Abstract
Protein α‐N‐terminal methylation is catalyzed by protein N‐terminal methyltransferases. The prevalent occurrence of this methylation in ribosomes, myosin, and histones implies its function in protein–protein interactions. Although its full spectrum of function has not yet been outlined, recent discoveries have revealed the emerging roles of α‐N‐terminal methylation in protein–chromatin interactions, DNA damage repair, and chromosome segregation. Herein, an overview of the discovery of protein N‐terminal methyltransferases and functions of α‐N‐terminal methylation is presented. In addition, substrate recognition, mechanisms, and inhibition of N‐terminal methyltransferases are reviewed. Opportunities and gaps in protein α‐N‐terminal methylation are also discussed.
Keywords: enzymes, inhibitors, methylation, proteins, reaction mechanisms
1. Early Discoveries in Protein α‐N‐Terminal Methylation
Emerging epigenetic modulators have continued to reshape our understanding of eukaryotic gene expression and regulation. Lys and Arg methylation are two well‐known epigenetic marks that play important roles in regulating chromatin dynamics and transcription activation. Protein lysine methyltransferases (PKMTs) and arginine methyltransferases (PRMTs) catalyze the covalent addition of a methyl group from S‐adenosylmethionine (SAM) to the ϵ‐amino group of the Lys side chain and guanidino group of the Arg side chain, respectively (Scheme 1). After the methyl group is transferred, SAM is transformed into S‐adenosylhomocysteine (SAH). Unlike PKMTs and PRMTs that methylate the side chain, protein N‐terminal methyltransferases (NTMTs) transfer a methyl group to the α‐amino group at the protein N terminus. Although protein α‐N‐terminal methylation has been a known post‐translational modification for over four decades, its emerging features have made it a relatively new addition to the list of players in these processes.
Scheme 1.
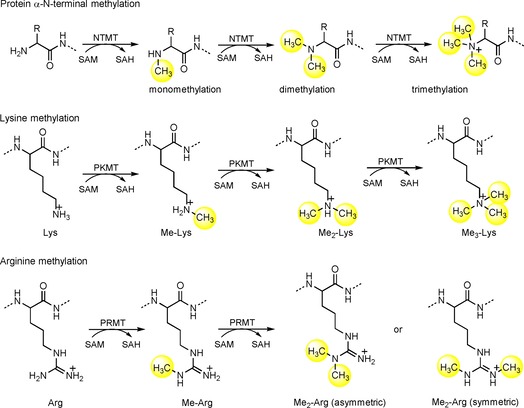
Schematic outline of protein methylation.
Protein α‐N‐terminal methylation was initially discovered in Escherichia coli on ribosomal subunits, including methylalanine (MeAla) on the α‐N terminus of L33 (MeAla‐Lys‐Gly) and S11 (MeAla‐Lys‐Ala), methylmethionine (MeMet) on L16 (MeMet‐Leu‐Gln), over four decades ago.1, 2, 3 Since then, many cases of α‐N‐terminal methylation have been reported on various proteins, such as dimethylproline (Me2Pro) on Crithidia oncopelti cytochrome c and Asterias rubens histone H2B, trimethylalanine (Me3Ala) on E. coli ribosomal protein L11, Tetrahymena histone 2B, and all vertebrate striated muscle light chains.4, 5, 6, 7 Because the aforementioned protein substrates are components of macromolecular complexes, α‐N‐terminal methylation has been inferred to mediate protein–protein interactions. In addition, eukaryotic N‐terminal methylated proteins were postulated to be involved in protein degradation on the basis that methylation might interfere with N‐terminal acetylation.8 However, knowledge of the physiological consequences of protein α‐N‐terminal methylation is still very limited. Recent identifications of eukaryotic protein α‐NTMTs have prompted increasing discoveries of new protein substrates;9, 10, 11, 12, 13 thus supporting that α‐N‐terminal methylation is a widespread post‐translational modification.
2. Discovery of Protein NTMTs
2.1. Prokaryotic protein NTMT
Protein L11 methyltransferase (PrmA) is responsible for catalyzing α‐N‐terminal methylation of the bacterial 50S ribosomal subunit protein L11.14, 15 It is conserved among bacteria, but absent from archaea.16 PrmA is a multifunctional methyltransferase (MTase) because it is able to modify both the α‐N‐terminal amine and ϵ‐amino groups of two different Lys residues.14, 16 PrmA consists of an N‐terminal domain for substrate recognition, a C‐terminal catalytic domain with a seven‐β‐strand structural fold, and a flexible linker helix (Figure 1 A).17 Structural studies revealed a wide range of domain movements of PrmA, as exemplified by the structure of PrmA bound to L11, in comparison with the apo form of PrmA (Figure 1 B).17 Such conformational changes are necessary for the recognition of multiple substrate sites. PrmA preferentially methylates free ribosomal protein L11 over an assembled 50S ribosomal subunit; therefore, methylation of L11 may facilitate the assembly of the large subunit.16 However, the role of L11 methylation remains a mystery because mutants and deletion of PrmA show no growth defects or any distinct phenotype in E. coli and Thermus thermophilus.15, 16 Investigation of those strains under different stress conditions may provide new insights into the function of N‐terminal methylation of L11.
Figure 1.
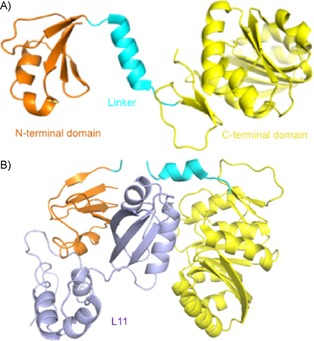
Representative crystal structures of PrmA. A) The N‐terminal domain, linker, and C‐terminal domain of apo‐PrmA (PDB ID: 2NXC) are colored in orange, cyan, and yellow, respectively. B) PrmA–L11 complex structure (PDB ID: 2NXN); L11 is given in purple.
2.2. Eukaryotic protein NTMTs that methylate an X‐P‐K/R motif
Yeast YBR261C/Tae1 protein was identified as the NTMT Ntm1 in Saccharomyces cerevisiae by Webb et al. in 2010.11 YBR261C recognizes the canonical X‐P‐K recognition motif and methylates ribosomal substrates Rp112ab and Rps25a/Rps25b. Meanwhile, YBR261C is able to methylate nonamer synthetic peptides, including PPKQQLSKY, which is derived from α‐N‐terminal Rps25a/b and A/S‐PKQQLSKY, with Ala or Ser replacing Pro.11 Previous chemical genetic profile analysis indicated that deletion of YBR261C in yeast abolished N‐terminal methylation, which consequently altered the ribosomal profile and led to defects in both translational efficiency and fidelity.11, 18 Overexpression of YBR261 validated its involvement in protein synthesis.18 In addition, α‐N‐terminal methylation has been detected in the yeast Rpt1 (PPKEDW) subunit of the 19S regulatory particle of 26S proteasome.19 If the PK sequence at the second and third positions was deleted from Rpt1, N‐terminal methylation of Rpt1 was abolished.19 With this PK deletion, yeast strains grow more slowly and are more sensitive to stress.19 Despite the implications of α‐N‐terminal methylation of Rpt1 on cell growth and stress tolerance in yeast,19 the molecular mechanism remains obscure. It is necessary to investigate how this methylation affects substrate recognition, ATPase activity, and the interactions of Rpt1 with other subunits of the 26S proteasome.
In 2012, dNTMT (CG1675) was identified as the enzyme for α‐N‐terminal methylation of H2B protein in Drosophila melanogaster.20 The N‐terminal methylation levels of H2B were increased during fly development and in the presence of cellular stress, such as heat and proliferation stress.20 dNTMT was mainly located in the nucleus, where the majority of chromatin‐bound H2B is methylated.19 dNTMT recognizes the N‐terminal sequence of D. melanogaster H2B (PPKTSG), which conforms to the canonical X‐P‐K recognition motif for its mammalian orthologs (X=A, P, or S). dNTMT methylation is not processive since monomethylated Pro was accumulated during the methylation reaction. A sequence search suggested about 36 proteins carrying a (M)‐A/P/S‐P‐K recognition motif in the predicted proteome of D. melanogaster.20 In addition, dART8, a PRMT for H3R2 methylation, negatively regulated H2B N‐terminal methylation;20 thus suggesting crosstalk between methylation on two histone tails.
Webb et al. first identified the human METTL11A/NTMT1 encoded by the METTL11A gene, along with the discovery of YBR261C/TAE1.11 Simultaneously, Macara et al. isolated the same protein from HeLa cell soluble nuclear extracts, and named it N‐terminal RCC1 methyltransferase (NRMT1) on account of its substrate regulator of chromosome condensation 1 (RCC1).10 The discovery of NTMT1/NRMT1 has led to rising reports on N‐terminal methylation existing in tumor suppressor retinoblastoma 1 (RB1), oncoprotein SET (also known as I2PP2A, TAF1α), damaged DNA‐binding protein 2 (DDB2), poly(ADP‐ribose) polymerase 3 (PARP3), and centromere proteins A and B (CENP‐A&B, Figure 2).10, 21, 22, 23, 24, 25 Crystal structures of NTMT1 in complex with peptide substrates and SAH revealed the structural basis for the preferred recognition motif X‐P‐K/R (X can be any amino acid, except D or E).26, 27 Substrate recognition is discussed in more detail in Section 2.4. The NTMT1 gene is expressed in all tissues, and the protein is expressed in most tissues, except spleen, liver, and fallopian tube tissue, according to the Human Protein Atlas (http://www.proteinatlas.org). Additionally, NTMT1 is overexpressed in tumor tissues of patients, including colorectal, melanoma, carcinoid, lung, and liver, according to ProteinAtlas (http://www.proteinatlas.org).28 Knockdown of NTMT1 results in mitotic defects and sensitizes etoposide and gamma irradiation in breast cancer cell lines such as MCF‐7 and LCC9, whereas NTMT1 knockout mice showed premature aging.29, 30 Both studies infer the function of NTMT1 in DNA damage repair.
Figure 2.
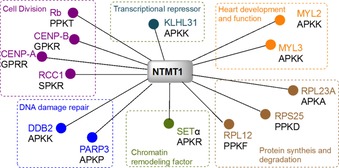
Validated substrates for NTMT1.
The NTMT1 homologue NTMT2/METTL11B shares over 50 % sequence similarity with NTMT1 (Figure 3 A).11 This homologue was suggested as another NTMT in 2010 and confirmed to be an NTMT by 2013.9 Both NTMT1 and NTMT2 recognize an X‐P‐K/R consensus sequence, in which X can be any amino acid, except D or E.9, 10, 11, 26, 27, 31 Although NTMT2 was originally proposed to be a monomethylase, recent studies indicated that it could also fully methylate both GPKRIA and PPKRIA peptides.9, 31 However, the physiological substrate of NTMT2 remains to be uncovered. NTMT2 is predominantly expressed in heart and skeletal muscle tissues,28 which suggesting the possibility of its role in a tissue‐specific context.
Figure 3.
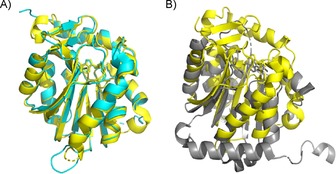
Structure alignment. A) Alignment of NTMT1 (yellow, PDB ID: 2EX4) with NTMT2 (cyan, PDB ID: 5UBB). B) Alignment of NTMT1 (yellow, PDB ID: 2EX4) with the C‐terminal domain of METTL13 (gray, PDB ID: 5WCJ).
YBR261C and NTMT1/2 are able to methylate nona‐ and hexamer peptides,11, 26, 31, 32, 33 respectively. In addition, eukaryotic protein NTMTs share an X‐P‐K/R motif. Therefore, it is reasonable to speculate that an N‐terminal linear sequence is sufficient for their recognition. Kinetic studies on protein substrates would shed light on how other sequences contribute to recognition and methylation transfer.
2.3. Eukaryotic protein NTMTs that methylate elongation factor 1A
In addition to α‐N‐terminal methylation on the classical X‐P‐K/R motif in eukaryotic cells, a novel N‐terminal methylation has been recently reported on eukaryotic elongation factor 1A (eEF1A) in both yeast and humans.12, 13 YLR285W, also named elongation factor methyltransferase 7 (Efm7), is a dual MTase that installs methyl groups at both N‐terminal Gly1 and Lys2 residues of yeast eEF1A protein.12 Lys2 is methylated only after trimethylation of Gly1.12 Yeast eEF1A starts with GKEKSHINV and is the only known substrate of Efm7, although there are 35 other yeast proteins with a G‐K sequence at their N termini.12 Unlike NTMT1/2, Efm7 was not able to methylate the synthetic decamer peptide GKEKSHINVV derived from the N terminus of eEF1A.12 Efm7 can methylate domain 1 (residues 1–238) of eEF1A, but to a smaller degree of trimethylation.12 In addition, methylation of eEF1A is increased in the presence of either GDP or GTP, which is known to bind to eEF1A and induce conformational changes.12 These data suggest that Efm7 substrate recognition may require the three‐dimensional structure, which is different from the classic linear X‐P‐K/R motif recognition by other eukaryotic protein NTMTs.
Recently, human MTase‐like protein 13 (METTL13) was identified as a dual MTase for both N‐terminal Gly1 and Lys55 of human eEF1A.13 Human eEF1A contains a very similar N‐terminal sequence, GKEKTHIN, with Thr substituted at the fifth position instead of Ser, as seen in yeast.12 METTL13 has two distinct MTase domains: N‐ and C‐terminal domains that appear to have different recognition preferences. Specifically, the C‐terminal domain is able to methylate peptides derived from the first 15 amino acids of eEF1A, whereas the N‐terminal domain is sufficient for methylation of Lys55.13 Structural alignment of the C‐terminal domain of METTL13 with NTMT1 displayed striking differences (Figure 3 B). So far, eEF1A is the only validated biological substrate for METTL13, although 49 potential substrates were suggested for METTL13.13 This reflects the high specificity of METTL13 for eEF1A.13 METTL13 is also called FEAT (faint expression in normal tissues, aberrant overexpression in tumors). As indicated by its alternative name, METTL13/FEAT is implicated in tumorigenesis in vivo by suppressing apoptosis.34 FEAT was observed in the cytoplasm, mitochondria, and nucleus of HeLa cells, as well as in the blood of cancer patients.35 On the other hand, the METTL13/FEAT protein was also implied as a tumor suppressor in bladder carcinoma by negatively regulating cell proliferation, migration, and invasion in bladder cancer cells.36
2.4. Substrate recognition
The α‐N‐terminal methylation has been reported on various N‐terminal sequences in prokaryotic proteins. However, all identified N‐terminal methylation on eukaryotic proteins is either a conserved X‐P‐K/R motif or specific to eEF1A (Table 1). The discussion herein focuses on substrate recognition and structure on three human NTMTs.
Table 1.
Protein NTMTs discussed herein.
| Protein | Organism | Substrate recognition |
PDB ID |
|---|---|---|---|
| PrmA | E. coli., | A‐K‐A/G/K | 3CJT, 3CJS, 3CJR, |
| T. thermophilus | M‐L/M/K‐G/Q | 3EGV, 2NXC, 2NXN | |
| dNTMT (CG1675) | D. melanogaster | X‐P‐K | |
| YBR261C/TAE1 | S. cerevisiae | X‐P‐K | |
| NTMT1/NRMT1/ | Homo sapiens | X‐P‐K/R | 2EX4, 5E1B, 5E1M, |
| METTL11A | 5E1O, 5E1D, 5E2B, 5E2A, 5CVD, 5CVE |
||
| NTMT2/NRMT2/ | H. sapiens | X‐P‐K/R | 5UBB, 6DUB |
| METTL11B | |||
| Efm7/YLR285W | S. cerevisiae | GKEKSH | |
| METTL13/FEAT | H. sapiens | GKEKTH | 5WCJ |
Crystal structures of NTMT1–SAH peptide substrate ternary complexes reveal that NTMT1 contains a seven‐strand β‐sheet and five α helixes, which is a Rossmann‐fold MTase (Figure 3).26, 27 In addition, NTMT1 has two unique structural components: an N‐terminal extension containing three helixes and a pair of β‐hairpins.26 Peptide substrates bind in a similar manner into a negatively charged channel.26 The first three residues (X‐P‐K/R) insert into a defined binding pocket, which explains the unique specificity of NTMT1 in methylation of the α‐N‐terminal amine versus the ϵ‐amine of Lys.26
Proteins starting with S/P/A/G‐P‐K/R have been confirmed to be methylated in vivo. Furthermore, NTMT1 is able to methylate X‐P‐K/R peptides (X is any amino acid, except D or E) in vitro.26 This expanded consensus implies that there are about 300 possible substrates for NTMT1. Among them, CENP‐A/B, DDB2, PARP3 have subsequently been confirmed (Figure 2). The tolerance for the first position of the substrate is because of its backbone hydrogen bond with an Asn168 residue of NTMT1 and a spacious binding pocket surrounding the side chain of the first residue X.10, 11, 26 The importance of Pro2 is revealed by its stacking interaction with Trp136.11, 26 Pro2 was reported to be replaceable by other residues, including Ala, Glu, Met, Asn, Gln, Gly, and Ser.23, 37 However, Dong et al. demonstrated that substitution of Pro2 with Ile, Gln, Glu, or Ser abolished its interaction with NTMT1.26 Basic residues, including Lys3 or Arg3, are preferred for the peptide substrate, in which electrostatic interactions are formed with key residues Asp177 and Asp180 of NTMT1 (Figure 4).10, 26, 37 Mutation of D180 to lysine abolished the enzyme activity.26 Cocrystal structures also indicated that NTMT1 had no significant preference for non‐ or monomethylated substrates because they had the same orientation to interact with NTMT1.26 This is consistent with comparable kinetic parameters of NTMT1 methylation with both substrates, as well as the distributive mechanism of NTMT1 methylation.26, 32, 33
Figure 4.
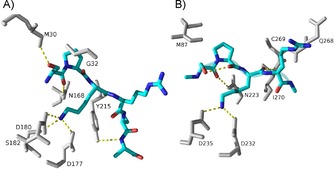
Substrate binding (SPKR, blue) in the active site with NTMT1 (A, gray; PDB ID: 5E1B) and NTMT2 (B, gray; PDB ID: 6DUB).
NTMT2 shares a comparable substrate recognition mode to that of NTMT1. A structural comparison of the substrate‐binding region of NTMT2 with that of NTMT1 revealed a similar set of residues (Figure 4).31 In NTMT2, key substrate‐engaging residues are Asn223, Trp191, Asp232, and Asp235, which are in agreement with Asn168, Trp136, Asp177, and Asp180 in NTMT1.26, 31 NTMT2 is able to methylate XPKRIA hexapeptides,31 although its physiological substrate has yet to be identified. However, NTMT2 exhibits a different product specificity from that of NTMT1. NTMT2 is able to fully methylate G/P‐PKRIA, but it predominantly installs one methyl group for other X‐PKRIA peptides.31 Residue Asn89 of NTMT2 serves as a gatekeeper to govern the product specificity.31 Besides those residues involved in substrate binding, multiple mutations have been reported in human cancer samples (Cosmic Catalogue of Somatic Mutations in cancer; https://cancer.sanger.ac.uk/cosmic/). Among them, the N209I endometrial and P211S lung cancer mutants decreased the trimethylation level of RCC1, whereas the Q144H lung cancer mutant increased the trimethylation level of RCC1 with minimal levels of mono‐ and dimethylated RCC1.38 For NTMT2, the V224L breast cancer mutant showed marginal methylation activity for methylation states.38 Those data infer that methylation levels may play different roles in different cancers. Meanwhile, it would be worth investigating how those mutations affect the kinetic parameters of the enzyme.
The C‐terminal domain of dual MTase METTL13 is responsible for the α‐N‐terminal methylation of eEF1A.13 Although yeast Efm7 is not able to methylate the decamer peptide that is derived from yeast N‐terminal eEF1A, METTL13 can methylate the 15‐mer peptide derived from human N‐terminal eEF1A. A peptide array study suggested an N‐terminal consensus sequence [G/A/P]‐[K/RF/Y/Q/H]‐E‐[K/R/Q/I/H/L] for METTL13, which implies 49 candidate substrates in human proteome.13 However, only peptide corresponding to the N terminus of eEF1A was substantially methylated by METTL13 through validation.13 The crystal structure of its core MTase domain (residues 470–699; PDB ID: 5WCJ) in complex with SAH reveals a Rossmann fold‐like structure.13 A docking study of the hexamer peptide GKEKTH suggested a hydrogen bond between the carboxylate O atom of Gly1 and side chain of Asn647.13 Such an interaction is conserved in both NTMT1 and ‐2.13, 26, 31 However, the unique interaction of Asp577 with the α‐amino group of Gly1 is required for enzymatic activity because the activity of the D577A mutant decreased by about half.13 It is imperative to obtain a cocrystal structure of METTL13 in complex with the eEF1A substrate to elucidate its substrate recognition and high specificity for eEF1A. In addition, structural information about full‐length METTL13 would shed light on how both N‐ and C‐terminal domains orient to favor methylation at the α‐N terminus and Lys55 of eEF1A, respectively. A wide range of domain movements are speculated to occur to induce conformational changes in METTL13.
3. Functions of α‐N‐Terminal Methylation
The α‐N‐terminal amines (pK a=6–8) are less basic in comparison with the side chain aliphatic amines (pK a≈10.5). Consequently, methylation of α‐N terminus alters not only the hydrophobicity and steric hindrance, but also charge state under the physiological condition. Early reported N‐terminal methylated proteins such as myosin light chain LC‐1, histone H2B, and cytochrome c‐557 are involved in large macromolecular complexes.39 Therefore, protein N‐terminal methylation has been proposed to regulate protein–protein interactions in complex macromolecular structures. N‐terminal methylation was also suggested to be involved in protein stability since methylation interplays with another predominant acetylation at the N terminus. Additionally, Hershko et al. demonstrated that a free α‐N‐amine group is required for ubiquitin‐mediated protein degradation in comparison with chemical methylated protein.40 The relevance of α‐N‐terminal methylation in protein–DNA interactions has been unveiled in interactions of RCC1 and CENP‐A/B with chromatin, and DDB2 with DNA damage foci.21, 23, 24, 41 In addition, the level of α‐N‐terminal methylation increases in response to a variety of extracellular stimuli, including increased cell density, heat shock, and arsenite treatment;20, 24 thus suggesting its potential as a new epigenetic mark.
3.1. Functions of methylated proline
Me2Pro was present in the relatively free‐moving N‐terminal region of C. oncopelti cytochrome c557 as a novel N‐terminal protein modification.42 The observation of only one CαH resonance of Me2Pro at δ=4.03 ppm in the NMR spectrum suggested that dimethylation might limit the rate of interconversion between cis and trans conformations.43 Thus, relatively rigid Me2Pro could yield specific folding for the interaction with other partners, including proteins and DNA. The occurrence of Me2Pro was also found in starfish histone H2B. The N‐terminal methylation of yeast 26S proteasome subunit Rpt1 (starts with Pro‐Pro‐Lys) is involved in cell growth or stress tolerance to oxidant and canavanine stress.19 Heat shock and arsenite treatments induced a rapid increase in Me2Pro of histone H2B and a shift of methylation sites of H3 in D. melanogaster, which correlated with chromatin remodeling and gene inactivation.44 The N‐terminal end of H2B was inferred to interact preferentially with DNA rather than histone,44 which suggested that this methylation could regulate both protein–protein and protein–DNA interactions.
3.2. Functions of methylated alanine
Accumulating evidence revealed the widespread occurrence of α‐N‐Me3Ala in a highly mobile N‐terminal region (about 40 residues) of myosin alkali 1 (A1) and LC2 light chains in vertebrate striated skeletal and cardiac muscles.6 The disappearance of the NMR signal for N‐methyl protons of Me3Ala in the presence of actin implied its role in the interaction of the myosin light chain with the C‐terminal region of actin.6, 43 Such interactions are weakened by increased ionic strength,6, 43 which suggests the involvement of electrostatic interactions in this case. Hayashibara and Miyanishi demonstrated that the positively charged α‐N‐Me3Ala was critical for the actin–A1 interaction because it lowered both V max and K M of the actin‐activated ATPase activity of the globular head of myosin A1.45 Such interaction results in a higher binding affinity for myosin to actin and a slower turnover rate of actin‐activated ATPase activity.45 Hence, trimethylation of Ala1 may have a suppressive effect on mobility between actin and myosin filaments.
Meanwhile, trimethylation of Ala1 can have a positive effect on DNA damage repair. DDB2 possesses an N‐terminal sequence of Ala‐Pro‐Lys, which undergoes α‐N‐methylation, with trimethylation being the predominant form.23 DDB2 recruits DDB1‐CUL4A‐based E3 ligase to initiate the DNA repair process. The α‐helical N‐terminal domain (102 residue) of DDB2 is important to mediate interactions with DDB1 and damaged DNA.46 α‐N‐Methylated DDB2 enhances its nuclear localization, recruitment to cyclobutene pyrimidine dimers, and ATM activation; thus indicating the function of N‐terminal methylation in UV‐damaged DNA repair.23
3.3. Functions of methylated serine
RCC1 is pivotal in regulating nuclear transport, mitotic spindle formation, and nuclear envelope assembly through its dynamic association with chromatin.47 Chen et al. identified that α‐N‐terminal methylation of the first Ser residue was important for RCC1 interacting with chromosomes, which was critical for mitotic spindle assembly and function.21, 48 In addition, RCC1 mutant K3Q or knockdown of NTMT1 abolished N‐terminal methylation of RCC1 and led to multipolar spindle formation and mitotic defects.21 Further studies by Hao and Macara revealed that α‐N‐methylation might enhance the association of RCC1 with chromatin through electrostatic interactions with DNA.49 Hitakomate et al. provided strong support for the importance of an α‐N‐methylated tail for stable association of RCC1 with interphase chromatin.50
3.4. Functions of methylated glycine
Human centromere plays an essential role in chromosome segregation to ensure genome stability. Trimethylation of Gly has a positive effect on the functions of both CENP‐A and ‐B proteins. CENP‐A, a centromere‐specific histone H3 variant, contains an N‐terminal Gly‐Pro‐Arg motif, which is subject to α‐N‐methylation in cells.25 This methylation is independent of other posttranslational modifications of the CENP‐A tail.25 CENP‐A replaces H3 in centromeric nucleosomes and is indispensable for recruitment and assembly of some components to the centromere and kinetochore.51 The N‐terminal tail of CENP‐A was proposed to direct proper deposition of CENP‐B at centromeres and stabilize its binding to centromeres through direct interaction.52 α‐N‐Trimethylation is implied to be essential for CENP‐A in maintaining chromosome segregation fidelity because methylation of CENP‐A is required for cell survival, localization of CENP‐T and CENP‐I, and formation of the bipolar spindle.41 Loss of CENP‐A methylation caused defects in chromosome segregation and cell death in the presence of p53. Methylation in CENP‐A demonstrated different effects in the absence of p53,41 which suggested a link between p53 and α‐N‐methylation.
CENP‐B contains an N‐terminal DNA‐binding domain that binds specifically to a 17‐bp CENP‐B box in centromeric α‐satellite DNA, and a C‐terminal dimerization domain. Interaction of the CENP‐B box with DNA supports faithful chromosome segregation through direct interaction with CENP‐A and CENP‐C.53 CENP‐B possesses a Gly‐Pro‐Lys sequence at its N terminus and it is primarily trimethylated in cells under stressed conditions. The α‐N‐trimethylation of chromatin‐bound CENP‐B is predominant and increases the binding of CENP‐B to the centromeric DNA.24
4. Mechanism and Inhibitors
Protein MTases generally promote a nucleophilic substitution reaction to transfer a methyl group from cofactor SAM to their substrates. NTMT1 was proposed to follow a common SN2 reaction mechanism.26, 27 NTMT1‐catalyzed methylation follows a random sequential Bi Bi mechanism, which involves the formation of a ternary complex with either substrate binding to NTMT1 first.32 Two highly conserved Asp180 and His140 act as general bases to facilitate deprotonation of the α‐amino group of the N terminus to attack SAM to transfer the methyl group.26 Mutant H140A lost the catalytic activity, but retained binding affinity to the peptide substrate.26 NTMT1 is known to be a trimethylase that catalyzes mono‐, di‐, and trimethylation.32, 33 During the process of multiple methylations, the substrate can be released and rebind to NTMT1, which proceeds through a distributive mechanism for multiple methylations.32, 33
The biological significances of NTMT1 in cell mitosis, chromatin segregation, and damaged DNA repair, along with its implications in cancer and aging, have stimulated interest in discovering NTMT1 inhibitors. Potent and specific inhibitors are critical to probe the function of each individual protein. Bisubstrate analogues that simultaneously target both binding sites are proven to be an effective strategy to obtain potent and selective inhibitors for many enzymes with two binding sites.54, 55, 56 Because NTMT1 forms a ternary complex during catalysis, a bisubstrate strategy has been applied to design and synthesize bisubstrate inhibitors by covalently linking a SAM analogue with a peptide substrate to mimic the transition state.57, 58 NTMT1 bisubstrate inhibitors contain three components: an N‐adenosyl‐l‐methionine (NAM) that replaces the sulfonium ion of SAM with a nitrogen atom, a hexapeptide derived from the N‐terminal sequence of NTMT1 substrate, and a linker (Scheme 2). Compound NAM‐TZ‐SPKRIA (IC50=(0.81±0.13) μm) contains a triazole linker and incorporates a SPKRIA peptide derived from the N terminus of RCC1, whereas NAM‐C3‐GPRRRS (IC50=0.94±0.16 μm) links a GPKRRS peptide derived from CENP‐A through a propylene group.57, 58 NAM‐TZ‐SPKRIA showed less than 50 % inhibition against PKMT G9a and PRMT1 at 50 μm.57 NAM‐C3‐GPRRRS showed no significant inhibition at 30 μm against either G9a or PRMT1.58 Kinetic analysis revealed that inhibitor NAM‐TZ‐SPKRIA engaged with both substrate binding sites.57 The potency of such bisubstrate inhibitors corroborate the Bi Bi mechanism of NTMT1 methylation. Despite the stability and cellular permeability of the above bisubstrate inhibitors, they provide a foundation to discover small‐molecule inhibitors for NTMT1.
Scheme 2.
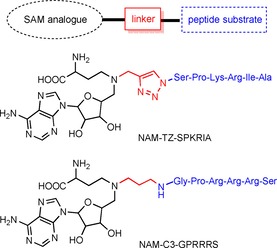
NTMT1 inhibitors.
5. Gaps and Opportunities in Protein α‐N‐Terminal Methylation
Evolutionary conservation of N‐methylation and the recognition motif of NTMT are low. It remains a puzzle whether cellular N‐terminal methylation plays differing roles in different organisms or if similar functions are exerted through different pathways. Recognition motifs of centromere‐related human substrates of NTMT1, including RCC1 and CENP‐A/B, are retained in mammals, but are missing in lower organisms.27 The recognition sequence of histone 2B protein for N‐terminal methylation is conserved from protozoans to insects, but not in mammals.27 For example, human H2B is eight residues longer than tetrahymena H2B, which masks the APKK motif.
Although an X‐P‐K/R motif search suggested about 300 proteins that may undergo α‐N‐terminal methylation, profiling a full spectrum of physiological protein substrates would shed light on pathways mediated by α‐N‐terminal methylation. Despite an increased number of reports on α‐N‐terminal methylation roles, the functions of N‐terminal methylation are still underexplored for those validated NTMT1 protein substrates, including the tumor suppressor RB1 and oncoprotein SET. Meanwhile, no recognition motif has been identified to read protein N‐terminal methylation. Revealing the molecular basis for recognizing α‐N‐terminal methylation could benefit the discovery of α‐N‐terminal methylation functions, as well as potential therapeutic applications involving α‐N‐terminal methylation in cancer and aging.
The reversibility of α‐N‐methylation is still under debate. Lys methylation used to be believed to be irreversible, but the discovery of histone demethylases, including lysine‐specific demethylase 1 and jmjc domain‐containing enzymes, demonstrate the dynamic character of histone methylation. Therefore, it is rational to hypothesize that α‐N‐terminal methylation might be reversible as well. Although predominant N‐terminal acetylation has led to a belief of the interplay between N‐terminal methylation and acetylation, rare acetylation of the X‐P‐K/R motif with Pro at the second position favors the likelihood of reversibility. Future studies are needed to determine if α‐N‐terminal methylation is reversible.
Besides the specific X‐P‐K/R recognition motif of α‐N‐terminal methylation in eukaryotes, the new discovery of N‐terminal methylation on eEF1A demonstrates a completely different recognition preference. This shifts our curiosity to question if there are any extra α‐N‐terminal methylation sites and/or writers. In summary, the identification of possible new writers, readers, and erasers would be important to illuminate the comprehensive function and regulation of the α‐N‐terminal methylation network. Finally, cell‐potent and druglike inhibitors are required for the NTMT family to enable the research community to reveal their physiological and pharmacological functions.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Rong Huang obtained her Ph.D. degree in bioorganic chemistry under the supervision of Prof. Richard F. Borch at Purdue University in 2006. After postdoctoral training with Prof. Philip A. Cole at Johns Hopkins University, she started her independent career as Assistant Professor of Medicinal Chemistry at Virginia Commonwealth University in 2011. Since 2017, she has been Associate Professor of Medicinal Chemistry and Molecular Pharmacology at Purdue University. Research in her lab focuses on the mechanism, recognition, and inhibition of protein α‐N‐terminal methylation and acetylation to identify and validate novel therapeutic targets.

Acknowledgements
This work was funded by the National Institutes of Health R01GM117275 (R.H.), U01CA214649 (R.H.), and Purdue University. The author acknowledges Andy Hudmon for critical comments and Huang laboratory members for helpful feedback.
R. Huang, ChemBioChem 2019, 20, 976.
References
- 1. Brosius J., Chen R., FEBS Lett. 1976, 68, 105–109. [DOI] [PubMed] [Google Scholar]
- 2. Wittmann-Liebold B., Pannenbecker R., FEBS Lett. 1976, 68, 115–118. [DOI] [PubMed] [Google Scholar]
- 3. Chen R., Brosius J., Wittmann-Liebold B., J. Mol. Biol. 1977, 111, 173–181. [DOI] [PubMed] [Google Scholar]
- 4. Martinage A., Briand G., Van Dorsselaer A., Turner C. H., Sautiere P., Eur. J. Biochem. 1985, 147, 351–359. [DOI] [PubMed] [Google Scholar]
- 5. Nomoto M., Kyogoku Y., Iwai K., J. Biochem. 1982, 92, 1675–1678. [DOI] [PubMed] [Google Scholar]
- 6. Henry G. D., Trayer I. P., Brewer S., Levine B. A., Eur. J. Biochem. 1985, 148, 75–82. [DOI] [PubMed] [Google Scholar]
- 7. Trayer I. P., Trayer H. R., Levine B. A., Eur. J. Biochem. 1987, 164, 259–266. [DOI] [PubMed] [Google Scholar]
- 8. Van Damme P., Arnesen T., Gevaert K., FEBS J. 2011, 278, 3822–3834. [DOI] [PubMed] [Google Scholar]
- 9. Petkowski J. J., Bonsignore L. A., Tooley J. G., Wilkey D. W., Merchant M. L., Macara I. G., Schaner Tooley C. E., Biochem. J. 2013, 456, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schaner Tooley C. E., Petkowski J. J., Muratore-Schroeder T. L., Balsbaugh J. L., Shabanowitz J., Sabat M., Minor W., Hunt D. F., Macara I. G., Nature 2010, 466, 1125–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Webb K. J., Lipson R. S., Al-Hadid Q., Whitelegge J. P., Clarke S. G., Biochemistry 2010, 49, 5225–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hamey J. J., Winter D. L., Yagoub D., Overall C. M., Hart-Smith G., Wilkins M. R., Mol. Cell. Proteomics 2016, 15, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jakobsson M. E., Małecki J. M., Halabelian L., Nilges B. S., Pinto R., Kudithipudi S., Munk S., Davydova E., Zuhairi F. R., Arrowsmith C. H., Jeltsch A., Leidel S. A., Olsen J. V., Falnes P. Ø., Nat. Commun. 2018, 9, 3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demirci H., Gregory S. T., Dahlberg A. E., Jogl G., Structure 2008, 16, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanet A., Plumberidge J. A., Guerin M. F., Alix J. H., Mol. Microbiol. 1994, 14, 947–958. [DOI] [PubMed] [Google Scholar]
- 16. Cameron D. M., Gregory S. T., Thompson J., Suh M. J., Limbach P. A., Dahlberg A. E., J. Bacteriol. 2004, 186, 5819–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demirci H., Gregory S. T., Dahlberg A. E., Jogl G., EMBO J. 2007, 26, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alamgir M., Eroukova V., Jessulat M., Xu J., Golshani A., BMC Genomics 2008, 9, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura Y., Kurata Y., Ishikawa A., Okayama A., Kamita M., Hirano H., Proteomics 2013, 13, 3167–3174. [DOI] [PubMed] [Google Scholar]
- 20. Villar-Garea A., Forne I., Vetter I., Kremmer E., Thomae A., Imhof A., Nucleic Acids Res. 2012, 40, 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T., Muratore T. L., Schaner-Tooley C. E., Shabanowitz J., Hunt D. F., Macara I. G., Nat. Cell Biol. 2007, 9, 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai X., Rulten S. L., You C., Caldecott K. W., Wang Y., J. Proteome Res. 2015, 14, 2575–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cai Q., Fu L., Wang Z., Gan N., Dai X., Wang Y., J. Biol. Chem. 2014, 289, 16046–16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dai X., Otake K., You C., Cai Q., Wang Z., Masumoto H., Wang Y., J. Proteome Res. 2013, 12, 4167–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bailey A. O., Panchenko T., Sathyan K. M., Petkowski J. J., Pai P.-J., Bai D. L., Russell D. H., Macara I. G., Shabanowitz J., Hunt D. F., et al., Proc. Natl. Acad. Sci. USA 2013, 110, 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dong C., Mao Y., Tempel W., Qin S., Li L., Loppnau P., Huang R., Min J., Genes Dev. 2015, 29, 2343–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu R., Yue Y., Zheng X., Li H., Genes Dev. 2015, 29, 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uhlén M., Bjo E., Agaton C., Szigyarto C. A., Amini B., Andersen E., Andersson A., Angelidou P., Asplund A., Asplund C., et al., Mol. Cell. Proteomics 2005, 4, 1920–1932. [DOI] [PubMed] [Google Scholar]
- 29. Bonsignore L. A., Butler J. S., Klinge C. M., Schaner Tooley C. E., Oncotarget 2015, 6, 12248–12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonsignore L. A., Tooley J. G., Van Hoose P. M., Wang E., Cheng A., Cole M. P., Schaner Tooley C. E., Mech. Ageing Dev. 2015, 146, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong C., Dong G., Li L., Zhu L., Tempel W., Liu Y., Huang R., Min J., Commun. Biol. 2018, 1, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richardson S. L., Mao Y., Zhang G., Hanjra P., Peterson D. L., Huang R., J. Biol. Chem. 2015, 290, 11601–11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richardson S. L., Hanjra P., Zhang G., Mackie B. D., Peterson D. L., Huang R., Anal. Biochem. 2015, 478, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang H., Fu Z., Jiang X., Wang N., Wang F., Wang X., Zhang S., Wang Y., Yan X., Guan W., Zhang C.-Y., Zen K., Zhang Y., Chen X., Zhou G., BMC Cancer 2015, 15, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y., Kobayashi K., Mona M. M., Satomi C., Okano S., Inoue H., Tani K., Takahashi A., J. Transl. Med. 2016, 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., Zhang G., Kong C., Zhan B., Dong X., Man X., Sci. Rep. 2016, 6, 19261. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Petkowski J. J., Schaner Tooley C. E., Anderson L. C., Shumilin I. A., Balsbaugh J. L., Hunt D. F., Minor W., Macara I. G., Biochemistry 2012, 51, 5942–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shields K. M., Tooley J. G., Petkowski J. J., Wilkey D. W., Garbett N. C., Merchant M. L., Cheng A., Schaner Tooley C. E., Protein Sci. 2017, 26, 1639–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stock A., Clarke S., Clarke C., Stock J., FEBS Lett. 1987, 220, 8–14. [DOI] [PubMed] [Google Scholar]
- 40. Hershko A., Heller H., Eytan E., Kaklij G., Rose I. A., Proc. Natl. Acad. Sci. USA 1984, 81, 7021–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sathyan K. M., Fachinetti D., Foltz D. R., Nat. Commun. 2017, 8, 14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pettigrew G. W., Smith G. M., Nature 1977, 265, 661–662. [DOI] [PubMed] [Google Scholar]
- 43. Smith G. M., Pettigrew G. W., Eur. J. Biochem. 1980, 110, 123–130. [DOI] [PubMed] [Google Scholar]
- 44. Desrosiers R., Tanguay R. M., J. Biol. Chem. 1988, 263, 4686–4692. [PubMed] [Google Scholar]
- 45. Hayashibara T., Miyanishi T., Biochemistry 1994, 33, 12821–12827. [DOI] [PubMed] [Google Scholar]
- 46. Yeh J. I., Levine A. S., Du S., Chinte U., Ghodke H., Wang H., Shi H., Hsieh C. L., Conway J. F., Van Houten B., Rapić-Otrin V., Proc. Natl. Acad. Sci. 2012, 109, E2737–E2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Renault L., Kuhlmann J., Henkel A., Wittinghofer A., Cell 2001, 105, 245–255. [DOI] [PubMed] [Google Scholar]
- 48. Clarke P. R., Nat. Cell Biol. 2007, 9, 485–487. [DOI] [PubMed] [Google Scholar]
- 49. Hao Y., Macara I. G., J. Cell Biol. 2008, 182, 827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hitakomate E., Hood F. E., Sanderson H. S., Clarke P. R., BMC Cell Biol. 2010, 11, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shuaib M., Ouararhni K., Dimitrov S., Hamiche A., Proc. Natl. Acad. Sci. USA 2010, 107, 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fachinetti D., Diego Folco H., Nechemia-Arbely Y., Valente L. P., Nguyen K., Wong A. J., Zhu Q., Holland A. J., Desai A., Jansen L. E. T., Cleveland D. W., Nat. Cell Biol. 2013, 15, 1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fachinetti D., Han J. S., McMahon M. A., Ly P., Abdullah A., Wong A. J., Cleveland D. W., Dev. Cell 2015, 33, 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dowden J., Hong W., Parry R. V., Pike R. A., Ward S. G., Bioorg. Med. Chem. Lett. 2010, 20, 2103–2105. [DOI] [PubMed] [Google Scholar]
- 55. Osborne T., Weller Roska R. L., Rajski S. R., Thompson P. R., J. Am. Chem. Soc. 2008, 130, 4574–4575. [DOI] [PubMed] [Google Scholar]
- 56. Foyn H., Jones J. E., Lewallen D., Narawane R., Varhaug J. E., Thompson P. R., Arnesen T., ACS Chem. Biol. 2013, 8, 1121–1127. [DOI] [PubMed] [Google Scholar]
- 57. Zhang G., Richardson S. L., Mao Y., Huang R., Org. Biomol. Chem. 2015, 13, 4149–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang G., Huang R., RSC Adv. 2016, 6, 6768–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]


