Abstract
Objective:
Metabolic derangements in sepsis stem from mitochondrial injury and contribute significantly to organ failure and mortality; however, little is known about mitochondrial recovery in human sepsis. We sought to test markers of mitochondrial injury and recovery (mitochondrial biogenesis) non-invasively in peripheral blood mononuclear cells (PBMCs) from patients with sepsis and correlate serial measurements with clinical outcomes.
Design:
Prospective case-control study
Setting:
Academic Medical Center and Veterans Affairs Hospital
Patients:
Uninfected control patients (n=20) and septic ICU patients (n=37)
Interventions:
Blood samples were collected once from control patients and serially with clinical data on days 1, 3, and 5 from septic patients. Gene products for HMOX1, NRF1, PPARGC1A, and TFAM, and mitochondrial DNA (mtDNA) ND1 and D-loop were measured by qRT-PCR. Pro-inflammatory cytokines were measured in plasma and neutrophil lysates.
Measurements and Main Results:
Median (IQR) APACHE II and SOFA scores were 21 (8) and 10 (4), respectively, and 90-day mortality was 19%. Transcript levels of all four genes in PBMCs was significantly reduced in septic patients on day 1 (P<0.05), while mtDNA copy number fell and plasma D-loop increased (both P<0.05), indicative of mitochondrial damage. D-loop content was directly proportional to TNF-α and HMGB1 cytokine expression. By day 5, we observed transcriptional activation of mitochondrial biogenesis and restoration of mtDNA copy number (P<0.05). Patients with early activation of mitochondrial biogenesis were ICU-free by 1 week.
Conclusions:
Our findings support data that sepsis-induced mitochondrial damage is reversed by activation of mitochondrial biogenesis and that gene transcripts measured non-invasively in PMBCs can serve as novel biomarkers of sepsis recovery.
Keywords: mitochondrial biogenesis, sepsis, biomarkers, mononuclear leukocytes
INTRODUCTION
Metabolic derangements in sepsis in experimental animal studies have been reliably linked to sepsis-induced mitochondrial injury and dysfunction (1–16). Mitochondrial damage in turn stimulates mitochondrial biogenesis, a highly regulated nuclear-encoded genetic program that promotes maintenance of healthy mitochondrial mass, is linked to anti-inflammatory, anti-oxidant, and anti-apoptotic cellular defenses, and is essential for organ recovery and survival in sepsis (1, 2, 4–9, 17). In critically ill patients with sepsis, respiration and mitochondrial biogenesis signaling are impaired based on analysis of skeletal muscle biopsies (1, 4, 18–20). However, the ability to measure mitochondrial biogenesis non-invasively, i.e., in peripheral blood cells, has not been demonstrated convincingly, and would make monitoring of ongoing mitochondrial damage or recovery easier. Using peripheral blood mononuclear cells (PBMCs), we sought to measure several key molecular markers of mitochondrial damage and recovery. PBMCs are mitochondria-rich with high rates of respiration (21), and therefore are prime candidates in circulating blood to provide reliable mitochondrial quality control information. We further sought to test whether that early transcriptional activation of mitochondrial biogenesis in PBMCs would predict better patient outcomes. Here we show that 1) PBMCs display signs of mitochondrial damage in patients with sepsis that is proportional to circulating early pro-inflammatory cytokine levels; 2) restoration of mitochondrial copy number occurs by day 5 through activation of mitochondrial biogenesis; 3) subjects with early activation of the mitochondrial biogenesis program (day 1) were ICU-free by 1 week; and 4) these events can be fit by multivariate analysis of markers to inflammation, mitochondrial damage, and mitochondrial recovery.
MATERIALS AND METHODS
Study Enrollment
The study was IRB-approved by Duke University (#Pro00014467) and the Durham Veteran Affairs Medical Center (DVAMC) (#01456). Subjects provided written informed consent prior to enrollment. Control subjects without acute illness were recruited from DVAMC outpatient clinics. Septic subjects were enrolled within 36 hours of admission to the ICUs of Duke University Hospital, Duke Regional Hospital, or DVAMC. Sepsis was defined by the presence of ≥2 systemic inflammatory response syndrome (SIRS) criteria (22), a known/suspected source of infection, and evidence of damage in ≥1 organ systems (23, 24). Subjects were excluded that were <18 years old, pregnant, considered moribund, neutropenic not due to sepsis (absolute neutrophil count <1000/mm3), had active leukemia/lymphoma, had prior bone marrow transplantation, or had illnesses or took medications known to be toxic to mitochondria (e.g., HIV infection, mitochondrial disorder, doxorubicin, azathioprine, tacrolimus, cyclosporine).
Data Collection and Analysis
Clinical data was collected prospectively for each subject. Subjects were deemed “ICU-free” if they were alive and discharged from the ICU by 1 week. Whole blood was collected once from control subjects and serially from septic subjects on study days 1, 3, and 5. Buffy coat (neutrophils), PBMCs, and plasma fractions were isolated by gradient centrifugation. qRT-PCR was performed for key nuclear-encoded genes that regulate transcriptional activation of mitochondrial biogenesis (HMOX1, NRF1, PPARGC1A, and TFAM) (9, 25–27), and for mitochondrial DNA content (MT-ND1 and D-loop). TNF-α and HMGB1 protein levels were determined by dot blots. Additional details regarding sample/data collection, RNA/protein studies, and statistical analyses are provided in the Supplemental Methods.
RESULTS
Twenty control subjects and forty septic patients were enrolled in the study. Three septic patients were subsequently disqualified from the analysis because they met exclusion criteria (bone marrow transplantation, n=1; drugs toxic to mitochondria, n=2) leaving thirty-seven patients in the final sepsis group. The clinical characteristics are shown in the Supplemental Table.
We performed qRT-PCR to determine the effects of sepsis on genetic activation of mitochondrial biogenesis in PBMCs. Compared with day 1 gene levels, septic subjects displayed significantly increased levels of HMOX1, NRF1, PPARGC1A and TFAM mRNA (all P<0.05) by day 3, indicative of activation of mitochondrial biogenesis (Supplemental Figure 1). By day 5, mRNA levels remained significantly increased for PPARGC1A and TFAM (both P<0.05). Compared with control mRNAs, PPARGC1A mRNA levels in the sepsis group were significantly depressed on study day 1 (P<0.05), suggesting early repression of mitochondrial quality control during sepsis.
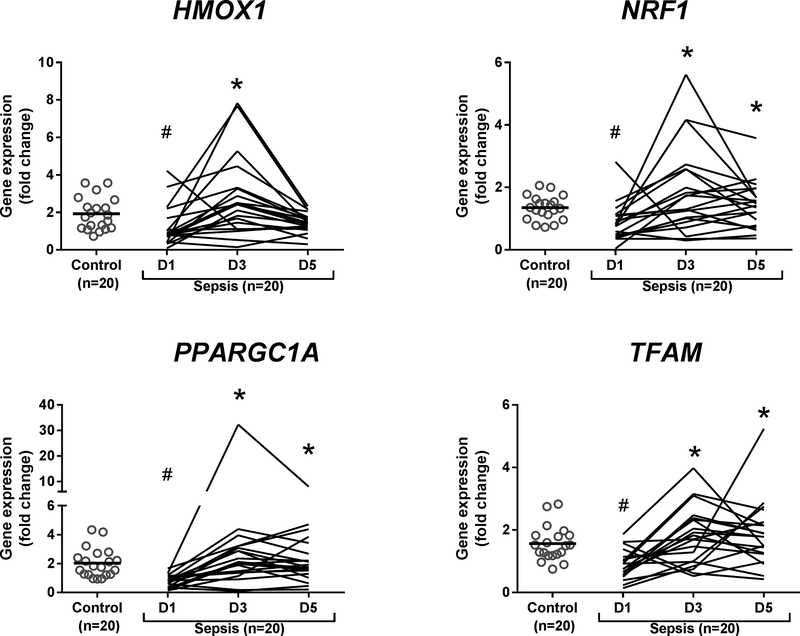
Due to the decline in sepsis group size over time from discharge (n=6), death (n=3), or refusing additional phlebotomy (n=8), and subsequent reduction in statistical power, we analyzed serial mRNA levels in the twenty septic patients that provided samples at all three time points (Figure 1). We found a statistically significant decrease in levels of all four transcripts at day 1 in the sepsis group compared with controls (P<0.05), further supporting an early sepsis-mediated disruption of mitochondrial quality control. Compared with day 1 mRNA levels, all four gene products had increased significantly by day 3 (P<0.05). For three genes (NRF1, PPARGC1A, and TFAM), mRNA levels remained significantly elevated at day 5 (P<0.05).
Figure 1.
Gene expression studies (qRT-PCR) in PBMCs in control subjects and septic subjects (paired samples only) on Days (D) 1, 3, and 5. Grey open circles represent individual control subjects. Black horizontal bars show median values. Line graphs represent individual septic subjects for which samples were collected at each time point (e.g. paired samples). Statistical analysis is Kruskal-Wallis ANOVA with Dunn’s post-hoc testing. *P<0.05 denotes statistically significant comparisons relative to D1, and # P<0.0.05 denotes statistically significant comparisons relative to control.
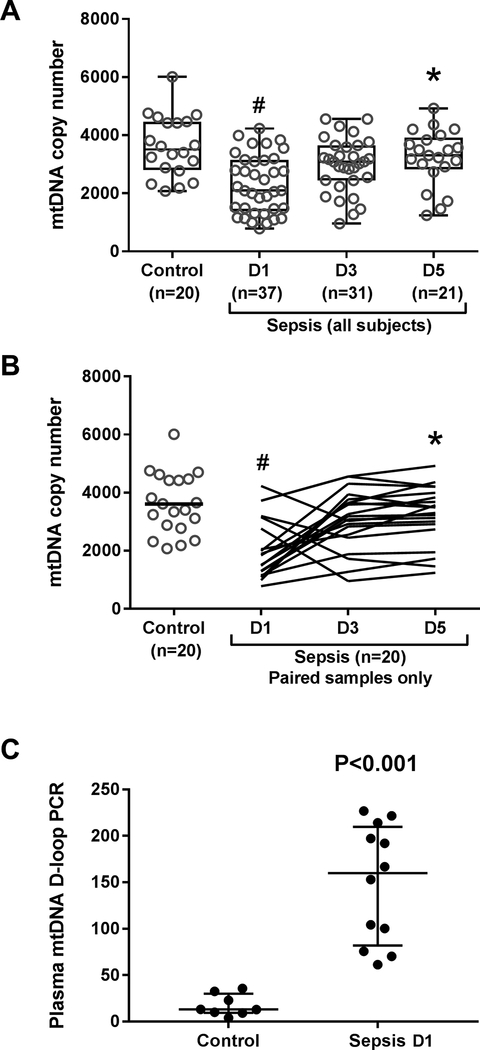
To determine whether these mRNA changes were associated with altered mitochondrial content, mtDNA copy number was measured by qRT-PCR. Compared with the control subjects, mtDNA copy number was significantly decreased on day 1 in the sepsis group (P<0.05) consistent with sepsis-induced mitochondrial damage and possibly loss of mitochondrial mass (Figure 2A). By day 5, however, mtDNA copy number had recovered and was significantly higher than at day 1 (P<0.05), consistent with induction of mitochondrial biogenesis. Similar findings were observed in the subset of twenty septic subjects with paired samples (Figure 2B). Together these data indicate sepsis produces mitochondrial damage in PBMCs that is reversed by activation of the mitochondrial biogenesis program.
Figure 2.
Mitochondrial (mt) DNA copy number measured by qRT-PCR of MT-ND1 gene in PBMCs in control and septic subjects. (A) mtDNA copy number in all collected control and sepsis samples (grey open circles). (B) mtDNA copy number in each control subject (grey open circles) and each septic subject (black line graphs) for which samples were collected at each time point (e.g. paired samples). Black horizontal bars show median values. Statistical analysis is Kruskal-Wallis ANOVA with Dunn’s post-hoc testing. *P<0.05 denotes statistically significant comparisons relative to D1, and #P<0.0.05 denotes statistically significant comparisons relative to control. (C) Circulating mitochondrial DNA (mtDNA) D-loop measured by PCR in plasma of control (n=8) and septic subjects (n=12). All samples were collected on study day 1. Black bars show median and interquartile range. Statistical analysis is Mann-Whitney U test.
To confirm that the sepsis-mediated reduction in mtDNA copy number was due to mtDNA damage rather than unopposed mitophagy, we measured plasma mtDNA D-loop DNA, a small segment of non-coding mtDNA that is highly sensitive to oxidant damage (28). Compared with controls, septic patients had significantly higher plasma D-loop levels on day 1 indicative of greater damage and leakage of fragmented mtDNA (P<0.001) (Figure 2C).
To investigate why increased mtDNA damage was observed in sepsis, we measured TNF-α, a cytokine known to damage mitochondria (29), and HMGB1, a DNA chaperone and damage-associated molecular pattern (DAMP) protein (30), in both neutrophil lysates and plasma of control and septic patients at study day 1. Compared with controls, septic patients displayed significantly increased expression of TNF-α and HMGB1 in both neutrophil lysates and plasma (P<0.05) (Supplemental Figure 2A-B). In fact, TNF-α and HMGB1 protein expression measured in both neutrophil lysates and in plasma was linearly correlated (Spearman r=0.7577, P<0.0001 and goodness-of-fit r2=0.5722, P<0.0001; Spearman r=0.5293, P=0.0164, and goodness-of-fit r2=0.3134, P=0.01, respectively) (Supplemental Figure 2C-D). Moreover, higher TNF-α and HMGB1 expression in neutrophil lysates, and higher HMGB1 expression in plasma were highly correlated with increased plasma mtDNA D-loop DNA levels (all Spearman r>0.6, goodness-of-fit r2>0.5, and P≤0.0003) (Supplemental Figure 2E-H). Finally, we measured correlations in TNF-α protein expression between neutrophil lysates and plasma and found a poor relationship, indicating a minimal neutrophil contribution to circulating TNF-α levels. However, we found a highly significant correlation for HMGB1 between neutrophils and plasma (Spearman r=0.6947, P<0.0007 and goodness-of-fit R2=0.4879, P<0.0006) (Supplemental Figure 3) implicating neutrophils as a major source for this plasma DAMP. Overall, these findings indicate that the pro-inflammatory TNF-α and HMGB1 proteins produced during sepsis are directly proportional to each other and to D-loop content in plasma, supporting a DAMP cascade.
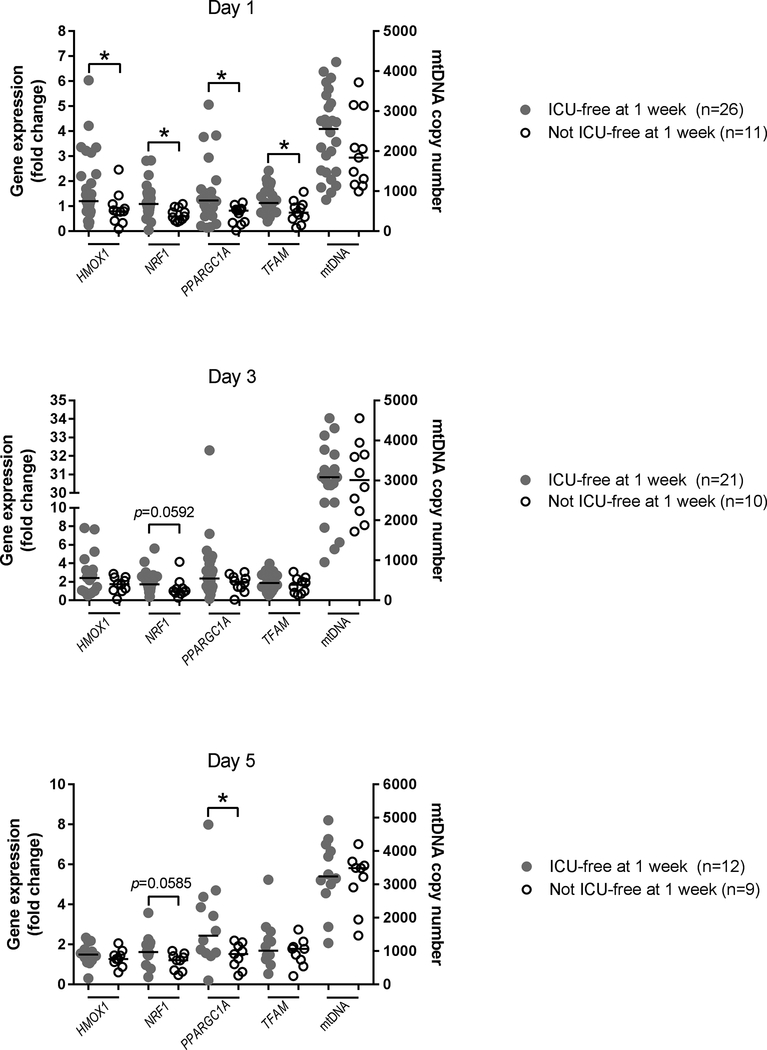
We then sought to determine whether mRNA levels in septic patients were associated with differences in our pre-specified clinical endpoints. No correlation was found between gene product levels and 90-day mortality, APACHE II score, or SOFA score, but in subjects that were not ICU-free by 1 week, day 1 PBMC levels of HMOX1, NRF1, PPARGC1A, and TFAM were significantly reduced (P<0.05) (Figure 3). This group also displayed significantly lower PPARGC1A transcript levels on day 5 (P<0.05). We found no significant differences between the groups for absolute mtDNA copy number, but these data collectively indicate that early activation of mitochondrial biogenesis by day 1 is associated with ICU-freedom by 1 week.
Figure 3.
Intensive care unit (ICU) freedom by 1 week as a function of PBMC gene expression on Days 1, 3, and 5. Closed grey circles depict individual subjects with sepsis that were discharged from the ICU within 1 week. Open black circles depict individual subjects with sepsis that had prolonged ICU stays (≥ 1 week) or early death (within 1 week of presentation). Left Y-axis shows gene expression (qRT-PCR) for HMOX1, NRF1, PPARGC1A, and TFAM. Right T-axis shows qRT-PCR of mtDNA copy number. Black horizontal bars depict median values. Statistical analysis is Mann-Whitney U test. *P<0.05 denotes statistically significant comparisons. Trends (P<0.1) are depicted on the graphs.
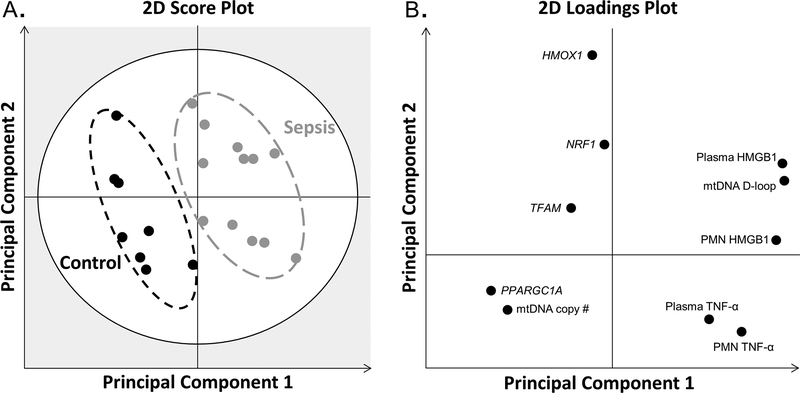
To better model the relationship between cytokines, mtDNA damage, and mitochondrial biogenesis, we performed multivariate PLS-DA (Figure 4). The PLS-DA score plot shows the profiles at study day 1 of control and septic subjects. While there was slight within-group heterogeneity, there were no outliers and the profiles produced distinct clusters separated by group (Figure 4A). The loading plot depicts associations between the measured variables and the clusters of study subjects in the score plot (Figure 4B). The loading plot shows an association between the septic subjects and increased mtDNA D-loop and pro-inflammatory cytokines (in both plasma and neutrophil lysates), and an association between control subjects and increased mtDNA copy number and levels of TFAM and PPARGC1A mRNA. Furthermore, the loading plot shows that 1) TNF-α levels (both plasma and neutrophil) vary inversely with TFAM, HMOX1, and NRF1 mRNA levels; and 2) mtDNA D-loop and HMGB1 vary inversely with mtDNA copy number and PPARGC1A gene levels. The multivariate modeling supports that the pro-inflammatory cytokine milieu produced during sepsis is associated with mitochondrial damage and suppression of transcriptional activation of mitochondrial biogenesis.
Figure 4.
Partial least-squares discriminant analysis. (A) 2D score plot shows the profiles of control (black circles) and septic (grey circles) subjects are distinct and separate. The grey zone in the score plot depicts outliers with 95% confidence. (B) The corresponding 2D loading plot depicts associations between clusters and the measured variables. The two principal components explain 85% of the variation of the model.
DISCUSSION
This is the first human study to measure transcriptional activation of the mitochondrial biogenesis program in sepsis 1) non-invasively in peripheral blood; and 2) serially over time. We found that septic patients display signs of mitochondrial damage with a reduction in PBMC mitochondrial copy number and an elevation in circulating D-loop DNA at study day 1 that rises linearly with pro-inflammatory cytokine expression. We also show that activation of mitochondrial biogenesis in PBMCs leads to restoration of mitochondrial DNA copy number by study day 5, and that subjects with the most PBMC transcripts are ICU-free by 1 week. These findings support PBMCs as a clinically useful, non-invasive measure for biomarkers of mitochondrial homeostasis that may predict recovery in septic patients.
Over the last two decades, preclinical and clinical studies have provided substantial evidence for sepsis-induced organ failure arising from mitochondrial injury and dysfunction (1, 3–18). This mitochondrial injury and impairment in oxidative phosphorylation triggers activation of mitochondrial biogenesis, a highly coordinated bi-genomic program to restore mitochondrial mass and homeostasis (7). We measured mRNA levels of HMOX1, NRF1, PPARGC1A, and TFAM, genes whose products allow activation of mitochondrial biogenesis, in highly aerobic human PBMCs (21), and found a temporal rise and fall in their levels. This is the first timeline for transcriptional activation of mitochondrial biogenesis in humans in response to sepsis-induced mitochondrial injury (7) and supports earlier work in rodents and cells (5–9, 17, 25–27).
Previously, skeletal muscle biopsy studies have contributed significantly to our understanding of changes in mitochondrial structure and function during sepsis (1, 4, 15, 18, 19), but biopsies are invasive, and recently, noninvasive tests in peripheral blood have been sought that could offer clues to the patient’s mitochondrial health. Earlier studies using whole blood transcriptomics (11, 31–34) demonstrated global defects in mitochondrial bioenergetics in experimental S. pneumoniae pneumonia in baboons or in humans with sepsis that correlated with organ failure severity and mortality (11, 32, 33). Furthermore, impairment of oxidative phosphorylation in lymphocytes sub-populations may contribute to secondary immuno-exhaustion frequently encountered in sepsis (20, 32, 35). Prior studies of PBMCs from septic patients demonstrated alterations in mitochondrial membrane potential (36), pyruvate metabolism (37), and respiration (35, 38–43). These alterations were further shown to 1) normalize during sepsis recovery (35, 36, 39, 40), which is consistent with restoration of respiratory capacity by mitochondrial biogenesis; and 2) vary inversely with survival, organ recovery, or both (36, 37, 39, 40, 43). We also found a significant association between markers for maintenance of mitochondrial homeostasis and improved sepsis outcomes. Specifically, transcriptional activation of the mitochondrial biogenesis program on study day 1, as evidenced by higher HMOX1, NRF1, PPARGC1A, and TFAM mRNA levels, was significantly associated with ICU exit by 1 week. Additionally, PPARGC1A mRNA levels on day 5 were statistically associated with ICU-freedom by 1 week. While our findings on days 3 and 5 are limited by the gradual reduction in sample size as subjects either recover (discharged) or die from their illness, our overall findings in PBMCs: 1) support invasive work showing that early transcriptional activation of mitochondrial biogenesis in patients with sepsis is associated with clinical recovery; and 2) support prior work in PBMCs examining mitochondrial function by providing mechanistic insight into how mitochondrial dysfunction becomes normalized.
Mitochondrial biogenesis restores oxidative metabolism and mitochondrial content after sepsis-induced injury with impaired respiration (5–9). Similarly, PBMCs in septic subjects here displayed reduced mtDNA content on day 1 and restored mtDNA content by day 5. Despite this, we did not detect a significant relationship between absolute PBMC mtDNA content and clinical outcomes. Two other studies similarly found low mtDNA content in peripheral leukocytes of patients with sepsis (2, 44), however in contrast to our results, found a significant correlation with survival (2) or illness severity (44). This difference may be explained by different methodologies among studies, for instance, enrolling subjects with a mixture of shock syndromes not limited to sepsis (2); measuring mtDNA from whole blood, which contains abundant neutrophils and platelets with differing mitochondrial content and respiratory capacity compared with PBMCs (2, 21, 44); and reporting mtDNA content relative to putative nuclear housekeeping genes that may be altered during sepsis (2, 5, 44, 45). Moreover, mtDNA concentrations themselves do not inform mitochondrial function, (e.g. reactive oxygen species (ROS) leak), or mitophagy, the regulated loss of mitochondrial mass, both highly relevant to sepsis recovery (46, 47). These limitations suggest the usefulness of mtDNA content alone as a sepsis biomarker may require partner biomarkers.
Although we measured the D-loop segment specifically because it is sensitive to oxidative damage (28), and oxidation of the D-loop may be a signal to activate mitochondrial biogenesis (48), unregulated oxidant production during sepsis causes mtDNA leakage into circulation as a DAMP, and further inflammation (49–51). It is known that the pro-inflammatory cytokines TNF-α and HMGB1, both elevated in plasma and neutrophil lysates are toxic to mitochondria (29, 30) but it is unknown if the increased HMGB1 protein content in neutrophil lysates localizes to specific organelles or cell membrane receptors (e.g. TLR4). The latter is a cause of neutrophil extracellular trap (NET) formation (52), secondary inflammasome activation (53), and further mtDNA damage (54). We cannot rule out that some fraction of the mtDNA measured in plasma was generated by NET formation rather than by oxidative damage and extrusion (55), but D-loop detection is probably reliable. A reduction in mitochondrial reserve is likely responsible for the metabolic derangements found in severe sepsis, but this may be amenable to targeted pharmacologic therapy (56).
When we incorporated these cytokines into a multivariate model with mtDNA damage and mitochondrial biogenesis markers, which depicted close associations between HMGB1 and D-loop, the cytokine levels varied inversely with mtDNA copy number and the mitochondrial biogenesis coactivator gene PPARGCA1. These findings advance prior work in lower animals and cells to human sepsis (5–9, 17, 25–27).
Some of the study limitations have been mentioned, but we note that because patient enrollment began before the Sepsis-3 guidelines (24), our sepsis definition still incorporated SIRS criteria, which have been removed by Sepsis-3 and may be absent in up to 12% of septic patients (57). It is therefore likely that we excluded some subjects meeting Sepsis-3 criteria; however, in keeping with Sepsis-3, 95% of enrolled subjects had a qSOFA score of ≥ 2. Second, our analyses at the later time points (days 3 and 5) are subject to selection bias because the patient group size decreased as patients recovered, died, or refused blood sampling. Also, changes in mitochondrial DNA content observed over time may not be unique to sepsis but more broadly to critical illness. A non-septic critically ill control group would answer this question. We also did not measure proteins for mitochondrial dynamics (fusion/fission), biogenesis, or mitophagy and therefore did not draw conclusions about overall mitochondrial quality control. Finally, we did not distinguish PBMC sub-populations, and thus cannot exclude a contribution by cell populations differing mitochondrial contents.
CONCLUSION
We report that mitochondrial biogenesis gene expression markers can be measured in PBMCs, follow a distinctive timeline of activation, and may be clinically useful to predict the likelihood of recovery from sepsis. We also report a multivariate human sepsis model of mtDNA damage and recovery that supports experiments in lower mammals. Future work is needed to validate these findings and to develop further understanding of mitochondrial quality control in the restoration of mitochondrial homeostasis in PBMCs and other readily retrievable cell types during sepsis.
Supplementary Material
Clinical Characteristics
Blood sample and clinical data collection, molecular studies (RNA and protein), and statistical methods.
Gene expression studies (qRT-PCR) in PBMCs in control subjects and septic subjects (all collected samples) on Days (D) 1, 3, and 5. Grey open circles represent individual subjects. Black horizontal bars show median values. Statistical analysis is Kruskal-Wallis ANOVA with Dunn’s post-hoc testing. *P<0.05 denotes statistically significant comparisons relative to D1, and #P<0.0.05 denotes statistically significant comparisons relative to control.
TNF-α and HMGB1 analyses in neutrophil lysates (PMN) and plasma of control (black circles) and septic (grey circles) subjects. (A-B) TNF-α and HMGB1 protein measured in (A) PMN and (B) plasma. (C-D) Spearman correlation (r) and goodness-of-fit (R2) linear regression of TNF-α and HMGB1 proteins measured in (C) PMN and (D) plasma. (E-F) Spearman correlation (r) and goodness-of-fit (R2) linear regression of plasma mtDNA D-loop qRT-PCR and TNF-α protein measured in (E) PMN and (F) plasma. The correlation and regression analyses for (F) were not statistically significant. (G-H) Spearman correlation (r) and goodness-of-fit (R2) linear regression of plasma mtDNA D-loop qRT-PCR and HMGB1 protein measured in (G) PMN and (H) plasma.
TNF-α and HMGB1 protein expression in plasma compared with neutrophil lysates (PMN) from control (black circles) and septic (red circles) subjects. Spearman correlation (r) and goodness-of-fit (R2) linear regression analyses for HMGB1 were statistically significant.
ACKNOWLEDGEMENTS
The authors thank Martha Salinas for her technical work, and Rhonda Wilder and Elizabeth Mocka for their assistance with subject enrollment and phlebotomy.
Conflicts of Interest and Source of Funding: Bryan Kraft receives funding from the National Institutes of Health (K08HL130557) and has received honoraria from Shionogi Pharmaceuticals and La Jolla Pharmaceutical Company. Lingye Chen has no financial support or conflicts to disclose. Hagir Suliman receives funding from the National Institutes of Health (R01-HL135239-01A1). Claude Piantadosi receives funding from the National Institutes of Health (R01-HL135239-01A1) and the Office of Naval Research (ONR N00014011240). Karen Welty-Wolf received funding from the National Institutes of Health (R01GM084116).
Copyright form disclosure: All authors received support for article research from the National Institutes of Health (NIH). Dr. Kraft’s institution received funding from the NIH/National Heart, Lung, and Blood Institute, and the National Institute of General Medical Sciences, and he received funding from Shionogi Pharmaceuticals (Advisory Board). Drs. Chen and Welty-Wolf’s institutions received funding from the NIH.
REFERENCES
- 1.Brealey D, Brand M, Hargreaves I, et al. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet 2002;360(9328):219–223. [DOI] [PubMed] [Google Scholar]
- 2.Cote HC, Day AG, Heyland DK. Longitudinal increases in mitochondrial DNA levels in blood cells are associated with survival in critically ill patients. Critical care 2007;11(4):R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer M The role of mitochondrial dysfunction in sepsis-induced multi-organ failure. Virulence 2014;5(1):66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carre JE, Orban JC, Re L, et al. Survival in critical illness is associated with early activation of mitochondrial biogenesis. American journal of respiratory and critical care medicine 2010;182(6):745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartz RR, Fu P, Suliman HB, et al. Staphylococcus aureus sepsis induces early renal mitochondrial DNA repair and mitochondrial biogenesis in mice. PloS one 2014;9(7):e100912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry AD, Suliman HB, Bartz RR, et al. Peroxisome proliferator-activated receptor gamma co-activator 1-alpha as a critical co-activator of the murine hepatic oxidative stress response and mitochondrial biogenesis in Staphylococcus aureus sepsis. The Journal of biological chemistry 2014;289(1):41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haden DW, Suliman HB, Carraway MS, et al. Mitochondrial biogenesis restores oxidative metabolism during Staphylococcus aureus sepsis. American journal of respiratory and critical care medicine 2007;176(8):768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athale J, Ulrich A, Chou Macgarvey N, et al. Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice. Free radical biology & medicine 2012;53(8):1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacGarvey NC, Suliman HB, Bartz RR, et al. Activation of mitochondrial biogenesis by heme oxygenase-1-mediated NF-E2-related factor-2 induction rescues mice from lethal Staphylococcus aureus sepsis. American journal of respiratory and critical care medicine 2012;185(8):851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocheteau P, Chatre L, Briand D, et al. Sepsis induces long-term metabolic and mitochondrial muscle stem cell dysfunction amenable by mesenchymal stem cell therapy. Nat Commun 2015;6:10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Science translational medicine 2013;5(195):195ra195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kantrow SP, Taylor DE, Carraway MS, et al. Oxidative metabolism in rat hepatocytes and mitochondria during sepsis. Arch Biochem Biophys 1997;345(2):278–288. [DOI] [PubMed] [Google Scholar]
- 13.Crouser ED. Mitochondrial dysfunction in septic shock and multiple organ dysfunction syndrome. Mitochondrion 2004;4(5–6):729–741. [DOI] [PubMed] [Google Scholar]
- 14.Welty-Wolf KE, Simonson SG, Huang YC, et al. Ultrastructural changes in skeletal muscle mitochondria in gram-negative sepsis. Shock 1996;5(5):378–384. [DOI] [PubMed] [Google Scholar]
- 15.Simonson SG, Welty-Wolf K, Huang YT, et al. Altered mitochondrial redox responses in gram negative septic shock in primates. Circulatory shock 1994;43(1):34–43. [PubMed] [Google Scholar]
- 16.Fink M Cytopathic hypoxia in sepsis. Acta anaesthesiologica Scandinavica Supplementum 1997;110:87–95. [DOI] [PubMed] [Google Scholar]
- 17.Piantadosi CA, Withers CM, Bartz RR, et al. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. The Journal of biological chemistry 2011;286(18):16374–16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredriksson K, Hammarqvist F, Strigard K, et al. Derangements in mitochondrial metabolism in intercostal and leg muscle of critically ill patients with sepsis-induced multiple organ failure. American journal of physiology Endocrinology and metabolism 2006;291(5):E1044–1050. [DOI] [PubMed] [Google Scholar]
- 19.Fredriksson K, Tjader I, Keller P, et al. Dysregulation of mitochondrial dynamics and the muscle transcriptome in ICU patients suffering from sepsis induced multiple organ failure. PloS one 2008;3(11):e3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maestraggi Q, Lebas B, Clere-Jehl R, et al. Skeletal Muscle and Lymphocyte Mitochondrial Dysfunctions in Septic Shock Trigger ICU-Acquired Weakness and Sepsis-Induced Immunoparalysis. Biomed Res Int 2017;2017:7897325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer PA, Ravi S, Chacko B, et al. A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: implications for their use as bioenergetic biomarkers. Redox Biol 2014;2:206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Critical care medicine 1992;20(6):864–874. [PubMed] [Google Scholar]
- 23.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive care medicine 1996;22(7):707–710. [DOI] [PubMed] [Google Scholar]
- 24.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA : the journal of the American Medical Association 2016;315(8):801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999;98(1):115–124. [DOI] [PubMed] [Google Scholar]
- 26.Suliman HB, Carraway MS, Welty-Wolf KE, et al. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. The Journal of biological chemistry 2003;278(42):41510–41518. [DOI] [PubMed] [Google Scholar]
- 27.Larsson NG, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 1998;18(3):231–236. [DOI] [PubMed] [Google Scholar]
- 28.Rothfuss O, Gasser T, Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res 2010;38(4):e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, et al. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. The Journal of biological chemistry 1992;267(8):5317–5323. [PubMed] [Google Scholar]
- 30.Gdynia G, Keith M, Kopitz J, et al. Danger signaling protein HMGB1 induces a distinct form of cell death accompanied by formation of giant mitochondria. Cancer research 2010;70(21):8558–8568. [DOI] [PubMed] [Google Scholar]
- 31.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature 2005;437(7061):1032–1037. [DOI] [PubMed] [Google Scholar]
- 32.Davenport EE, Burnham KL, Radhakrishnan J, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med 2016;4(4):259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss SL, Cvijanovich NZ, Allen GL, et al. Differential expression of the nuclear-encoded mitochondrial transcriptome in pediatric septic shock. Critical care 2014;18(6):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraft BD, Piantadosi CA, Benjamin AM, et al. Development of a novel preclinical model of pneumococcal pneumonia in nonhuman primates. American journal of respiratory cell and molecular biology 2014;50(5):995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng SC, Scicluna BP, Arts RJ, et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nature immunology 2016;17(4):406–413. [DOI] [PubMed] [Google Scholar]
- 36.Adrie C, Bachelet M, Vayssier-Taussat M, et al. Mitochondrial membrane potential and apoptosis peripheral blood monocytes in severe human sepsis. American journal of respiratory and critical care medicine 2001;164(3):389–395. [DOI] [PubMed] [Google Scholar]
- 37.Nuzzo E, Berg KM, Andersen LW, et al. Pyruvate Dehydrogenase Activity Is Decreased in the Peripheral Blood Mononuclear Cells of Patients with Sepsis. A Prospective Observational Trial. Annals of the American Thoracic Society 2015;12(11):1662–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belikova I, Lukaszewicz AC, Faivre V, et al. Oxygen consumption of human peripheral blood mononuclear cells in severe human sepsis. Critical care medicine 2007;35(12):2702–2708. [DOI] [PubMed] [Google Scholar]
- 39.Japiassu AM, Santiago AP, d’Avila JC, et al. Bioenergetic failure of human peripheral blood monocytes in patients with septic shock is mediated by reduced F1Fo adenosine-5’-triphosphate synthase activity. Critical care medicine 2011;39(5):1056–1063. [DOI] [PubMed] [Google Scholar]
- 40.Weiss SL, Selak MA, Tuluc F, et al. Mitochondrial dysfunction in peripheral blood mononuclear cells in pediatric septic shock. Pediatr Crit Care Med 2015;16(1):e4–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merz TM, Pereira AJ, Schurch R, et al. Mitochondrial function of immune cells in septic shock: A prospective observational cohort study. PloS one 2017;12(6):e0178946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrabou G, Moren C, Lopez S, et al. The effects of sepsis on mitochondria. The Journal of infectious diseases 2012;205(3):392–400. [DOI] [PubMed] [Google Scholar]
- 43.Nucci LA, Santos SS, Brunialti MK, et al. Expression of genes belonging to the interacting TLR cascades, NADPH-oxidase and mitochondrial oxidative phosphorylation in septic patients. PloS one 2017;12(2):e0172024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyle A, Burn DJ, Gordon C, et al. Fall in circulating mononuclear cell mitochondrial DNA content in human sepsis. Intensive care medicine 2010;36(6):956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cummings M, Sarveswaran J, Homer-Vanniasinkam S, et al. Glyceraldehyde-3-phosphate dehydrogenase is an inappropriate housekeeping gene for normalising gene expression in sepsis. Inflammation 2014;37(5):1889–1894. [DOI] [PubMed] [Google Scholar]
- 46.Ghanta S, Tsoyi K, Liu X, et al. Mesenchymal Stromal Cells Deficient in Autophagy Proteins Are Susceptible to Oxidative Injury and Mitochondrial Dysfunction. American journal of respiratory cell and molecular biology 2017;56(3):300–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang AL, Ulrich A, Suliman HB, et al. Redox regulation of mitophagy in the lung during murine Staphylococcus aureus sepsis. Free radical biology & medicine 2015;78:179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pastukh VM, Gorodnya OM, Gillespie MN, et al. Regulation of mitochondrial genome replication by hypoxia: The role of DNA oxidation in D-loop region. Free radical biology & medicine 2016;96:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakahira K, Kyung SY, Rogers AJ, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med 2013;10(12):e1001577; discussion e1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schafer ST, Franken L, Adamzik M, et al. Mitochondrial DNA: An Endogenous Trigger for Immune Paralysis. Anesthesiology 2016;124(4):923–933. [DOI] [PubMed] [Google Scholar]
- 51.Schumacker PT, Gillespie MN, Nakahira K, et al. Mitochondria in lung biology and pathology: more than just a powerhouse. American journal of physiology Lung cellular and molecular physiology 2014;306(11):L962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tadie JM, Bae HB, Jiang S, et al. HMGB1 promotes neutrophil extracellular trap formation through interactions with Toll-like receptor 4. American journal of physiology Lung cellular and molecular physiology 2013;304(5):L342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kahlenberg JM, Carmona-Rivera C, Smith CK, et al. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. Journal of immunology 2013;190(3):1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu J, Nagasu H, Murakami T, et al. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proceedings of the National Academy of Sciences of the United States of America 2014;111(43):15514–15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousefi S, Mihalache C, Kozlowski E, et al. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell death and differentiation 2009;16(11):1438–1444. [DOI] [PubMed] [Google Scholar]
- 56.McCall CE, Zabalawi M, Liu T, et al. Pyruvate dehydrogenase complex stimulation promotes immunometabolic homeostasis and sepsis survival. JCI Insight 2018;3(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaukonen KM, Bailey M, Pilcher D, et al. Systemic inflammatory response syndrome criteria in defining severe sepsis. The New England journal of medicine 2015;372(17):1629–1638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical Characteristics
Blood sample and clinical data collection, molecular studies (RNA and protein), and statistical methods.
Gene expression studies (qRT-PCR) in PBMCs in control subjects and septic subjects (all collected samples) on Days (D) 1, 3, and 5. Grey open circles represent individual subjects. Black horizontal bars show median values. Statistical analysis is Kruskal-Wallis ANOVA with Dunn’s post-hoc testing. *P<0.05 denotes statistically significant comparisons relative to D1, and #P<0.0.05 denotes statistically significant comparisons relative to control.
TNF-α and HMGB1 analyses in neutrophil lysates (PMN) and plasma of control (black circles) and septic (grey circles) subjects. (A-B) TNF-α and HMGB1 protein measured in (A) PMN and (B) plasma. (C-D) Spearman correlation (r) and goodness-of-fit (R2) linear regression of TNF-α and HMGB1 proteins measured in (C) PMN and (D) plasma. (E-F) Spearman correlation (r) and goodness-of-fit (R2) linear regression of plasma mtDNA D-loop qRT-PCR and TNF-α protein measured in (E) PMN and (F) plasma. The correlation and regression analyses for (F) were not statistically significant. (G-H) Spearman correlation (r) and goodness-of-fit (R2) linear regression of plasma mtDNA D-loop qRT-PCR and HMGB1 protein measured in (G) PMN and (H) plasma.
TNF-α and HMGB1 protein expression in plasma compared with neutrophil lysates (PMN) from control (black circles) and septic (red circles) subjects. Spearman correlation (r) and goodness-of-fit (R2) linear regression analyses for HMGB1 were statistically significant.