Abstract
Members of the SidE effector family from Legionella pneumophila represent a new paradigm in the ubiquitin world. These enzymes catalyze ubiquitination of target proteins via a mechanism different from conventional E1-E2-E3 biochemistry and play important roles in L. pneumophila virulence. They combine mono-ADP-ribosylation and phosphodiesterase activities to attach ubiquitin onto substrates, in great contrast to the orthodox pathway. A series of recent structural and mechanistic studies have shed new light into the action of these enzymes. Herein we summarize the key insights into these proteins’ structure and function, emphasizing their modular nature, and discuss the biochemical implications of these proteins as well as areas of further exploration.
Keywords: ubiquitination, bacteria, ADP-ribosylation, signaling, Legionella, effectors
Ubiquitination and the SidE family
The post-translational modification known as ubiquitination, vital for cellular function and signaling in eukaryotic organisms, is defined as the attachment of the small protein ubiquitin (Ub) through its C-terminus onto the lysine residue of a protein substrate via an isopeptide bond [1]. Ubiquitin is attached to the substrate by the coordinated, ATP-driven action of three enzymes termed as the Ub activating E1, Ub conjugating E2, and Ub E3 ligase that catalyze a cascade of reactions involving activation and sequential transfer of Ub, ultimately resulting in isopeptide linkage of Ub with target Lys residues. Addition of a single Ub via this E1-E2-E3 cascade results in monoubiquitination, which is often further elaborated by successive addition of more Ub groups to produce polyubiquitin chains in which the monomeric Ub units are isopeptide linked via one of the seven internal Lys residues, or the N-terminal Met of a preceding unit with the C-terminus of a succeeding one. Chains formed between different residues represent distinct biological signals specifying distinct biological outcomes, forming the foundation of a complex signaling network based on Ub modification [2]. This modification plays a major role in a variety of cellular functions, including proteostasis, immunity, and trafficking among others. Due to this major signaling role in cellular immunity, the Ub system is often the target of interference and manipulation by invading pathogens seeking to evade host responses [3]. This mode of host-pathogen interplay has been validated through the discovery of a wide array of bacterial effectors that interact with the Ub system [4]. It is to be noted that bacteria lack a Ub system of their own; nevertheless, effectors in bacterial genomes have been found to perform actions including deubiquitination [5,6], Ub ligation [7], and Ub modification [8].
Intriguingly, some bacterial proteins have evolved to interact with Ub through unconventional mechanisms never before seen in nature. The most striking example of this in the literature to date is the noncanonical ubiquitination of host substrates by the Legionella pneumophila (L.p.) SidE effector family. The SidE family contains four large, modular, and highly conserved proteins SidE, SdeA, SdeB, and SdeC that are required for optimal Legionella virulence. While the importance of this effector family in virulence has been known for over a decade, their biochemical function remained elusive until recently. A key study reported that these four proteins catalyzed ubiquitination of host substrates using an unprecedented mechanism which differs significantly from the established route of ubiquitination in eukaryotes and represents the first and thus far only known example of ubiquitination occurring independently of the well-known E1-E2-E3 cascade [9]. The initial discovery of this new mechanism has since sparked intense worldwide research efforts in the past two years which have yielded a bounty of biological and biochemical insights into this process [10,11].
In this Review, we discuss the structural basis of this novel ubiquitination process, focusing on newly presented crystallographic data on the SidE proteins and their substrates (Table 1), and provide an overview of their separate domains and their interplay [12–16]. We also examine how these enzymes recognize their substrates and provide possible future directions and applications of this process, including its relevance to Legionella pathogenesis.
Table 1:
Summary of crystal structures in PDB of SdeA and homologs, with bound ligands and residue numbers.
| Protein | Construct | Domains | Ligand(s) | PDB ID |
|---|---|---|---|---|
| SdeA | 1–193 | DUB | Ubiquitin-VME | 5CRA |
| SdeA | 1–193 C118A | DUB | 5CRB | |
| SdeA | 1–163 | DUB (partial) | 5CRC | |
| SidE | 222–1057 | mART, PDE, CC (partial) | 5ZQ2 | |
| SidE | 222–1057 | mART, PDE, CC (partial) | Ubiquitin (R42A) | 5ZQ5 |
| SidE | 222–1057 | mART, PDE, CC (partial) | Ubiquitin (R42A), NAD+ | 5ZQ7 |
| SidE | 222–1057 | mART, PDE, CC (partial) | Ubiquitin (R42A), ADPR | 5ZQ6 |
| SidE | 222–589 | PDE | Ubiquitin | 5ZQ3 |
| SidE | 222–589 | PDE | Ubiquitin, ADPR | 5ZQ4 |
| SdeA | 231–1190 | mART, PDE, CC (partial) | 5YIM | |
| SdeA | 231–1190 | mART, PDE, CC (partial) | Ubiquitin | 5YIK |
| SdeA | 231–1190 | mART, PDE, CC (partial) | Ubiquitin, NADH | 5YIJ |
| SdeA | 211–910 | PDE, mART | 6B7Q | |
| SdeD | 6B7P | |||
| SdeD | 1–341 | PDE | Ubiquitin | 6B7M |
| SdeD | 1–341 | PDE | ADPR-Ubiquitin, Ubiquitin | 6B7O |
| SdeA | 213–907 | PDE, mART | 6G0C | |
| SdeA | 765–905 | mART (partial) | 5YSJ | |
| SdeA | 765–905 E860/E862A | mART (partial) | 5YSK | |
| SdeA | 765–905 E860/E862A | mART (partial) | NAD+ | 5YSI |
Early work and characterization of SidE enzymatic activity
Of all the known pathogenic bacteria, L.p. has the largest repertoire of effectors, maintaining an arsenal of over 330 individual proteins that is delivered into the host cell through its Dot/Icm secretion system [17,18]. While many of these effectors appear to be functionally redundant, a strain of L.p. lacking the SidE family exhibits impaired growth of the bacteria within host cells [19]. Further, expression of SidE proteins in yeast and mammalian cells leads to cytotoxic effects [20].
In pursuit of biochemical functions of this effector family, it was first noticed that the first roughly 200 residues of the SidE proteins harbored a deubiquitinase (DUB) domain (hereafter referred to as SdeADUB), capable of removing isopeptide-linked Ub from protein substrates as well as Ub chains with a distinct preference for Lys-63 linked polyubiquitin chains [21]. The structure of SdeADUB revealed a core fold that was first characterized in the yeast desumoylase Ulp1 and human deneddylase (DEN1) [22]. This so-called Ulp1 fold is also shared by other bacterial DUBs and deneddylases in the CE-clan of prokaryotic proteases [23,24]. However, a strain of L.p. lacking the SidE family regained full virulence when made to express SdeA with an inactive DUB mutation (C118A). This finding suggested that the DUB domain was not needed for bacterial virulence in the host systems used for these experiments.
In pursuit of further biochemical characterization, SidE proteins were shown to possess an unprecedented, all-in-one ubiquitinating activity outside the DUB domain, an activity that requires NAD+ and a putative catalytic motif typically found in bacterial mono-ADP ribosyltransferase (mART) toxins [9]. Indeed, Ub is ADP ribosylated at Arg42 forming ADPR-Ub as a reactive intermediate [9]. Further biochemical characterization revealed a two-step mechanism for SidE enzymes [10,11]. The ADPR-Ub intermediate produced in the first step is further processed by a novel phosphodiesterase (PDE) domain leading to phosphor-ribosyl (PR) linked Ub conjugation to target Ser residues. This review will focus on the ubiquitinating activity of these proteins, which is distinct from the DUB activity and important for bacterial virulence.
The SidE ligase catalytic machinery
The core of the SidE family ubiquitination machinery comprises two catalytic units that work in sequence to catalyze PR-linked Ub attachment onto substrate Ser residues (Figure 1). In stark contrast to the conventional ATP-driven E1-E2-E3 cascade, the SdeA ubiquitination catalysis requires the nucleotide cofactor NAD+. Overall, the underlying chemistry is strikingly different between the SidE all-in-one ubiquitination machinery and the three-enzyme eukaryotic system. The SidE ligase machinery carries out Ub activation using a mono-ADP ribosyl transferase reaction whereby the ADP ribose group of the nucleotide cofactor is covalently added to Arg42 of Ub forming ADPR-Ub. This enzymatic chemistry follows the same mechanism as seen in the well-known cholera toxin (CT) group of bacterial toxins that modify host proteins through mono-ADP ribosylation of Arg [25]. The similarity in enzymatic chemistry is due to sharing of the characteristic RSE catalytic motif with the bacterial toxins (Arg for positioning of NAD+ in the active site, Ser for stabilizing NAD+ in its binding pocket and Glu of the Glu(Gln)-X-Glu triad for promoting nucleophilic attack by the acceptor Arg), examples of which include Iota toxin from Clostridium perfringens [25], HopU1 of Pseudomonas syringae [26], and scabin from Streptomyces scabies [27]. The SidE mART domain (SidEmART) spans approximately between residues 600–900 of the protein. An important distinguishing feature of SidEmART is its ability to selectively accept Ub as the substrate for ADP ribosylation, which implies the presence of Ub recognition elements as a part of the mART catalytic unit. The toxin mARTs catalyze ADP ribosylation resulting in a stable modification of the host protein, whereas SidEmART catalyzes the formation of a species that undergoes further reaction.
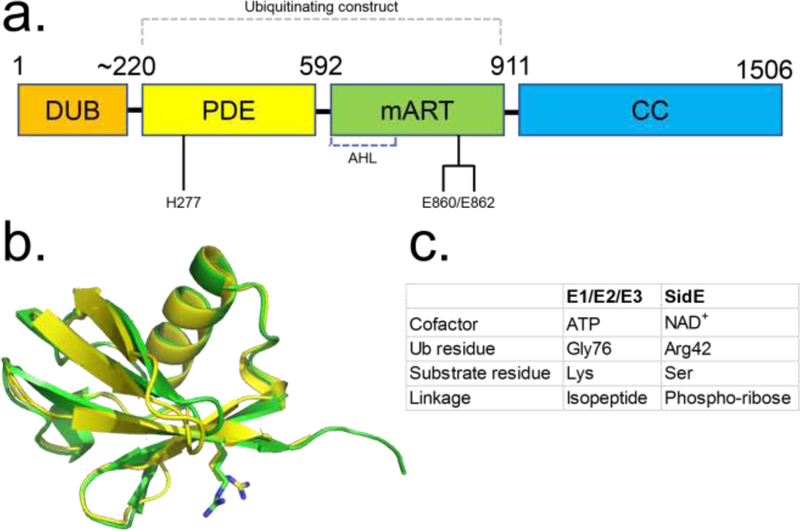
Figure 1.
Diagram depicting the general domain organization of SidE proteins. (A) Key catalytic residues are indicated (Residue numbers pertain to SdeA). (B) Structural alignment of Ub (green ribbon) and NEDD8 (yellow) with R42 residues shown in sticks. (C) Comparison of key aspects of E1-E2-E3 and SidE-mediated ubiquitination reactions. Abbreviations: AHL, α-helical lobe; CC, coiled-coil; DUB, deubiquitinase; mART, mono-ADP ribosyltransferase; PDE, phosphodiesterase; Ub, ubiquitin.
Between the DUB and mART domains lies a distinct region that shares sequence similarity with another Legionella effector, the nucleotide binding protein known as lpg1496 of unknown function, and also with other known bacterial phosphodiesterases (PDEs) [10]. This PDE domain lies approximately between residues 200–600 of the protein (SidEPDE). Outside the L.p. genome, the closest similarity of this domain is with a cyclic GMP dinucleotide PDE which shares 23% sequence identity, including the key conserved catalytic residues - a pair of His’s and a Glu (His277, His407 and Glu340, SdeA numbering) [28]. Despite being referred to as a PDE the actual reaction catalyzed by this domain from the point of view of ubiquitination chemistry is akin to a phosphotransferase activity (rather than a hydrolase activity) wherein a substituted phosphate group as phospho-ribosyl-Ub is transferred from the ADPR-Ub donor to a substrate hydroxyl group. Mechanistically this is similar in part to the catalytic activity of certain bacterial His kinases [29,30]. In fact, the enzymatic chemistry in SidEPDE catalysis does involve a substituted phospho-His intermediate where a His is phosphorylated transiently before the attack of the substrate Ser residue [14].
While the sequential activity of both domains is required for substrate ubiquitination, each domain can function independently of the other; with in vitro experiments showing that the mART domain by itself is capable of producing ADPR-Ub, and the PDE domain is able to use the intermediate in trans to ubiquitinate substrates or produce PR-Ub [12]. However, close interactions between the two domains as found in the full-length core construct, SidECore (defined as the SidE segment encompassing the PDE and mART domains) are required for maximal activity [12,14]. Conserved residues at the interface of the mART and PDE domains stabilize inter-domain packing that provides better catalytic efficiency of SidECore.
Beyond the mART domain (residues 900 to end), lies a C-terminal coiled-coil (CC) domain that ultimately is not required for catalytic activity in vitro but may play a role ultimately in localization of the enzymes in host cells, it also carries the translocation signals recognized by the Dot/Icm protein secretion system. It has been shown that a portion of the CC domain is used in interactions with the components of the translocation machinery [15], presumably for injection of the effector into the host cytoplasm.
Overview of structural data
Four independent studies have been thus far published in 2018 reporting crystal structures of different core constructs of SidE proteins, mainly SdeA, either in its apo form or in complex with various ligands, including NAD+ (and NADH), Ub (and its R42A mutant), and the intermediate ADPR-Ub (Table 1) [12–15]. Additionally, a more recent paper of the crystal structure of the mART domain has contributed additional insights into the mechanism of catalysis by the SidE ligase [16]. The crystal structures confirmed the biochemical prediction of SidE proteins possessing distinct mART, PDE, and CC domains (Figure 2) [10]. In addition to core constructs, some of these structures included parts of the CC domain, which appears to be conformationally flexible relative to the PDE-mART core adopting different orientations in a length-different manner (different truncations of the CC domain were captured but not the entire predicted domain). Although not absolutely essential for ubiquitination activity it may play a role in relative positioning of the PDE domain with respect to the mART domain thereby supporting appropriate interdomain packing for optimal activity. In one construct, the SidE CC domain forms a domain-swapped dimer in crystal and in solution [12]. Nevertheless, the PDE and mART domains are invariantly placed next to each other in the structure. The active sites of the two domains face in different directions with a substantial spatial separation indicating the independence of the two catalytic centers (Figure 2b).
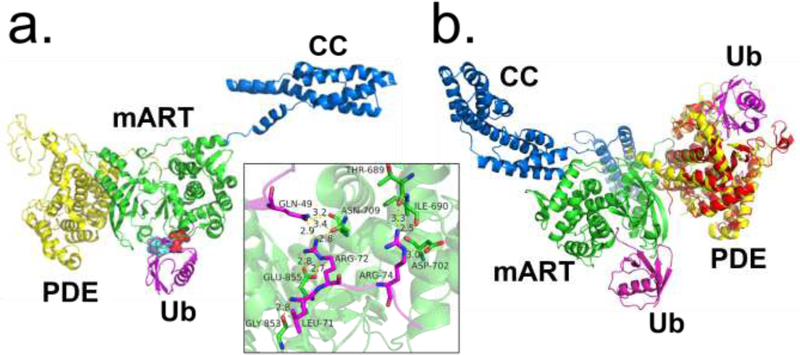
Figure 2.
Ubiquitin recognition by the SidE ligase. (A) Structure of SidE in complex with ubiquitin (Ub) (magenta) at mART site with key interacting residues of Ub highlighted. The hydrophobic patch of Ub is depicted in cyan, and key Arg residues depicted in red. Inset depicts polar/charge interactions between the SidE mART domain and Ub. (B) Structure of SdeA in complex with Ub at mART site (magenta) aligned with structure of PDE domain of SdeD (red) in complex with ADPR-Ub (magenta). The SdeD L.p. effector has a domain homologous to the SidE PDE domain with the same catalytic residues but lacks an mART domain. Abbreviations: L.p., Legionella pneumophila; mART, mono-ADP ribosyltransferase; PDE, phosphodiesterase; ADPR-Ub, ADP ribosylated ubiquitin.
mART domain and the alpha-helical lobe
The structure of SdeA mART domain features two distinct lobes typically seen in bacterial mART enzymes, a primarily α-helical lobe (AHL, between approximately residues 600–750) and a main lobe (mART core) comprising a β-sandwich structure [25]. The AHL in other bacterial mART proteins lies in close proximity to the core creating a binding pocket for NAD+ which is lined with residues from both lobes. In the crystal structures of two SdeA constructs lacking CC domain, the AHL is observed protruding out from the remainder of the mART core connected by a flexible hinge in what seems to be an unproductive arrangement of the two lobes. Since these constructs are catalytically active AHL likely moves closer to the mART core during catalysis. However, in constructs containing parts of the CC region the AHL is observed in a canonical arrangement as seen in bacterial mARTs. Inclusion of the CC domain does seem to stabilize a productive form of mART domain in solution, with the full-length SdeA construct showing higher activity than the constructs lacking the CC region [14]. The key catalytic residues of the RSE motif are present in the main lobe, which shares substantial sequence similarity with the bacterial mART toxin, HopU1 (approx. 21%) [26]. In productive arrangement the NAD+ binding pocket is located within a cleft between the two lobes. Three co-crystal structures (NAD+ and NADH bound with mART domain) reveal key contacts between the nucleotide and residues lining the NAD+ binding pocket [12,15,16]. (Figure 3, Key Figure) NAD+ is held in a position poised for reaction, with the nucleotide adopting a strained-ring conformation in which the nicotinamide group (NAM) is forced to stack against the AMP group. Similar strained conformation of NAD+ has been observed as a key catalytic feature in mART structures of other bacterial toxins, where the strain conformation has been proposed to facilitate the nucleophilic attack of the substrate Arg residue through an SN1 mechanism, described as SN1-strain alleviation mechanism [25]. Ub has been captured bound to the mART domain in two co-crystal structures, also with bound nucleotide [12,15]. Interestingly, Ub captured in the co-crystal structures appear to represent a pre-catalytic substrate recognition state with the actual nucleophile Arg42 facing away from the C1′ ribose attack center (the ribose group attached to NAM in NAD+, riboseNic, Figures 3a and 3b) with a distance of 11□Å between the NωH group and the C1′ atom. Instead, Arg72 is closer to the C1′-ribose center as if it were the true nucleophile. Arg72 does play an important role in substrate recognition, however during the actual reaction it must be repositioned to allow Arg42 to take its place proximal to the reaction center. It thus appears that Ub recognition might proceed in at least two distinct steps, an initial engagement, which places Arg72 proximal to the C1-riboseNAM atom, followed by a rearrangement to bring the nucleophilic Arg42 adjacent to the ribose moiety. Additional structural work will be required to capture the correct catalytic state of Ub in its binding site in SdeA mART domain.
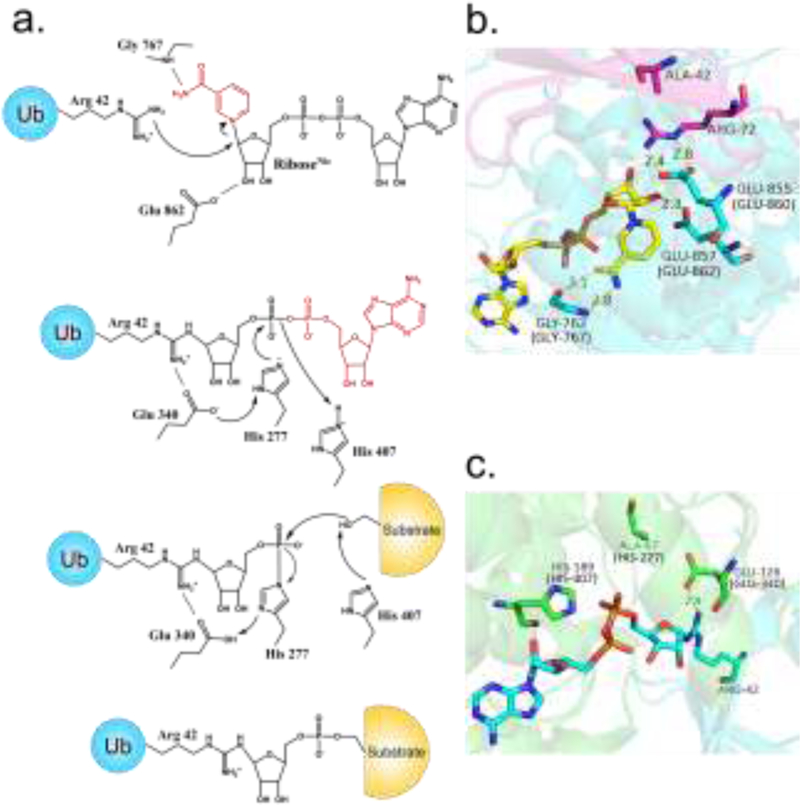
Figure 3, Key Figure.
The proposed mechanism of SidE-catalyzed ubiquitination. (A) Key residues are included (residue numbers pertain to SdeA). (B) Structure of SidE bound to NAD+ and Ub R42A, depicting a likely pre-catalytic state analogous to ADP ribosylation of R42, corresponding to the scheme in A. (C) Structure of SdeD bound to ADPR-Ub, analogous to the phospho-transfer step of the proposed mechanism catalyzed by the PDE domain. (SdeA residue numbers are included in parentheses). Abbreviations: ADPR-Ub, ADP ribosylated ubiquitin; Nic, nicotinamide; Ub, ubiquitin.
Ubiquitin recognition by SidE mART
The Ub bound structures reveal an overall position for Ub that is consistent with reaction with NAD+ for the ADP ribosylation reaction [12,15]. Ub approaches the mART center by facing the NAD+ cleft and is perched on a concave surface contributed by both the AHL and main lobes. The AHL lobe plays a critical role in Ub recognition alongside the catalytic motif of the main lobe, with major interactions occurring between Ub C-terminal tail residues Arg72 and Arg74 and two acidic pockets in SdeA comprised of the catalytic Glu860/Glu862 (main lobe) and the AHL Asp691/Asp707, respectively. Additional interactions of the Ub’s I44 patch (a hydrophobic patch of three side chains Leu8/Ile44/Val70, widely used in Ub recognition by eukaryotic enzymes in the Ub pathway) with a complementary hydrophobic patch on the main-lobe (Val822/Ile826/Phe827) contribute to the interface and the observed orientation of Ub in its binding site (Figure 2a). As noted earlier, the actual catalytic state would require a reorientation of Ub to bring R42 to face the NAD+ binding pocket. Mutation of Arg72 or Arg74 results in significant loss of mART activity and the interaction involving Arg72 may serve as a determinant of selectivity of SidE mART for Ub over NEDD8. In fact, mutation of the Ala72 to Arg in NEDD8 results in appreciable ADP ribosylation of NEDD8 [10]. Thus, interactions with Arg72 may contribute to substrate selection by SidE mART domain.
The last two Gly residues (Gly75-Gly76) and the C-terminal carboxylate group are not engaged and are solvent exposed [12,15] which explains previous biochemical results showing Ala substitution at the Gly75-Gly76 segment and addition of another Ub group, as in di-Ub, are tolerated in mART function [9,31].
PDE domain
The structure of the PDE domain reveals discernible similarities with other PDE enzymes at the same time revealing unique features required for a phospho-transfer reaction. A key feature specific to the SidE family is a cap lobe, which is necessary for Ub binding and critical for activity. In SdeA, it had been known that mutation of His277 or His407 caused a loss of PDE activity [10]. When His407 was changed to Asn, reaction with Ub and NAD+ caused the accumulation of a stable intermediate between His277 and the phosphate group of PR-Ub [14]. These observations strongly suggest that H277 acts as a nucleophile to attack ADPR-Ub, further supported by a hydrogen bonding interaction with nearby Glu340 in the crystal structure with the latter serving to stabilize the deprotonated form of the imidazole group (the nucleophile) [15]. A possible double role for H407 has been proposed: first, as a proton donor to promote the departure of the AMP group of ADPR-Ub leading to the H277-PR-Ub intermediate, and second, as a mediator of the attack of a substrate Ser residue on the same intermediate (Figure 3a) [13,14]. This proposed mechanism up to AMP departure is reminiscent of many bacterial histidine kinases [29]; in SidE catalysis the His-PR bond is further subjected to an additional nucleophilic attack by a substrate Ser residue.
Ub (ADPR-Ub) recognition
One study resolved the structure of the PDE domain of SidE (222–589) in complex with Ub, revealing that a stretch of N-terminal residues Lys6-Thr9 of Ub were a major site of interaction [12]. Another structure of the PDE domain bound to ADPR found that it also forms extensive contacts with its ADP moiety, with the orientation of the catalytically relevant His residues in agreement with the proposed mechanism above (Figure 3c). A co-crystal structure of ADPR-Ub bound to a related Legionella effector SdeD also agreed with these results [13]. SdeD, while sharing a similar PDE domain including the same three catalytic residues (His-His-Glu) with the SidE family, lacks DUB, mART, and CC domains, and therefore its role in a biological setting remains unclear. It is noteworthy that the Ub binding sites of the mART and PDE domains are not near one another (Figure 2b); and further studies will be required to examine the mechanism by which ADPR-Ub moves to the PDE domain for the second step of catalysis.
CC domain
An enigmatic region of the SidE proteins has been the C-terminal domain. Interestingly, structural data have revealed that it likely plays a major role in AHL stabilization. A comparison of crystal structures of SidE family members shows that the AHL is positioned away from the mART core in structures where the CC domain is absent [12–15]. (Figure 4a) This conformation is likely unfavorable for Ub binding. Biochemical analysis shows that the CC domain is required for optimum mART activity, possibly via stabilization of the AHL in a productive arrangement with respect to the mART core, supported by structural data showing that a combination of electrostatic and hydrophobic interactions hold together the AHL and the CC domain (Figure 4b), as well as another set of interactions between the mART core and the CC domain (Figure 4c) [12,15]. However, further studies will be required to determine the full extent of CC-mART and possible CC-PDE domain interactions.
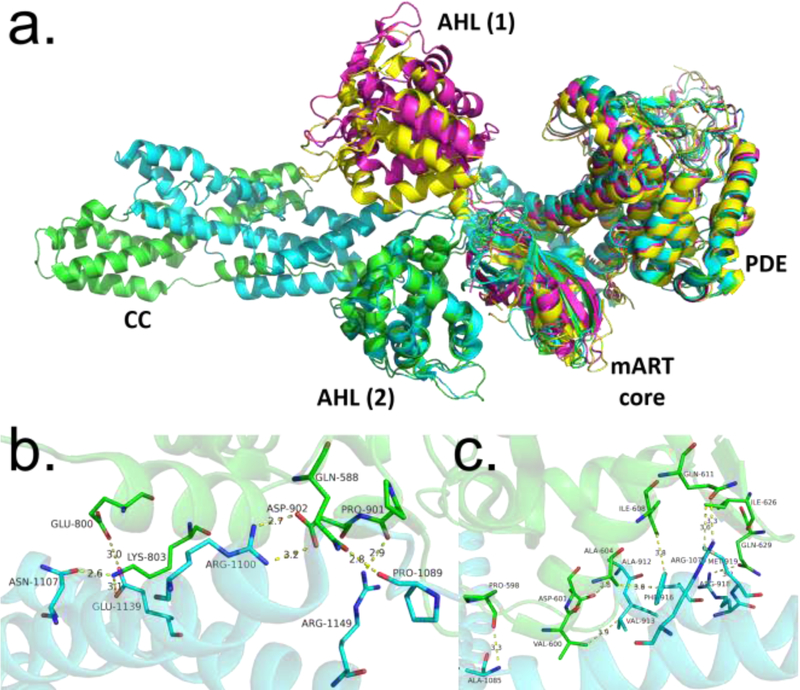
Figure 4.
Comparison of SidE and SdeA structures. (A) Structural alignment of various constructs of SidE proteins (green: SidE222−1057, cyan: SdeA231−1190, magenta: SdeA211−910, yellow: SdeA213–907), with domains labeled. AHL (1) and AHL (2) refer to two different orientations of the AHL domain observed in crystal structures of the SidE constructs, with the latter corresponding to a productive orientation. (B) Interactions between mART core and CC domain of SdeA. (C) Interactions between mART AHL and CC domain. Abbreviations: AHL, α-helical lobe; CC, coiled-coil; mART, mono-ADP ribosyltransferase.
Another role for the CC domain was investigated through binding assays, which showed that SdeA1101−1350 was able to bind tightly to IcmS-IcmW-DotL; a Legionella adaptor protein complex involved in secretion of effectors [15,32,33].
Interestingly, one study resolved a crystal structure of SdeA with two Ub moieties bound to the CC domain [15]. No previous evidence exists of the CC domain directly able to bind Ub, therefore the biological and biochemical relevance of this phenomenon will need to be revisited in a further study. The CC domain appears to also a play a role in promoting dimer formation of SidE and some SdeA constructs, the significance of which is not known. Two of the studies found that SidE222−1057 and SdeAFL constructs form dimers in solution [12,14], where a third study found that SdeA231–1190 is a monomer [15]. The basis for this conflict in SdeA may be truncation of the CC domain, as a truncated SidE construct behaved as a monomer in solution compared to the longer construct.
Substrate recognition
Recently, great advances have been made in our understanding of how SidE proteins identify their protein substrates. Initially, five substrates had been identified: Rab1a, Rab30, Rab33b, Rab6a, and Rtn4 [9,11]. Additionally, the SidE proteins self-ubiquitinate. Two other proteins, Rab5a and Rac1, were intriguingly found not to be ubiquitinated despite considerable structural similarity to the four other Rab substrates [9]. Mass spectrometric analysis identified Ser154 of Rab33b as a site of ubiquitination [10]. However, mutation of Ser154 to Ala failed to prevent ubiquitination, suggesting the presence of additional sites. New data have shown that for substrate ubiquitination, SidE proteins do not appear to have a structural preference [12,14]. Instead, they are able to ubiquitinate Ser residues that are part of any unstructured region that can fit into the PDE groove containing the catalytic site. For example, Ser residues near the N-terminus of the known substrate proteins were found to be major targets. This hypothesis was further validated by the fact that one can transform a non-substrate protein like Rac1 into a robust substrate by adding a flexible Ser-containing region [12]. Such manipulation even allowed structurally dissimilar proteins such as Ub and GST (glutathione S-transferase) to be ubiquitinated by SdeA. According to this model, the originally identified Ser154 of Rab33b is likely targeted due to its location in the flexible G4 loop region of the protein [34], but the primary site is the N-terminus, which presents a larger unstructured region, with many more Ser residues. Further, by screening Rtn4 peptides, hydrophobic residues around the target Ser appeared to improve ubiquitination efficiency [14]. These new insights suggest that SidE proteins’ localization rather than structure will dictate their true biological targets. The ability to add a monoubiquitination site onto a wide variety of proteins or peptides through a flexible Ser-containing region may also prove a valuable chemical biology tool in future studies.
Concluding Remarks
One eminent feature of the hundreds of effectors of L. pneumophila is that few of them are necessary for virulence in commonly used host systems [35]. Members of the SidE family display clear functional redundancy because the bacterium displays a defect in intracellular growth in protozoan hosts only when all members of the family were deleted, and such a defect can be complemented by any member of the family [19,20]. Deletion of sidEs caused a delay in the conversion of its vacuolar membrane into an ER-like compartment, which is the result of ubiquitination of multiple proteins involved in ER structure and function. The fact that SidEs attack structurally diverse proteins associated with the ER such as Rab small GTPases and reticulon 4 (Rtn4) suggests a large repertoire of their cellular targets [11]. Further studies are necessary to obtain a more complete list of substrates of SidEs by taking advantage of Ub variants that cannot be used by the canonical mechanism (see Outstanding Questions). Ubiquitination of Rtn4 results in the formation of ER tubules and its enrichment on the membrane of the bacterial vacuole, but the effects on the activity Rab proteins are less clear because modified Rab33b only is marginally defective in GTP hydrolysis [9]. Thus, it is unclear how ubiquitination induced by SidEs changes the function of their targets and how such changes benefit the bacterium. Another intriguing question is whether ubiquitination driven by ADP-ribosylation also exists in eukaryotes and other pathogens, or it is exclusive to the SidE family. Indeed, eukaryotes including humans possess a family of ADP-ribosylating enzymes named ectoARTs that contain the RSE motif, and which are related to the RSE bacterial mART toxins [9]. The better-studied poly (ADP-ribose) polymerase (PARP) family in eukaryotes is also related, albeit more distantly [36]. However, it has yet to be determined whether these eukaryotic enzymes act only alone or are part of a novel ubiquitination scheme. Answers to these questions will surely expand our current appreciation of the role of ubiquitin in cell signaling and disease development.
Highlights.
The SidE family of Legionella pneumophila effectors can ubiquitinate host protein substrates on Ser residues via a novel mechanism considerably different from the canonical E1-E2-E3 pathway.
X-ray crystallographic data of these proteins reveal a unique 3 domain arrangement and identify Ub and cofactor binding sites.
Biochemical studies have provided insights into the SidE mechanism, and also discovered that flexible regions of proteins containing Ser residues can serve as ubiquitination targets.
Outstanding Questions.
Theoretically any protein with a flexible, Ser-containing region may be a substrate for ubiquitination by SidE effectors. What, then, are the primary host protein targets of SidE? What is the biological outcome of PR-linked ubiquitination? Are other hydroxyl-containing residues such as Thr or Tyr targeted?
How does the intermediate ADPR-Ub move from the mART to the relatively distant PDE domain to undergo phosphotransfer?
What is the biological function of SdeD, a homologous effector altogether lacking an mART or CC domain?
Does the SidE ubiquitination mechanism exist as a eukaryotic cellular process?
Acknowledgement
We kindly acknowledge the NIH (1R01GM126296) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A and Ciechanover A (1998) The Ubiquitin System. Annual Review of Biochemistry 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Komander D and Rape M (2012) The Ubiquitin Code. Annual Review of Biochemistry 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 3.Bhavsar AP et al. (2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449, 827–834 [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y and Zhu Y (2015) Diversity of bacterial manipulation of the host ubiquitin pathways. Cellular Microbiology 17, 26–34 [DOI] [PubMed] [Google Scholar]
- 5.Mevissen TET and Komander D (2017) Mechanisms of Deubiquitinase Specificity and Regulation. Annual Review of Biochemistry 86, 159–192 [DOI] [PubMed] [Google Scholar]
- 6.Ronau JA et al. (2016) Substrate specificity of the ubiquitin and Ubl proteases. Cell Research 26, 441–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huibregtse J and Rohde JR (2014) Hell’s BELs: Bacterial E3 Ligases That Exploit the Eukaryotic Ubiquitin Machinery. PLoS Pathog 10, e1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui J et al. (2010) Glutamine Deamidation and Dysfunction of Ubiquitin/NEDD8 Induced by a Bacterial Effector Family. Science 329, 1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu J et al. (2016) Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhogaraju S et al. (2016) Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 167, 1636–1649. e13 [DOI] [PubMed] [Google Scholar]
- 11.Kotewicz KM et al. (2017) A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host & Microbe 21, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y et al. (2018) Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE. Cell 173, 1231–1243. e16 [DOI] [PubMed] [Google Scholar]
- 13.Akturk A et al. (2018) Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector. Nature 557, 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalayil S et al. (2018) Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 557, 734–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Y et al. (2018) Structural basis of ubiquitin modification by the Legionella effector SdeA. Nature 557, 674–678 [DOI] [PubMed] [Google Scholar]
- 16.Kim L et al. (2018) Structural and Biochemical Study of the Mono-ADP-Ribosyltransferase Domain of SdeA, a Ubiquitylating/Deubiquitylating Enzyme from Legionella pneumophila. Journal of Molecular Biology 430, 2843–2856 [DOI] [PubMed] [Google Scholar]
- 17.Luo Z-Q and Isberg RR (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A 101, 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensminger AW (2016) Legionella pneumophila, armed to the hilt: justifying the largest arsenal of effectors in the bacterial world. Curr. Opin. Microbiol 29, 74–80 [DOI] [PubMed] [Google Scholar]
- 19.Bardill JP et al. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Molecular Microbiology 56, 90–103 [DOI] [PubMed] [Google Scholar]
- 20.Havey JC and Roy CR (2015) Toxicity and SidJ-Mediated Suppression of Toxicity Require Distinct Regions in the SidE Family of Legionella pneumophila Effectors. Infect. Immun 83, 3506–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheedlo MJ et al. (2015) Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc. Natl. Acad. Sci. U.S.A 112, 15090–15095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reverter D et al. (2005) Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J. Mol. Biol 345, 141–151 [DOI] [PubMed] [Google Scholar]
- 23.Rawlings ND et al. (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruneda JN et al. (2016) The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases. Mol Cell 63, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsurumura T et al. (2013) Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex. PNAS 110, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong B et al. (2011) Structure Function Analysis of an ADP-ribosyltransferase Type III Effector and Its RNA-binding Target in Plant Immunity. J. Biol. Chem 286, 43272–43281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons B et al. (2016) Scabin, a Novel DNA-acting ADP-ribosyltransferase from Streptomyces scabies. J. Biol. Chem 291, 11198–11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinaldo S et al. (2015) Structural Basis of Functional Diversification of the HD-GYP Domain Revealed by the Pseudomonas aeruginosa PA4781 Protein, Which Displays an Unselective Bimetallic Binding Site. Journal of Bacteriology 197, 1525–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klumpp S and Krieglstein J (2002) Phosphorylation and dephosphorylation of histidine residues in proteins. European Journal of Biochemistry 269, 1067–1071 [DOI] [PubMed] [Google Scholar]
- 30.Matte A et al. (1998) How do kinases transfer phosphoryl groups? Structure 6, 413–419 [DOI] [PubMed] [Google Scholar]
- 31.Puvar K et al. (2017) Ubiquitin Chains Modified by the Bacterial Ligase SdeA Are Protected from Deubiquitinase Hydrolysis. Biochemistry 56, 4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cambronne ED and Roy CR (2007) The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog 3, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak M-J et al. (2017) Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat Microbiol 2, 17114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muratcioglu S et al. (2015) GTP-Dependent K-Ras Dimerization. Structure 23, 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu J and Luo Z-Q (2017) Legionella and Coxiella effectors: strength in diversity and activity. Nat. Rev. Microbiol 15, 591–605 [DOI] [PubMed] [Google Scholar]
- 36.Cohen MS and Chang P (2018) Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nature Chemical Biology 14, 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]