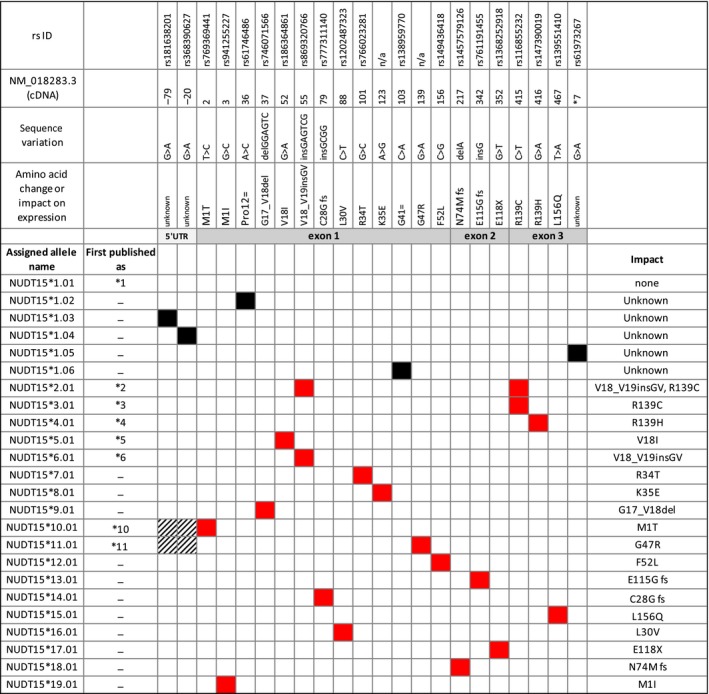
Allele nomenclature summary. The graph contains a graphical summary of all NUDT15 allelic variants described to date. Of the published variants now designated NUDT*1 through NUDT*19, eight have been published using star nomenclature (*1‐*6, *10, and *11); those designated *7 to *9 were named as such post hoc. Six novel haplotypes were designated *14 to *19. Red boxes indicate SNVs that confer an amino acid change; black boxes indicate SNVs in noncoding regions of unknown functional consequence or synonymous SNPs in coding regions. Gray shaded boxes indicate the gene regions not covered by sequencing. SNV rs IDs, their positions on the cDNA reference sequence, nucleotide changes, and impact (amino acid change, frameshift (fs), or stop codon (X)) within a haplotype are shown in the top panels and the right‐hand column. An extended figure with additional information, including references, and PharmVar IDs is provided as Supplemental Material .
There are substantial racial and ethnic differences in the population frequencies of NUDT15 variants. For example, alleles containing p.R139C (NUDT15*2 and NUDT153) are predominantly found among Asians, but have also been detected in US Hispanics, especially those with high Native American ancestry. In contrast, the allele containing a five‐nucleotide deletion in exon 1 (NUDT15*9) is almost exclusively found in patients with European ancestry.4
Why Nomenclature Is Needed
Clinical pharmacogenetic testing can be performed by numerous platforms and approaches, including targeted genotyping, sequencing, and array‐based technologies. Molecular genetic testing results typically use the Human Genome Variation Society (HGVS) nomenclature system for reporting sequence variants and the International System for Human Cytogenetic Nomenclature system for reporting copy number variants. Although haplotype nomenclature is available within the HGVS framework, results of pharmacogenetic testing for genes involved in drug metabolism, such as CYPs, are most commonly reported using the star allele (*) nomenclature system. In addition to the identified variants and/or haplotypes, the interpretation of these results includes variant allele function and inferred phenotype, which can vary between testing laboratories. As such, it is imperative to establish clearly defined haplotype nomenclature as new pharmacogenes are discovered and subsequently offered by clinical testing laboratories and included in dosing guidelines.
Because of its pivotal role in thiopurine metabolism, clinical laboratories will likely incorporate preemptive NUDT15 genotyping in concert with TPMT to avoid potentially life‐threatening toxicity in patients carrying TPMT and/or NUDT15 risk alleles. Although nomenclature databases focusing on haplotype definitions exist for TPMT, UGTs, and NATs, there are no centralized resources for other important pharmacogenes, including NUDT15, DPYD, or drug transporters. Standardized allele nomenclature will allow all professionals “to speak the same language” and facilitate the communication and dissemination of allelic variation. As discussed by Kalman et al.,5 there is a need to increase transparency and standardization of clinical pharmacogenetic result reporting as well as the dissemination of research findings. To that end, PharmVar is now developing nomenclature for important non‐CYP pharmacogenes, such as NUDT15, with an initial focus on genes with available CPIC guidelines.
Some of the NUDT15 allelic variants were assigned star allele numbers when published (NUDT15*1‐NUDT15*6, NUDT15*10, and NUDT15*11), whereas others were reported without a star allele designation or designations were assigned post hoc (NUDT15*7‐ NUDT15*9) (Figure 1 ). The absence of a centralized and widely accepted naming system not only makes it difficult to facilitate standardized test reporting, but also to interpret test results and compare research findings. Furthermore, reports from different clinical laboratories may not be consistent and may lead to confusion regarding dosing recommendations. Consequently, standardized nomenclature, ideally established when allelic variation is first described, and subsequent consistent use of agreed‐on terminology are pivotal for clinical reporting of test results to reduce potential errors in drug selection and dosing.
PharmVar Nomenclature for NUDT15
Experts representing global research, clinical testing, and implementation interests were recruited from PharmVar members to serve on the NUDT15 expert panel and tasked to establish a formal nomenclature, according to PharmVar allele designation principles. The panel also included a Pharmacogenomics Knowledge Base/CPIC representative to ensure that the nomenclature is consistent with CPIC guidelines and to facilitate dissemination to a greater audience through the PharmGKB knowledgebase as well as other databases, such as ClinGen. The panel met via teleconferences and communicated by email. A literature search was performed on the PubMed database (beginning of PubMed to July 13, 2018, keyword “NUDT15”) to compile records for published allelic variants. Most of the 38 articles only tested for p.R139C; there were only three articles describing allelic variants (Figure S1 ).
Because of the relatively few known allelic variants and the fact that most haplotypes are characterized by a single defining nucleotide variant, the review of haplotypes with star allele names was straightforward. Discussions centered mostly on the alleles initially published as NUDT15*2 and NUDT15*3, carrying either p.R139C alone or p.R139C and p.G17‐V18ins in cis. NUDT15*2 and NUDT15*3 share c.415C>T (p.R139C), which causes a severe decrease in in vitro function assays and predisposes carriers to a high risk of developing toxicity when exposed to thiopurines. Hence, the question was raised whether c.415C>T dictates the function of both alleles and, therefore, the allele published as NUDT15*2 should be designated as a suballele of NUDT15*3 to be consistent with PharmVar criteria for allele designation. According to those criteria, all haplotypes/alleles that carry a sequence variation causing no function are listed under the same star number (see ALLELE DESIGNATION CRITERIA under the SUBMISSIONS menu tab). The panel also discussed concerns regarding a potential merge of the two alleles into a single star allele and suballele designation because this may cause confusion with existing literature and among the clinical laboratories that already offer NUDT15 testing, an argument that ultimately drove the decision to keep these two designations separate. More important, this issue of NUDT15 star allele revision underscores the need to develop systematic haplotype nomenclature when variants are initially reported.
Figure 1 provides an overview of all variants published or submitted to PharmVar at the time this report was prepared. Allele frequencies can be accessed on the PharmGKB website at http://www.pharmgkb.org/page/nudt15RefMaterials. Function is assigned on the basis of in vitro activity data and/or thermal stability (details are available through the NUDT15 READ ME document on the PharmVar NUDT15 gene page and Figure S1 ). Allelic variants containing a frameshift or premature stop codon are predicted to be nonfunctional. The panel's recommendation was reviewed and approved by the PharmVar Steering Committee.
PharmVar maps all coordinates of NUDT15 allelic variation to the most recent genomic and transcript reference sequences, GRCh37 and GRCh38 (Table 1 ). The NUDT15 gene page which was released in September 2018 includes READ ME and CHANGE LOG documents that provide important information, including details regarding allele functionality; these documents will be updated as new information becomes available. All updates will also be coordinated with PharmGKB to provide unified information to the pharmacogenomics community. Furthermore, PharmVar will be announcing the incorporation of standardized NUDT15 nomenclature to stakeholders via blogs, social media, and targeted announcements/email communications to promote the use of standardized nomenclature because testing is being offered by an increasing number of laboratories.
Novel NUDT15 Allelic Variants
Since the inception of the NUDT15 expert panel in April 2018, six novel previously unpublished allelic variants containing nonsynonymous sequence variations were submitted, reviewed, and designated NUDT15*14 through NUDT15*19 (Figure 1 ). An additional five haplotypes were designated as NUDT15*1 suballeles because of the presence of single‐nucleotide variants in noncoding regions or harboring synonymous single‐nucleotide variants. It remains unknown whether these variants have any functional consequences.
PharmVar strongly encourages submissions by investigators before publication (submission requirements and details of how to submit are provided at http://www.PharmVar.org). All information is regarded as confidential and will not be shared outside of the expert panel and Steering Committee. Designated haplotypes can be held for up to 6 months to allow timing of release on PharmVar and through article publication. PharmVar also welcomes direct submissions to make information available to the public independent of a publication.
Funding
This work was funded by the National Institutes of Health for Pharmacogene Variation (R24GM123930) (A.G., and N.A.M.) and PharmGKB (R24GM61374) (M.W.‐C, T.E.K.), the National Institutes of Health (P50GM115279 and R01GM118578) (J.J.Y.), the European Commission Horizon 2020 UPGx grant (668353), Robert Bosch Stiftung (Stuttgart, Germany) (M.S. and E.S.), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (401.820/2014‐9) (G.S.‐K.).
Conflict of Interest
A.J.T.'s efforts are supported in part by RPRD Diagnostics, an independent clinical laboratory offering pharmacogenetic testing services, including NUDT15; and S.A.S. is a paid employee of Sema4, a clinical laboratory that performs pharmacogenetic testing, including NUDT15. None of the other authors declare any conflicts of interest.
Supporting information
Figure S1. NUDT15 nomenclature.
Acknowledgments
We are grateful to the members of the Pharmacogene Variation Steering Committee for critically reviewing the manuscript.
References
- 1. Gaedigk, A. et al The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moriyama, T. et al NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 48, 367–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Relling, M.V. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin. Pharmacol. Ther. 105, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang, J.J. et al Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. 33, 1235–1242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalman, L.V. et al Pharmacogenetic allele nomenclature: international workgroup recommendations for test tesult teporting. Clin. Pharmacol. Ther. 99, 172–185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. NUDT15 nomenclature.


