Abstract
A mechanistic understanding of the yeast telomere requires an integrated understanding of telomere chromatin structure (telosomes), telomeric origins-of-replications, telomere length homeostasis, and telosome epigenetics. Recent molecular and genetic studies of the yeast telosomal components, Rap1, Rif1, Rif2, the Mre11 complex, and Tel1ATM, promise to increase our insight into the coordination between these processes. We propose an intricate relationship between these multiple components that has resulted in increased appreciation of the multiple levels of telomere length control and their differentiation from DSB repair. The mre11A470 motif (A470-A482) alleles have also opened new avenues to the exploration of telosome structure and function.
Keywords: telomere, DSBs, ORIs, anti-checkpoint, telosome
The Basic Characteristics of the Yeast Telosome
Eukaryotic linear chromosomes face a problem in genomic stability, termed the end-replication problem: the expected loss of termini as a function of semi-conservative replication. Most eukaryotes solve this problem using the ribonucleoprotein reverse transcriptase telomerase (see Glossary), which uses its RNA component as template for G+T-rich telomere (see Glossary) repeats (poly G1-3T in Saccharomyces cerevisiae; abbreviated TG) [1]. The TG single-stranded DNA also forms telomeric non-Watson crick G-T Hoogsteen base pairs [2]. Duplex repeats are formed after both primer synthesis (by polymerase α) and subsequent semi-conservative replication (by polymerases δ and ε), and are packaged in a non-nucleosomal chromatin structure called the telosome [3]. However, as in most eukaryotes, an essential equilibrium between positive and negative regulators of telomere length is maintained to generate a constant average length.
The major telosome component is Repressor/Activator protein 1 (Rap1, see Glossary) [4]. Telosomal Rap1 regulates telomere length in two ways: through binding to the telomeric tract consensus sequences embedded within the telomere tract, and through association of the Rap1 C-terminus (RCT) with two negative length regulators, Rif1 and Rif2 (Rap1 interacting factors 1 and 2; see Glossary) [5–7]. Most of the functions of Rif1 are mediated through its associated protein phosphatase 1, PP1 (see Glossary), abbreviated Rif1/PP1 (see Glossary). Telosomes also associate with end-binding factors, including the Mre11 DNA damage response (DDR, see Glossary) complex, Mre11/Rad50/Xrs2 (Figure 1A) [8]. Rif1 and Rif2 also compete for RCT association with the subtelomeric heterochromatin binding factors, Sir2, Sir3 and Sir4 (Silent information regulators 2, 3, and 4) [1]. One of these, Sir4, participates in an additional positive role in telomere addition [9, 10]. Both the Rif1 protein and the Mre11 complex (see Glossary) are conserved through evolution. However, the variation in Rif1 and Mre11 complex functions at telomeric and non-telomeric sequences highlights the functional flexibility of telomere length regulators.
Figure 1: Rif1 and Tel1 Play Roles in Telomere Replication Timing and at the Telomere ‘Anti-Checkpoint’.
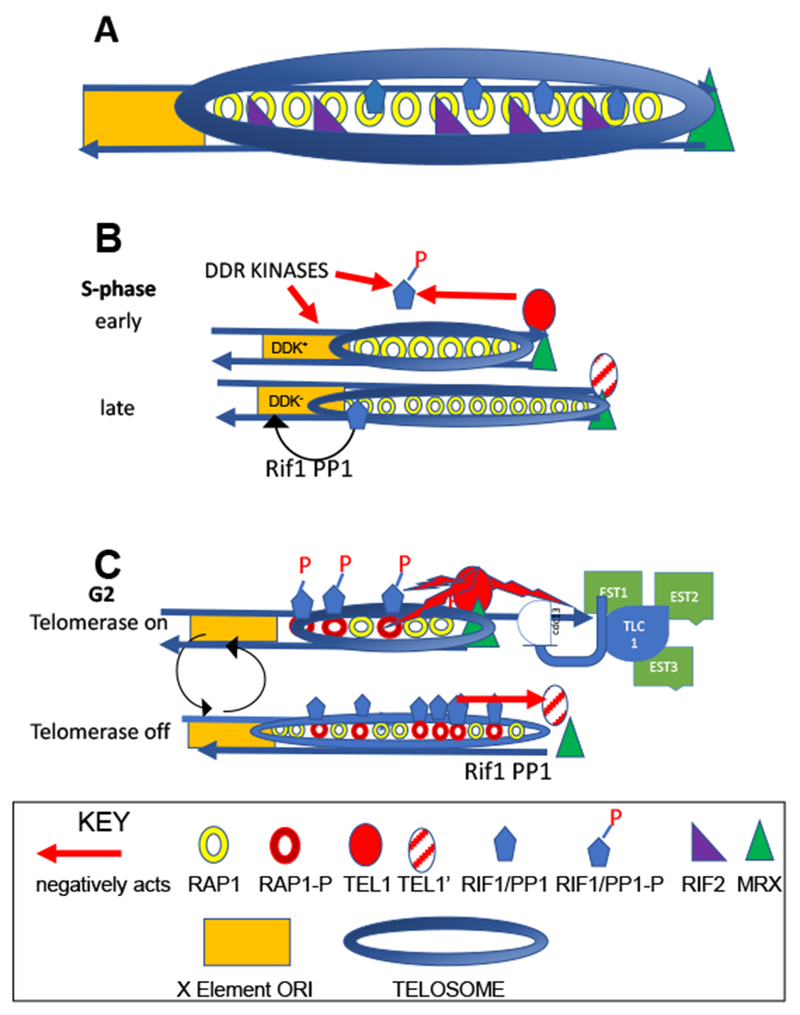
A. The telosome is the non-nucleosomal chromatin that binds both duplex telomeric DNA and several hundred base pairs of telomere-proximal subtelomeric DNA (in this case the X element). The telosome is made up of at least two primary components with additional exchangeable proteins. The first component is the telomere binding protein Rap1 that binds telomeric DNA through a duplication of a c-myb domain. Rap1 binds to a consensus motif every 20 bp of the wild type ~300 bp telomere tract and interacts with other telomeric regulators via its sites within the conserved C-terminal domain. The heterochromatin protein Sir3 and the Sir4/Sir2 heterodimer associations with the C-terminal domain are not shown.
B. The Tel1ATM kinase and other DNA damage response (DDR) kinases the Dbf4-stimulated kinase (DDK) kinases stimulate early replication of short or transiently-deleted wild-type telomeres through phosphorylation and inactivation of late-activating ORI components within the subtelomeric X element, including Rif1/PP1. In wild-type length telomeres, Rif1/PP1, among other proteins, stimulates late ORI firing due to the presence of a de-phosphorylated PP1, that serves to de-phosphorylate kinases (Rif1/PP1 arrow), including Tel1ATM that binds with lower affinity to MRX (striped red circle) and DDK that cannot activate the MCM complex, required for ORI activity.
C. The anti-checkpoint maintains a telomere length equilibrium between the addition and deletion of telomere sequences to form a stable equilibrium. Tel1ATM and the Mre11 complex bind to the shortest telomeres (red, green respectively). At the telomere, Tel1ATM activates the telomerase holoenzyme (Est1-3, Cdc13 and the telomerase RNA) and, when coupled with a second kinase that phosphorylates Cdc13, recruits the telomerase complex to single-stranded DNA. Tel1ATM also phosphorylates Rap1 and PP1, allowing Rif1/Rap1 binding and PP1 inactivation, and preventing dephosphorylation of Tel1ATM, respectively (top). As the telomere elongates, Rif1 acts through PP1 to dephosphorylate and reduce the binding of Tel1ATM to the Mre11 complex (bottom). These activities lead to a cycle of elongation and sequence loss due to deletion and replicative attrition. Although not shown, Rif2 also prevents the Tel1ATM/telosome association.
Telosomes are likely to form alternative structures depending on the associated RCT binding proteins. Crystal structure analysis of the telosome is consistent with a “Velcro structure” composed of the overlapping Rap1, Rif1 and Rif2 proteins [6].
The signature length distribution of the telomere tract in most strains of yeast has a mean telomere length of 300 base pairs with an average variation of 50 bp. Shorter telomeres are formed by replicative attrition, nucleolytic resection, and intrachromatid deletions [1, 11], while longer telomeres are the result of telomerase addition onto the shortest telomeres that are telomerase-extendible [1, 12]. For most experiments, short and long telomere lengths are defined by cloned telomeres of 80 bp and 250 bp, respectively.
Rif1, the Mre11 complex, and Tel1ATM (the yeast homolog of ataxia telangiectasia mutated {ATM}), act on short telomeres, both to regulate the temporal control of telomere replication and to create a telomere length homeostasis (see [13]). The first system regulates the timing of the subtelomeric semi-conservative origins of replication (ORIs) [14] in response to differing telomere lengths. The second system, telomere length homeostasis, uses the same regulators to maintain telomere length as part of the anti-checkpoint (see Glossary). The proposed telomere ‘anti-checkpoint’ involves the conversion of the DDR machinery into a process that protects the terminus from double-strand break (DSB) repair, checkpoint factors and checkpoint arrest [15]. We call the length homeostasis that is established while checkpoint activity is inhibited the anti-checkpoint equilibrium. The combination of replication timing and anti-checkpoint equilibrium confers the physiological telomeric state.
This commentary focuses on the unusual properties of Rif1, Rif2, Tel1ATM, and the Mre11 complex that raise intriguing questions regarding the characteristics of the physiological state and epigenetic heritability of the telosome. We also discuss studies that display the remarkable multi-functional nature of these factors.
Rif1/PP1/ORI Function Responds to Telomere Length
Rif1 has a common role at internal ORIs in S. cerevisiae (sc), Schizosaccharomyces pombe (sp), and human (h) cells [16–18]. As noted, scRif1 and telomere length are key regulators of the late-firing ORIs that lie in subtelomeric regions adjacent to yeast telosomes (Figure 1B) [14].The kinase activity of Tel1ATM stimulates early S phase replication of short 80 bp telomeres in wild type cells and truncated 160 bp telomeres in cells containing mutations in the catalytic subunit (Est2) of telomerase [19, 20]. In contrast, null mutations in the Tel1ATM gene produce short telomeres that are replicated later in S phase. Similarly, a mutation in the Xrs2 subunit of the Mre11 complex interferes with_the early replication of short telomeres. These data demonstrate a requirement for Tel1ATM - telomere recruitment by the Mre11 complex in the early replication of short telomeres. To this end, Tel1ATM and other DDR proteins target the phosphorylation of multiple S/Q sites, including, but not confined to scRif1/PP1, thereby inhibiting ORI negative regulators [20]. At wild-type length telomeres, scRif1/PP1 confers late replication through the active unphosphorylated PP1, (Figure 1B). PP1 counteracts the Dbf4-dependent kinase (DDK, see Glossary) phosphorylation of the Mini-chromosome Maintenance (MCM) complex that is essential for initiation of replication and replisome stability [21].
Human Rif1 (hRif1) has a further diverged ORI function that nonetheless bears some functional and temporal relationships to scRif1 at subtelomeric origins. Unlike scRif1 and spRif1, hRif1 is not recruited to normal telomeric tracts, and thus does not act through displacement of Tel1ATM [22, 23]. Rather, hRif1/PP1 acts to dephosphorylate and inhibit DDK [18, 24]. Based on hRif1 depletion studies, both mouse and human hRif1 also play a role in the temporal organization and late origins of replication, respectively [25, 26]. Interestingly, hRif1-PP1 also acts as a general ORI licensing factor, in part through the stabilization of the origin replication complex [18].
Anti-Checkpoint-Mediated Yeast Telomere Length Homeostasis
Experiments using the cloned 80 bp and 250 bp telomeres [13] [15] have demonstrated that the Tel1ATM DDR/checkpoint system is modified to maintain, rather than repair, telomeres, as part of a feedback loop between counteracting facilitators and inhibitors of telomerase, probably acting at a post-replicative step [27].
Three types of activities contribute to the telomere homeostasis and compensate for the smaller telomeres that are expected due to terminal primer loss and processing. First, multiple components of the Tel1ATM pathway, including the Mre11 complex and other end binding factors, process the terminus and stimulate telomerase activity preferentially on the shortest telomeres [12, 15, 28–30].
Second, site-specific phosphorylation of Rap1 by Tel1ATM and Mec1ATR have been reported to be essential for Rap1-Rif1 interaction, which in turn may interfere with Tel1ATM, and subsequent telomerase, activation [31]. This activity appears to be mediated through PP1 targeting and inactivation of Tel1ATM [27]. Conversely, Tel1ATM kinase can phosphorylate multiple S/Q sites in PP1 [32] in telomere length control, possibly as part of a regulatory loop. Extension of a short telomere is sufficient for anti-checkpoint activity to prevent cell-cycle arrest in cis and in trans.
Third, Rif2, a factor specific to yeast, binds directly to Rap1 on elongated telomeres and blocks Tel1ATM stimulation of telomerase, probably through Mre11 complex interference with Tel1ATM recruitment [33]. Both Rap1-Rif1 and Rap1-Rif2 complexes are likely to have a key role in feedback regulation of telomere elongation. As a consequence, the anti-checkpoint telomere equilibrium protects the telomere terminus from the destabilizing effects of end-to-end fusion, chromosome breakage, and translocation.
These intriguing results indicate that positive and negative regulators of short and long telomeres act on both ORI timing and telomere length homeostasis. Are these shared effects part of a larger regulatory scheme? One view is that short telomeres are normally a minority of overall telomere ends. The short telomeres may be ensured therefore of a coordinated Tel1ATM inactivation of Rif1-PPI that leads to an activation of early replication and telomerase activation. Whether this coordination is specifically established to initiate and maintain checkpoint resistance of short telomeres and chromosome stability remains unknown. If so, the mechanism may not be obligatory for all telomeres, since some subtelomeric ORIs may be dormant (see [34, 35]). In contrast, the telomere length equilibrium formed during checkpoint inhibition is likely to act at all telomeres, given their conserved structures. An experimental solution to the roles of pre- and post-replicative control in the telomere length control system awaits future progress.
Did Components of ‘Anti-Checkpoints’ Arise from ORI Regulatory Circuit?
In agreement with a previous proposal [14], we believe that, during yeast evolution, recruitment of Rif1 to telomeres by scRap1 and spTaz1 in S. cerevisiae and S. pombe [36], respectively, may have led to origin capture by telomeres that resulted in the shared factors among ORIs and telomeres. The spRifl activity to stabilize G-quartets [37] may extend to S. pombe telomeres that have DNA sequences consistent with the formation of these structures [32], although direct evidence is lacking. Rif1 capture may have been particularly selected for in organisms containing telomeres with imperfect repeats, such as S. cerevisiae and S. pombe. The presence of imperfect repeats would inhibit the formation of stable homology-driven end protective secondary structures. The common ancestor of S. pombe and S. cerevisiae may have had a strong selection for sequestration of Rif1 and PP1 at late-replicating subtelomeric ORIs (see [16]), thereby enabling telomeric chromatin to form a system of telomere homeostasis. Whether S. pombe has a telomere homeostasis or anticheckpoint system similar to S. cerevisiae awaits further testing and will help to support or deny this evolutionary model.
The Distinction Between Mre11 Complexes at Telomeric and Non-Telomeric DSBs
The Mre11 complex Mre11/Rad50/Xrs2 (NBS1 in mammals) belongs to the SMC (see Glossary)(structural maintenance of chromosomes) class of protein complexes [24]. In particular, Rad50 is structurally similar to SMC1 and SMC3 of cohesin {see Glossary) [24]. However, the Mre11 complex normally plays a central role in DNA end resection and DDR, and DNA is held together at DSBs. In contrast, cohesin confers sister chromatid association.
The eukaryotic Mre11 complex is composed of Mre11 and Rad50 homodimers containing Mre11/Rad50 association sites in each monomer. Mre11 also associates with Xrs2/NBS1 [38]. The ability of NBS1 to “stretch” across the Mre11 complex suggests the presence of additional contacts between Rad50 and NBS1. At double-strand breaks, both Mre11 and Rad50 bind DNA and participate in exo- and endo-nucleolytic processes in mitosis and meiosis, in conjunction with additional nucleases, depending upon the presence of either a blocked end or telomeric sequences. These activities have been defined through many genetic and biochemical analyses. In bacterial and archaeal species, only the Mre11/Rad50 portion of the complex is present. Yet, the high level of homology among all three has allowed interpretation of yeast mre11 and rad50 mutations [39, 40]. The crystal structures are being used to understand the Mre11 complex enzymatic reaction mechanisms at the structural level [41].
The Mre11 complex is normally associated with activation of the Tel1ATM DDR pathway [42]. At a non-telomeric DSB, Tel1ATM autophosphorylation and downstream Rad53 (hCHEK) phosphorylation subsequently leas to activation of a cascade of DNA processing enzymes. There is, however, some redundancy between the Tel1ATM pathway and the Mec1ATR (Mec1 is the yeast homolog of human ataxia telangiectasia related {ATR} pathway). 3’ overhangs are formed by the DNA nicking activity of MRX/N endo-nuclease, followed by, exonuclease resection by Mre11 and/or Sae2 3’ to 5’ exonuclease. A requirement for the Mre11 complex exonuclease depends on the sequence context, together with the presence of other DSB-specific nucleases [38]. When one or both ends contain a 3’ overhang, homologous recombination between sister chromatids is the primary mechanism of DNA repair. The recombination can involve a single-sided strand invasion, followed by replication to the end of the chromosome, using the sister chromatid as template, in a process termed break-induced replication (BIR) [43], or can involve two-sided recombination, such as gap repair. Both are error-free mechanisms, unlike those produced by non-homologous end joining that can produce both precise and imprecise fusions.
Two additional Mre11 complex activities, end-to-end interaction and nuclear membrane localization, have come to light in recent studies [24, 44]. Binding of N-terminal OB domain of Replication Factor A Subunit 1 (Rfa1, see Glossary) to a DSB is necessary for recruitment of the Mre11 complex and the interaction of the ends of the DSB [45, 46]. The same domain of Rfa1 is required for the recruitment of the Mre11 complexes that are implicated in a ‘cohesin-like’ sister chromatid association at both stalled forks and breaks [24, 47]. Following recruitment of Mre11 and terminal processing at DSBs, the two ends are held together until recombination begins [44]. The resected strands localize to the Mps3 (see Glossary) “SUN” domain at the nuclear membrane, with more unstable ends localized to nuclear pores [44].
The end-binding factors at telomeres and at typical DSBs have been investigated extensively. Regulated DNA breaks provide the most precise model system for comparison of the fate of DSBs with or without flanking telomeric sequencing at a single break. Most recently, this has been elegantly demonstrated by the fate of the two ends after digestion of a MAT allele gene with a site for a GAL-regulated-HO endonuclease following 0, 80 or 250 bp of telomeric (TG) sequence (Figure 2) [44]. After cleavage with HO, a non-TG end is present distal to the telomere sequences and the proximal end terminates with 0 bp (TG0), 80 bp (TG80), or 250 bp (TG250) of telomere sequences. This allows the direct comparison of a normal DSB and a TG-flanked DSB at the two ends of a single break.
Figure 2. The Asymmetric HO DSB Assay.
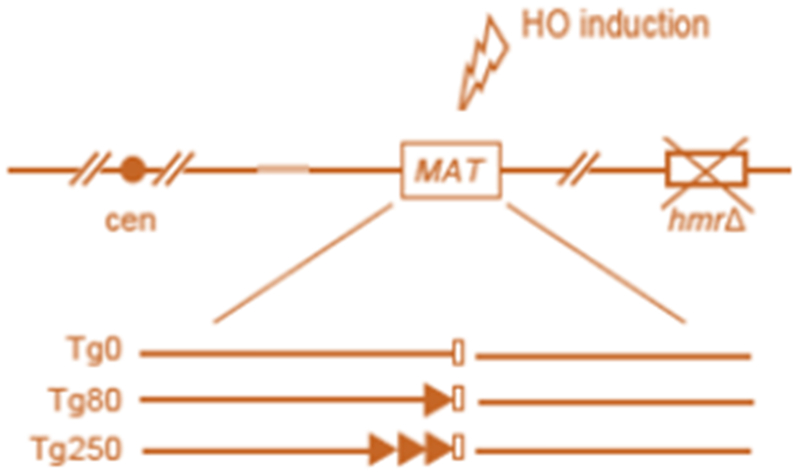
Reproduction of Figure 1A of Reference 44 demonstrating the cleavage by the HO endonuclease to produce a non—telomeric end and an end containing 0, 80 or 250 bp of telomeric sequence.
At TG-flanked HO sites, the non-telomeric end will undergo resection, which allows strand invasion into an available ectopic homologous sequence. This process may be preceded by association with Mps3. The TG side of the DSB does not undergo resection, due to its failure to recruit the Mre11 complex, leading to an asymmetric resection. In this case, a unidirectional strand invasion event, mediated by the resected non-TG side of the break, will generate a homologous non-reciprocal translocation. If there is no TG tract, there will be end resection on both sides. The binding of MRX and repair will ensue by either imprecise end joining or canonical homologous recombination.
At the terminal DSB containing 80 bp of telomeric sequence, the short tract is rapidly elongated by telomerase. The 3’ telomeric sequence overhangs can also be resolved by recombination in telomerase-negative cells. The 80 bp end is transiently associated with Mps3, dislocated by telomerase activity, and rebinds to the nuclear envelope [48, 49]. The longer 250 bp substrate, on the other hand, is not elongated by telomerase. Differential binding of the Mre11 complex to the two ends at a DSB, particularly if resection is blocked on one side by telomeric repeats, leads to loss of end-to-end association.
A critical difference between the behavior of 80 bp TG tracts at a DSB and at a telomere is the presence of Mre11 complexes at short telomeres, but not at the double-strand break. This difference may be the consequence of a greater role of the Mre11 complex with Tel1ATM recruitment and activation of telomerase than for resection [15]. We note that Mre11 complex inhibition at long telomeres may be due in part to the aforementioned Rap1-Rif1 and Rap1-Rif2 antagonism of Mre11 complex binding. Based on previous studies, additional enzymes are required for conversion to a productive telomere. These include Sae2, in conjunction with Sgs1 helicase, Dna2, and Exonuclease 1 [50]. In a telomeric context, Rfa1 has also been shown to remove other secondary structures and proteins from single-stranded telomeric ends [51]. Since the presence of different numbers of telomere sequences is likely to be related to the number of Rap1-Rif1 and Rap1-Rif2 complexes, and thus inversely to the number of the Mre11 complex molecules bound, this mechanism is consistent with the homeostatic regulation of telomere length. Further experiments are needed to resolve the difference in the behavior of TG flanking DSBs and at a bona fide telomere.
Mre11 Alleles Can Confer Altered Telosome Structures
The mre11A470T missense allele was identified in a suppressor screen for a regulated process of recombinational telomere loss, termed telomere rapid deletion [11]. This allele is part of a highly conserved motif that spans 13 amino acids from A470 to A482 (AVQEFVDKDEKDA; the Mre11A470 motif). The telomere phenotypes have been the sole effects of mre11A470 alleles rigorously tested to date, and clearly these are likely to extend to non-telomeric processes as well.
Four phenotypes have been tested only in the mre11A470T allele [8, 52]. First, telosomes in mre11A470T and MRE11 cells have distinct micrococcal nuclease patterns. Mre11A470T telosome release from chromatin requires greater times of digestion with micrococcal nuclease than Mre11 wild-type telosomes, consistent with an mre11A470T structurally less accessible (or more compact) complex.
Second, Mre11 associates, at least transiently, with the telosome. Based on ChIP analysis, wild-type telosomes can bind both wild-type and mutant Mre11. Associations of Mre11A470T with the telosome are also suggested by genetic interactions between mre11A470T and the rap1-5 allele of the RAP1 gene. The interactions lead to synergistic defects in viability and cell cycle length [8, 53]. Further quantitative ChIP characteristics of Mre11 and Mre11A470T association with wild-type and mutant telomeres studies are necessary to test whether Mre11 is an intrinsic structural component of the telosome.
Third, mre11A470T phenotypes extend to putative recombination events [52] [53]. such as the mre11A470T bypass of replicative senescence (see Glossary) in telomerase-negative cells. This process occurs through recombinational and/or replicative addition of 100bp TG1-3 tract to the Rad51-dependent class of BIR-dependent survivors [43]. The prevention of continual tract shortening is likely to lead to robust viability. One process that is consistent with such a bypass of senescence is the short-tract recombination of the TG1-3 tracts present at subtelomeric/subtelomeric junctions [54].
Fourth, the mitotic segregation pattern of haploids containing both the wild-type and mre11A470T alleles suggest that the pre-existing chromatin epigenetically defines the stable maintenance of telomere length and chromatin structure. Specifically, when a wild-type allele is integrated into the mre11A470T motif mutant, the strain maintains its original short telomere length and, in the single mre11A470T allele tested, its original telosome structure. Conversely, cells containing an integration of an mre11A470 motif allele into a wild-type strain maintain the wild-type telomere length and, at least for mre11A470T, the wild type telosome structure. At present it is unknown why the initial phenotype is maintained in this epigenetic fashion despite the presence of an equal amount of stable wild-type or mutant protein. One highly speculative model (Figure 3) posits a negative feedback regulation of the pre-existing telosome on the formation of the telosome having the opposing mre11 allele. Alternatively, wild type or mutant telosomes may elicit specific protein modifications on both sister chromatids, thereby favoring the continued formation of the same telosome form. Regardless of the mechanism of this epigenetic behavior it may well constitute an evolutionary barrier to the introduction of a telosome-Mre11 variant.
Figure 3: A Speculative Model for the Epigenetic Behavior of Wild-type and Mutant Telosomes.
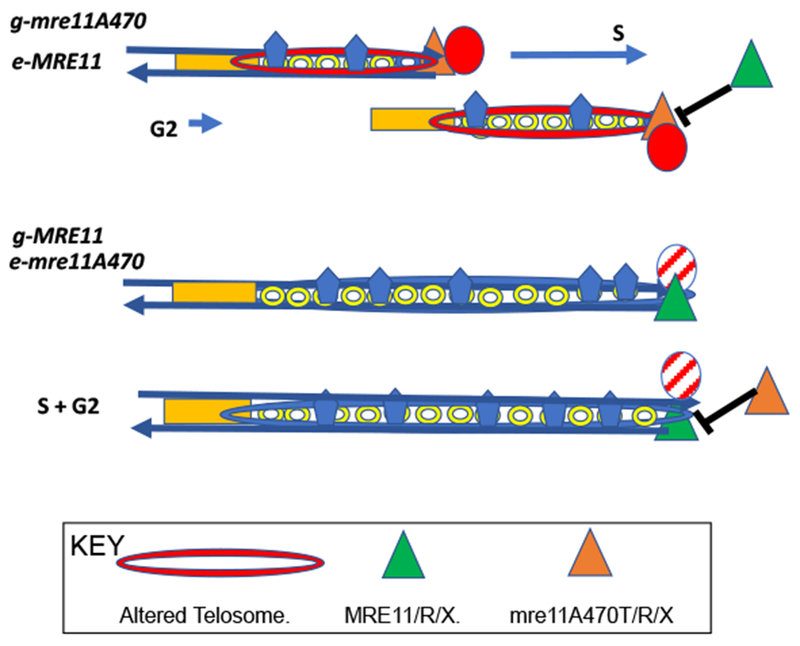
Phenotypes were determined after integration of the MRE11 allele as a single ectopic copy (e) in strains carrying an mre11A470 motif genomic allele (g) (g-mre11A470 e-MRE11). These cells led to mutant short telomeres. In contrast, the integration of mre11A470 motif allele (e) into MRE11 strains (g-MRE11 e-mre11A470) led to wild-type length telomeres. In the case of the specific mre11A470 allele tested, mre11A470T, telosome structure remained mutant in the g-mre11A470T e-MRE11 strains and g-MRE11 e-mre11A470T strains retained a wild-type telosome structure, despite the equal abundance of stable wild-type and mutant proteins in both cell types.
We propose a model based on a mutant telosome structure that has greater resistance to micrococcal nuclease (top, mutant; bottom, wild type), suggesting a more compact, tighter structure for mutant telosomes (red ovals). The central tenet of this model is the incompatibility of mutant Mre11A470T and wild-type Mre11 telosomes. Modified activities, epigenetic marking, or, a negative feedback loop, assist an S-phase microenvironment that favors formation of the mutant telomere. These epigenetic effects may play an evolutionary role in maintenance of the initial telosome after introduction of a modified form of the telosome.
The altered Mre11A470T telosome structure may be a reflection of a distortion in the association between Mre11/Rad50 binding domains. A mechanistic clue to the overall behavior of the mre11A470 alleles lies in the structure of the eukaryotic Chaetomium thermophilum (Ct) Rad50 “binding domain” (RBD) and Rad50/Nucleotide Binding Domain (NBD) which has a single Mre11/Rad50 interface [40]. In particular, the A470 motif is part of the Mre11 alpha helical bundle in the MRE11 RBD/RAD50NBD-ATPγS complex that associates at the C-terminal alpha helix of the Rad50 coiled-coil domain. Mutations in this complex, when extrapolated to the expected positions in yeast confer hypersensitivity to a number of DNA damaging agents, attesting to its functionality. Although this is one of a multiplicity of Mre11/Rad50 interacting regions, an alteration in this structure may disrupt or distort the conformation of this Mre11/Rad50 interface and the resulting phenotypes. Deletion studies of the suspected domain are needed to confirm or deny this hypothesis.
Concluding Remarks
The recent surge of experiments on the role of the yeast Rif1 and the Mre11 complex in yeast model systems has led to a markedly increased understanding of telomere homeostasis and the distinction between telomeric and non-telomeric DSBs. Clearly, the functions of Rif1 and Rif1 PP1 at both subtelomeric ORIs and telomere anti-checkpoints suggest the presence of linked activities acting on telomere dynamics, in which length is used as a mean of origin timing and as a substrate for telomerase extension. The activities of Mre11 have also been expanded beyond DDR to participation in DSB end association and nuclear substructure association. Interestingly, at DSBs, the Mre11 complex binds differentially to a non-TG sequence, giving rise to end resection. At the break having the TG sequences, the Mre11 complex does not bind, and short telomere sequences (TG80) are elongated by telomerase. This differs markedly from the role of the Mre11 complex at the telomere where it serves in the anti-checkpoint equilibrium through activation of both Tel1ATM and telomerase, to short-but not long-telomeres. In contrast, Rap1, Rif1 (and its associated PP1 phosphatase) and Rif2 play a role in the restriction of Mre11 complex/Tel1ATM recruitment. The phenotypes of mre11A470 motif alleles have extended the range of Mre11 functions to telosome structure and heritability. These data, coupled with structural studies of the telosome, will ultimately distinguish between differing mechanisms of Rap1, Rif1, Rif1, the Mre11 complex, and Tel1 in yeast telomere homeostasis (Outstanding Questions).
OUTSTANDING QUESTIONS.
DOES A FUNCTIONAL COORDINATION UNDERLY THE EFFECT OF SHORT TELOMERES ON ORI TIMING AND THE ANTI-CHECKPOINT REGULATION IN YEAST?
WHAT IS THE MECHANISM OF THE TELOMERE ANTI-CHECKPOINT?
WHAT ARE THE CAUSES OF THE DIFFERING CHARACTERISTICS OF TG TRACTS AT DSBs AND TELOMERES?
IS THE MRE11 COMPLEX AN INTRINSIC COMPONENT OF THE TELOSOME?
WHAT IS THE SOURCE OF THE TELOSOME RELATED EPIGENETIC INHERITANCE OF TELOSOME STRUCTURE AND TELOSOME TRAITS?
IS AN MRE11/RAD50 JUNCTION DEFECT RESPONSIBLE FOR THE mre11A470 PHENOTYPES?
HIGHLIGHTS.
RIF1/PPI REGULATES THE TIMING OF TELOMERE REPLICATION BASED ON TELOMERE SIZE
THE PREDOMINANT MECHANISM FOR TELOMERE SIZE HOMEOSTASIS IS THE EQUILIBRIUM BALANCE BETWEEN THE POSITIVE REGULATOR OF TELOMERASE, TEL1/MRX, AND THE NEGATIVE REGULATORS OF TELOMERE SIZE, RIF1/PP1, AND RIF2.
THE MRE11 COMPLEX HAS FUNCTIONS IN DDR, END-TO-END INTERACTION, AND NUCLEAR LOCALIZATION OF ENDS
TG SEQUENCES HAVE DIFFERENT MRE11 COMPLEX AND TEL1 REQUIREMENTS AT DSBS AND AT THE TELOMERE.
A SET OF MRE11A470 GAIN-OF-FUNCTIONALLELES ESTABLISH FUNCTIONS OF MRE11 OR REGULATORS OF MRE11 IN TELOSOME STRUCTURE, AND CHROMATIN HERITABILITY.
Acknowledgements
We would like to thank Dr. Astrid Engel and Ms. Bonnie Hoffman for critical review of the manuscript and assistance with the artwork. This study was funded by R01GM069943, and institutional funds from both the Department of Biochemistry and Molecular Biology and the Tulane Cancer Center.
Glossary
- Anti-checkpoint:
A system regulated by Tel1, the Mre11 complex, Rap1, Rif1, and Rif2 to prohibit the accumulation of checkpoint proteins in response to the telomere in cis and in trans.
- Cohesin
an SMC responsible for sister chromatids separation
- DDK:
Enzyme that phosphorylates the MCM complex that is essential for the initiation of replication
- DDR:
The checkpoint mechanisms mediated by Tel1 and Mec1 that are primarily elicited by double-strand breaks and/or collapsed or damaged forks and shortened telomeres, respectively. These proteins initiate a protein phosphorylation cascade of proteins necessary for the repair of the DNA damage
- Mps3:
The nuclear envelope protein that is responsible for association with resected double strand breaks
- Mre11 complex:
The SMC is composed of a dimer of Mre11, a dimer of Rad50, and, in eukaryotes, a monomer of NBS1 (Xbs2 in yeast). The complex is recruited for DSBs to initiate the Tel1 DDR and also serves to resect DSBs for processing. At the telomere, the complex is involved in recruiting Tel1 as a positive regulator of telomerase
- PP1 (Rap1/Rif1):
The phosphatase partner protein of Rif1 that both inactivate S/Q kinases and is inactivated by S/Q kinases forming regulatory loops in replication timing and telomere length homeostasis
- Rap1:
The major telomere binding protein in yeast that binds regulatory factors involved in telomere length homeostasis (Rif1, Rif2) and silencing (Sir2, Sir3, Sir4). Rap1 is also found as a promoter element in a multiplicity of genes
- Rif1:
The Rap1 associating protein that is a negative regulator of telomere length homeostasis and an antagonist of subtelomeric late firing origins of replication
- Rif2:
A protein specific to yeast that acts as a negative regulator of telomerase through interference with Tel1 association at the telomere
- Rfa1:
Subunit 1 of Replication Factor A, a multi-functional protein. In this context, Rfa A 1 is required for the loading of the Mre11 complex onto resected DSBs and for the protein- and DNA-structure clearing of telomeric single-stranded DNA
- Senescence:
In the absence of telomerase, telomere sequence in yeast is slowly lost leading to a dysfunctional telomere incapable of supporting viability.
- SMC:
Protein complexes with a high level of structural similarity that are responsible for the interaction between DNA, chromatids or chromosomes.
- Tel1ATM:
An S/Q kinase that activates a pathway similar to the higher eukaryotic sATM pathway that is responsible for repair of double-strand breaks and the activation of telomerase and early replication at short telomeres.
- Telomerase:
The ribonucleoprotein complex that contains a catalytic protein (Est2) and the RNA component that contains the template for the GT-rich sequence as the core enzyme, and additional regulators, Est1, Est3, and Cdc13 as the holoenzyme
- Telomere:
The termini of eukaryotic chromosomes that solve the intrinsic end-replication problem normally through activation of the ribonucleoprotein reverse transcriptase, telomerase
- Telosome:
The chromatin structure that is formed at telomere and regulates a multiplicity of telomere functions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wellinger RJ and Zakian VA (2012) Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191 (4), 1073–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lustig AJ (1992) Hoogsteen G-G base pairing is dispensable for telomere healing in yeast. Nucleic Acids Res 20 (12), 3021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright JH et al. (1992) Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev 6 (2), 197–210. [DOI] [PubMed] [Google Scholar]
- 4.Wright JH and Zakian VA (1995) Protein-DNA interactions in soluble telosomes from Saccharomyces cerevisiae. Nucleic Acids Res 23 (9), 1454–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy CF et al. (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6 (5), 801–14. [DOI] [PubMed] [Google Scholar]
- 6.Shi T et al. (2013) Rif1 and Rif2 shape telomere function and architecture through multivalent Rap1 interactions. Cell 153 (6), 1340–53. [DOI] [PubMed] [Google Scholar]
- 7.Wotton D and Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11 (6), 748–60. [DOI] [PubMed] [Google Scholar]
- 8.Baek IJ et al. (2017) The mre11 A470 alleles influence the hereditability and the segregation of telosomes in Saccharomyces cerevisiae. PLoS One 12 (9), e0183549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hass EP and Zappulla DC (2015) The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palladino F et al. (1993) SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75 (3), 543–55. [DOI] [PubMed] [Google Scholar]
- 11.Bucholc M et al. (2001) Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol Cell Biol 21 (19), 6559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teixeira MT et al. (2004) Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117 (3), 323–35. [DOI] [PubMed] [Google Scholar]
- 13.Hirano Y et al. (2009) Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 33 (3), 312–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattarocci S et al. (2016) Rif1: A Conserved Regulator of DNA Replication and Repair Hijacked by Telomeres in Yeasts. Front Genet 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribeyre C and Shore D (2012) Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 19 (3), 307–13. [DOI] [PubMed] [Google Scholar]
- 16.Hafner L et al. (2018) Rif1 Binding and Control of Chromosome-Internal DNA Replication Origins Is Limited by Telomere Sequestration. Cell Rep 23 (4), 983–992. [DOI] [PubMed] [Google Scholar]
- 17.Hayano M et al. (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26 (2), 137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraga SI et al. (2017) Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep 18 (3), 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooley C et al. (2014) Tel1ATM dictates the replication timing of short yeast telomeres. EMBO Rep 15 (10), 1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sridhar A et al. (2014) At short telomeres Tel1 directs early replication and phosphorylates Rif1. PLoS Genet 10 (10), e1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alver RC et al. (2017) Reversal of DDK-Mediated MCM Phosphorylation by Rif1-PP1 Regulates Replication Initiation and Replisome Stability Independently of ATR/Chk1. Cell Rep 18 (10), 2508–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L and Blackburn EH (2004) Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol 167 (5), 819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dave A et al. (2014) Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep 7 (1), 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeber A et al. (2016) RPA Mediates Recruitment of MRX to Forks and DoubleStrand Breaks to Hold Sister Chromatids Together. Mol Cell 64 (5), 951–966. [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki S et al. (2012) Rif1 regulates the replication timing domains on the human genome. EMBO J 31 (18), 3667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornacchia D et al. (2012) Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31 (18), 3678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedziora S et al. (2018) Rif1 acts through Protein Phosphatase 1 but independent of replication timing to suppress telomere extension in budding yeast. Nucleic Acids Res 46 (8), 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabourin M et al. (2007) Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27 (4), 550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hector RE et al. (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27 (5), 851–8. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi A and Shore D (2007) Early replication of short telomeres in budding yeast. Cell 128 (6), 1051–62. [DOI] [PubMed] [Google Scholar]
- 31.Yang CW et al. (2017) Telomere shortening triggers a feedback loop to enhance end protection. Nucleic Acids Res 45 (14), 8314–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J et al. (2018) A Heterochromatin Domain Forms Gradually at a New Telomere and Is Dynamic at Stable Telomeres. Mol Cell Biol 38 (15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martina M et al. (2012) A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres. Mol Cell Biol 32 (9), 1604–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis EJ and Vershinin AV (2005) Chromosome ends: different sequences may provide conserved functions. Bioessays 27 (7), 685–97. [DOI] [PubMed] [Google Scholar]
- 35.McCarroll RM and Fangman WL (1988) Time of replication of yeast centromeres and telomeres. Cell 54 (4), 505–13. [DOI] [PubMed] [Google Scholar]
- 36.Kanoh J and Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11 (20), 1624–30. [DOI] [PubMed] [Google Scholar]
- 37.Kanoh Y et al. (2015) Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat Struct Mol Biol 22 (11), 889–97. [DOI] [PubMed] [Google Scholar]
- 38.Paull TT (2018) 20 Years of Mre11 Biology: No End in Sight. Mol Cell 71 (3), 419–427. [DOI] [PubMed] [Google Scholar]
- 39.Lammens K et al. (2011) The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145 (1), 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seifert FU et al. (2016) Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J 35 (7), 759–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syed A and Tainer JA (2018) The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu Rev Biochem 87, 263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suetomi K et al. (2010) Effects of Saccharomyces cerevisiae mecl, tell, and mrel 1 mutations on spontaneous and methylmethane sulfonate-induced genome instability. Genes Genet Syst 85 (1), 1–8. [DOI] [PubMed] [Google Scholar]
- 43.McEachern MJ and Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75, 111–35. [DOI] [PubMed] [Google Scholar]
- 44.Marcomini I et al. (2018) Asymmetric Processing of DNA Ends at a Double-Strand Break Leads to Unconstrained Dynamics and Ectopic Translocation. Cell Rep 24 (10), 2614–2628 e4. [DOI] [PubMed] [Google Scholar]
- 45.Kaye JA et al. (2004) DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol 14 (23), 2096–106. [DOI] [PubMed] [Google Scholar]
- 46.Lobachev K et al. (2004) Chromosome fragmentation after induction of a doublestrand break is an active process prevented by the RMX repair complex. Curr Biol 14 (23), 2107–12. [DOI] [PubMed] [Google Scholar]
- 47.Tittel-Elmer M et al. (2009) The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J 28 (8), 1142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira HC et al. (2011) The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol 13 (7), 867–74. [DOI] [PubMed] [Google Scholar]
- 49.Schober H et al. (2009) Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23 (8), 928–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nugent CI et al. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol 8 (11), 657–60. [DOI] [PubMed] [Google Scholar]
- 51.Audry J et al. (2015) RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J 34 (14), 1942–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joseph IS et al. (2010) An mre11 mutation that promotes telomere recombination and an efficient bypass of senescence. Genetics 185 (3), 761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao H et al. (2014) The Ctf18RFC clamp loader is essential for telomere stability in telomerase-negative and mre11 mutant alleles. PLoS One 9 (2), e88633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aksenova AY et al. (2015) Expansion of Interstitial Telomeric Sequences in Yeast. Cell Rep 13 (8), 1545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]


