Abstract
The speed at which bacteria develop antimicrobial resistance far outpace drug discovery and development efforts resulting in untreatable infections. The World Health Organisation recently released a list of pathogens in urgent need for the development of new antimicrobials. The organisms that are listed as the most critical priority are all Gram-negative bacteria resistant to the carbapenem class of antibiotics. Carbapenem resistance in these organisms is typified by intrinsic resistance due to the expression of antibiotic efflux pumps and the permeability barrier presented by the outer membrane, as well as by acquired resistance due to the acquisition of enzymes able to degrade β-lactam antibiotics. In this perspective article we argue the case for reversing resistance by targeting these resistance mechanisms – to increase our arsenal of available antibiotics and drastically reduce antibiotic discovery times – as the most effective way to combat antimicrobial resistance in these high priority pathogens.
Keywords: antibiotic resistance, antibiotics, efflux pump inhibitor, membrane permeability, reversal of resistance, synergism
The current status of antimicrobial resistance
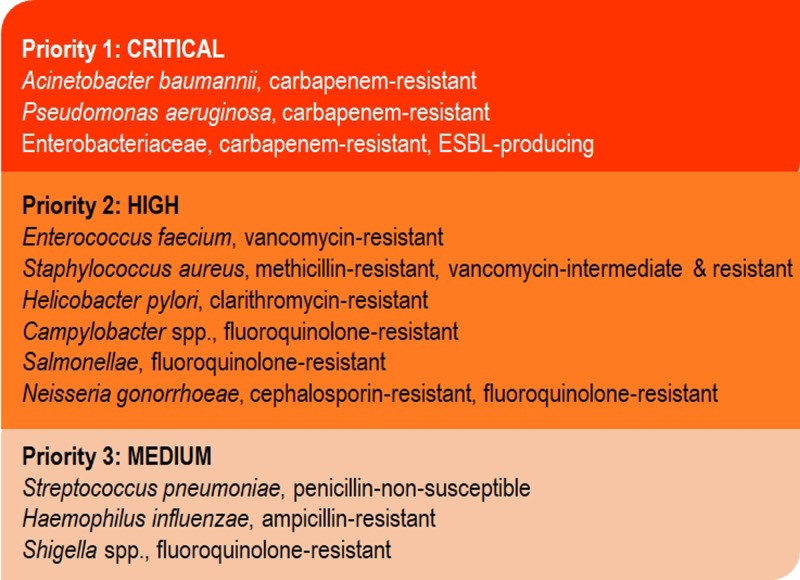
The World Health Organisation (WHO) recently published a list of antimicrobial-resistant (AMR) organisms for which the need of new therapies are the greatest (Figure 1). The most critical priority consists solely of Gram-negative organisms, specifically carbapenem-resistant Acinetobacter baumannii, carbapenem-resistant Pseudomonas aeruginosa and members of the family Enterobacteriaceae which are carbapenem-resistant and containing extended spectrum β-lactamases [1]. Infections caused by Gram-negative pathogens prove much harder to treat compared with Gram-positive organisms due to the very high intrinsic drug resistance of Gram-negatives. Intrinsic antibiotic resistance in these organisms is due to the presence of an outer membrane (OM) – which acts as a permeability barrier – and the expression of several drug efflux pumps [2–4]. Additionally, these organisms could also harbour acquired resistance mechanisms such as drug inactivation through β-lactamases that would render β-lactam antibiotics ineffective, or modification of the drug target so that the antibiotic can no longer efficiently act on that target [2] (Figure 2). Most multidrug-resistant organisms harbour several of these resistance mechanisms (e.g. [3]). However, antibiotic efflux is the predominant mechanism for aminoglycoside resistance in P. aeruginosa [5], fluoroquinolone resistance in Listeria monocytogenes [6] resistance to a variety of antibiotics in Burkholderia species [7] and linezolid resistance in a range of Gram-negative pathogens [8] even though the latter is available for the treatment of resistant Gram-positive bacteria. The combination of reduced OM permeability (through a lack of the OprD porin) and efflux pump expression were reported to be the main mechanisms of carbapenem resistance in P. aeruginosa [9].
Figure 1. The list of the most dangerous pathogens in need of antimicrobial drug development according to the WHO [1].
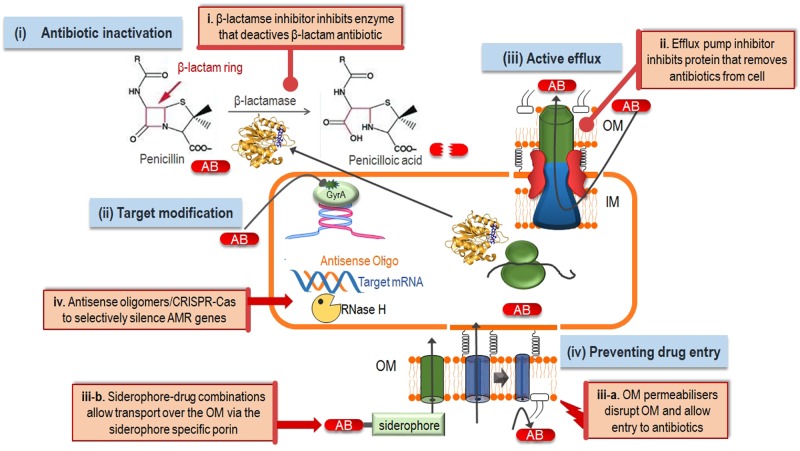
Figure 2. Antimicrobial resistance mechanisms in Gram-negative bacteria and ways to reverse resistance.
The four main mechanisms of antibiotic resistance in Gram-negative organisms (blue boxes) are (i) antibiotic inactivation, for example, the production of β-lactamase enzymes that hydrolyse the β-lactam ring thereby deactivating this class of antibiotics; (ii) target modification, for example modifications in the GyrA protein confers resistance to fluoroquinolones; (iii) active efflux, where drug efflux pumps remove the antibiotic from the bacterial cell thereby lowering antibiotic concentration to sub-toxic levels and (iv) prevention of drug entry through the OM by the expression of more selective porins, mutations in porins or loss of porins. Antibiotic resistance can be reversed by the addition of resistance breakers (orange boxes) such as (i) β-lactamase inhibitors to prevent antibiotic degradation; (ii) efflux pump inhibitors to allow the antibiotic to reach its target instead of being removed by the efflux pump; (iii-a) OM permeabilisers that destabilise the bacterial cell, thereby allowing antibiotics entry through the normally impenetrable OM; (iii-b) siderophore-drug conjugates which allow the antibiotic to breach the OM barrier by being transported through the siderophore specific porin and (iv) gene-silencing techniques to prevent expression of resistance determinants.
Very limited treatment options remain for infections caused by carbapenem-resistant Gram-negatives. Hence, the ability to reverse resistance in these organisms would be of immense clinical value.
Why is there such a shortfall in antibiotics?
Despite the well-recognised medical need for new antibiotics and the almost linear increase in antibiotic resistant infections, there has been a dramatic decrease in antibacterial drug discovery [10–12]. Many companies left the area with Novartis being the latest company to close down their anti-infective discovery pipeline (https://www.fiercebiotech.com/biotech/despite-looming-resistance-crisis-novartis-ducks-out-antibiotics-research).
The main reason for the lack of antibiotic development by pharmaceutical companies is the low return on investment [13]. Contributing factors to the lack of financial gain to be derived from antibiotic development are the short treatment times (1–7 days typically), stewardship (restricting the use of antibiotics) and short-lived efficacy before resistance start to develop (1–4 years) while the development of antibiotic is under the same stringent regulatory requirements as more lucrative drugs [14,15].
Antibiotic drug discovery pose many challenges e.g. GlaxoSmithKline used bioinformatic analysis of genomic information to identify new antibiotic targets and ran more than 70 high-throughput screening campaigns between 1995 and 2001 which did not yield a single agent to the antibiotics pipeline [16]. In fact, almost all of the antibiotics approved during the last 30 years are modifications of earlier classes of antibiotics to increase efficacy against resistant bacteria (data from the PEW charitable trusts, February 2018, https://www.pewtrusts.org/-/media/assets/2018/03/antibiotics_clinical_dev_table_february2018.pdf) [14,17] and with major drug companies exiting the field, antibiotic drug discovery now falls on academic institutions such as the Combating Antibiotic Resistant Bacteria Biopharmaceutical Accelerator (CARB-X) programme, the BEAM alliance [18], the Global Antibiotic Research & Development Partnership (GARDP) and CO-ADD community open source [19,20].
However, the trends in antibiotic development still suggest that the standard pharmaceutical and economic model will not be sufficient to address the lack of new antibiotics. While modifications of current classes of antibiotics constitutes a valuable approach, given the unique nature of these targets and consequent ability to develop agents with a high therapeutic index, we argue here the case for the development of compounds that could reverse resistance, hence reinvigorate antibiotics to which resistance has developed.
Reversal of resistance
Compounds that reverse resistance (generally termed antibiotic adjuvants, resistance breakers, antibiotic potentiators or chemosensitisers) possess no or little antimicrobial activity themselves, but when co-administered with an antibiotic, they potentiate the activity of the antibiotic [21–23]. Most resistance breakers act by inhibiting one of the following three resistance mechanisms: (i) inhibition of the β-lactamase enzymes that inactivate β-lactam antibiotics, (ii) inhibition of antibiotic efflux pumps, (iii) acting on the bacterial OM to breach the OM permeability barrier. (iv) A completely different kind of therapy, that is not based on small molecule adjuvants, is the use of antisense-mediated gene silencing [24] or the bacterial CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated) immune system [25] (Figure 2).
(i) Inhibitors of β-lactamase enzymes
β-Lactam antibiotics are among the most useful and frequently prescribed classes of antibiotics to treat bacterial infections. β-Lactams target the penicillin binding protein (PBP, a peptidyl transferase) which is a crucial enzyme needed for cell wall synthesis in bacteria. Bacteria can relatively quickly and effectively acquire resistance to β-lactams by the production of β-lactamase enzymes which cleave the β-lactam ring thereby rendering the antibiotic ineffective [2]. The use of β-lactamase inhibitors combined with a β-lactam antibiotic has been a successful strategy for overcoming β-lactamase-mediated resistance. β-Lactamase inhibitors are compounds (mostly stable β-lactams) that inhibit the β-lactamase enzymes and hence prevent antibiotic degradation (Figure 2) [26]. These inhibitors are already clinically used to great effect. For example, the combination of the β-lactam antibiotic amoxicillin and the β-lactamase inhibitor clavulanic acid is one of the most commonly prescribed antibiotics the community and hospitals [2]. A substantial amount of research and development have also been done in this field (e.g. reviewed recently in [27]. Therefore, this perspective piece would not elaborate on β-lactamase inhibitors other than an example of the newest development in the field and stating that their success is proof-of-principle that reversal of other resistance mechanisms are a viable option for treating multidrug-resistant infections.
New developments – Vabomere: a carbapenem and carbapemase inhibitor combination to treat complicated urinary tract infections
The carbapenem class of β-lactams are a particularly useful class of antibiotics, especially in the treatment of infections caused by multidrug-resistant Gram-negatives, as a result of their resistance to hydrolysis by numerous β-lactamases [28,29]. However, many Gram-negative organisms such as P. aeruginosa, A. baumannii, Escherichia coli, Klebsiella pneumoniae, etc. produce powerful carbapemases that inactivate nearly all β-lactams (including carbapenem antibiotics). The widespread dissemination of carbapemases is threatening the effectiveness of this class of antibiotics with very few treatment options remaining for serious Gram-negative infections [30]. A proven strategy to overcome resistance driven by β-lactamases is by co-administration of a β-lactamase inhibitor with the β-lactam antibiotic (Figure 2) [31–34]. Unfortunately, older inhibitors such as clavulanate are not effective against carbapemases. Vaborbactam is a novel, non-β-lactam that was specifically developed to inhibit KPC β-lactamases [35]. A combination of vaborbactam and meropenem displayed potent inhibitory activity against carbapenem-resistant Gram-negative bacteria [36,37]. In August 2017, the FDA approved the use of a meropenem–vaborbactam combination (Vabomere) for complex urinary tract infections caused by resistant Gram-negative organisms, confirming the huge potential of this method of resistance reversal when few other treatment options remain.
(ii) Efflux pump inhibitors
Antibiotic efflux pumps are membrane proteins that actively remove antibiotics from the bacterial cell thereby lowering on-target antibiotic concentrations to sub-toxic levels (Figure 2) [38–43]. These efflux pumps are able to recognise and expel a wide spectrum of antimicrobial compounds thereby conferring multidrug resistance on pathogens including resistance against common disinfectants and last line antibiotics such as colistin [44–50]. The promiscuous substrate specificity of efflux pumps also means that other chemicals including disinfectants could select for resistance against antibiotics. For instance, an antidepressant and the chemicals found in common weed killers have been shown to select for organisms with increased resistance to clinically used antibiotics such as fluoroquinolone through the expression of antibiotic efflux pumps [51–53]. Moreover, organisms can only acquire resistance in the presence of active efflux pumps [54] and enhanced efflux of antibiotics contributes to bacterial persistence during antibiotic and other stresses [55]. Hence, efflux pumps are very attractive targets for inhibition [43,56]. Efflux pump inhibitors (EPIs) could synergise with antibiotics and resensitise bacteria to these antibiotics. This would greatly extend the arsenal of available antibiotics and also extend the lifetime of antibiotics in currently clinical use (Figure 2).
Antibiotic efflux pumps in Gram-negative organisms are large macromolecular complexes that span the inner membrane (IM), the OM and the periplasm of Gram-negative pathogens [57–59]. These drug efflux complexes are tripartite assemblies consisting of an inner-membrane protein (IMP) of the resistance nodulation cell division (RND) family, an outer-membrane protein (OMP) and a periplasmic adapter protein (PAP), which connects the first two proteins (Figure 2). The IMP catalyses drug/H+ antiport and is the part of the complex responsible for drug selectivity. Although Gram-negative organisms have the ability to express different classes of drug efflux proteins, the RND-type of efflux systems are the only ones that confer clinical levels of resistance [41,60–62].
The best studied example of an EPI against the tripartite antibiotic efflux pump of Gram-negative organisms is phenylarginyl-β-naphthylamide (PAβN), a simple naphthylamide peptide which did not progress beyond clinical trials due to toxicity [63]. Recent activity in this field by our group and others led to the design and synthesis of several compounds with increased efficacy [64–67] and low cytotoxicity [68].
The current status of EPI discovery
Several compounds that are able to synergise with antibiotics against drug-resistant Gram-negative bacteria are described in the literature. However, the rate of translation of these promising compounds into EPIs for clinical application is still low. One of the foremost reasons for poor eventual performance of promising lead compounds is due to the lack of follow-through from first identification of a compound with synergistic effects to identification of target-specific activity, and then execution of a thorough investigation into its mechanism of action. One of the most significant problems in current screening campaigns for EPIs is that in many cases the synergism observed is actually due to off-target effect such as non-specific damage to the bacterial membrane [43]. This is an important issue, as it indicates that the compound could have similar activity against mammalian cells and hence would be cytotoxic. This was clearly the case for PAβN [63].
However, our and other groups has some success with functional and structural determination of tripartite efflux pumps [57,69–71] as well as their interaction with carbapenems [72,73]. This robust understanding of assembly and efflux mechanism combined with the first inhibitor-bound structures of RND-type efflux proteins [66,74] could form a solid platform for drug discovery and development aimed at reversing resistance through efflux inhibition.
New developments – an EPI success story for Gram-positive infections
As is the case with all antibiotic development, the development of EPIs for Gram-negative bacteria lags behind that of Gram-positive bacteria. Many compounds that inhibit the efflux pumps of Gram-positive bacteria have been discovered and are well-progressed along the path of clinical development. Specifically, EPIs that reverse resistance in Mycobacterium tuberculosis have already been shown to accelerate treatment with rifampin in murine models of infection [75] which have led to the initiation of a clinical trial (Annual Report of the National Institute for Research in Tuberculosis; http://www.nirt.res.in).
(iii) By-passing the permeability barrier
Another intrinsic mechanism of resistance in Gram-negative organism is the OM which is the first line of defence by acting as a formidable permeability barrier to prevent the entry of many antibiotics (Figure 2). The OM is an elaborate asymmetric bilayer consisting of phospholipids (inner leaflet) and lipopolysaccharides or lipo-oligosaccharides [76,77]. Large hydrophobic antibiotics can traverse the OM through passive diffusion which is a relatively slow process, while small hydrophilic compounds gain access through porins (used for uptake of nutrients) that are embedded in the OM [4,78–80]. Large hydrophobic antibiotics are excluded. Examples include compounds that are effective against Gram-positive bacteria such as vancomycin and teicoplanin.
The efficacy of antibiotics heavily relies on their ability to reach their intended targets at inhibitory concentrations. Efficient delivery of antibiotics to their bacterial targets are therefore an additional challenge in Gram-negative bacteria that should be taken into account in the development of antibiotic treatments against these infections [81]. Methods to quantify antibiotic concentrations in the periplasm or in the cytosol of bacterial cells [82–84] would therefore be a valuable tool in future antimicrobial drug discovery against Gram-negative pathogens. Additionally, the importance of porins in the uptake of antibiotic necessitate an in-depth understanding of the translocation process [78] that would facilitate the use of virtual screening techniques to search for new molecular scaffolds with enhanced permeation [85].
Several studies have reported that targeting of OM permeability can be an effective strategy for increasing antibiotic efficacy [22,86–88]. Some antibiotic screening campaigns use a ΔTolC mutant of E. coli as absence of the OMP TolC enhances drug sensitivity [89]. There are some merits in this approach as it would allow a higher rate of discovery of compounds with activity against various targets in Gram-negative bacteria; target delivery can then be the next step in the drug development pathway. A different approach is to deliver antibiotics in combinations with chemosensitisers that could breach the permeable barrier of the OM and so enhance antibiotic uptake.
OM permeabilisers
We have investigated the addition of ethylenediaminetetraacetic acid (EDTA) as chemosensitiser. EDTA is a well-known metal chelator that can cause OM permeabilisation [90] and is widely used to study e.g. bioenergetics in Gram-negative bacteria [91] and for dye-based methods to confirm IM integrity [67,68]. EDTA treatment leads to a release of LPS which is then compensated for by an increase in glycerophospholipids, resulting in patches of phospholipid bilayer with increased permeability to lipophilic compounds [76]. We have already showed that OM permeabilisation with sub-toxic concentrations of EDTA could enhance efficacy of an EPI by several fold [67] and that a Gram-positive selective new antibiotic also displayed Gram-negative activity in the presence of EDTA [92]. The safety profile of EDTA by itself is well-established as intravenous EDTA-chelation therapy is used to treat lead poisoning [93] and an EDTA chelation therapy regimen has been trailed to determine its safety and efficacy for individuals with prior heart attacks [94].
Other chemosensitisers are compounds that are used as antimicrobials such as silver, polymyxins, etc. but could be used as antibiotic adjuvants at sub-toxic levels to enhance permeation and subsequent efficacy of antibiotics [87,95]. Polymyxins are cationic cyclic lipopeptides that bind to LPS and so permeabilise the OM. These peptides re-emerged in clinics to treat multidrug-resistant Gram-negative infections. Polymyxin E, otherwise known as colistin, is now the last-resort treatment for infections caused by carbapenem-resistant pathogens [96,97]. The dose regime for polymyxins needs to be very carefully controlled due to their inherent nephrotoxicity. However, polymyxins could be used at concentrations far below their MIC to permeabilise the OM and synergise with other antimicrobials [95,98,99]. Alternatively, the non-cytotoxic polymyxin non-apeptide could be used as antibiotic adjuvant [86]. The octapeptins are another family of cyclic lipopeptides which were discovered about 40 years ago and, similar to the polymyxins, they have been largely ignored in the interim. Importantly though, octapeptin retains efficacy against polymyxin-resistant bacteria due to their interaction with both lipid A and phospholipids. Octapeptins also have a broader spectrum of activity that include Gram-positives and yeasts and displays a superior preclinical safety profile compared with the polymyxins [86,100,101]. Hence, in addition to their antibiotic activity octapeptin could also be ideal resistance breakers to be used as adjuvants to reverse resistance in the most critically important multidrug-resistant organisms. Pletzer et al. [102] reported that antibiofilm peptides also acted as resistance breakers and synergised with a range of antibiotics in an in vivo mouse model of infection with multidrug-resistant Gram-negatives bacteria such as K. pneumoniae, A. baumannii, P. aeruginosa and Enterobacter cloacae. At least part of this synergism was due to OM permeabilisation by the peptides.
Lipid modulation plays an important role in permeability, hence compounds that would alter the lipid composition or lipid content of the OM could be valuable chemosensitisers too. To this extent, a high-throughput analysis revealed the small molecule MAC13243 as membrane permeabiliser to facilitate increased influx of large antibiotics in E. coli. This molecule was identified as an inhibitor of the LolA, a periplasmic chaperone that traffics lipoproteins from the inner to the OM [103].
A ‘Trojan horse’ strategy of connecting antibiotics to iron-binding molecules (siderophores) and thereby utilising the inherent iron uptake machinery of Gram-negative bacteria to breach the OM barrier has been under investigation since the 1980s (Figure 2) [104,105]. Siderophores are high-affinity iron scavenging molecules excreted by pathogens to remove iron thereby allowing the organism to overcome iron limitation in the host. Gram-negative organisms use dedicated OM porins to allow entry to the siderophores. Importantly, the addition of antibiotics to siderophores did not seem to hamper their uptake through their respective porins. This very promising approach lead to the development of several siderophore-conjugated monobactam antibacterial agents with excellent in vitro activity against multidrug-resistant Gram-negative pathogens such as P. aeruginosa [106]. Unfortunately, further development of these particular combinations was hampered by a lack of in vivo activity and the quick development of resistance against the first candidate conjugates [107,108].
New developments – Trojan horses to deliver antibiotics over the OM
Siderophore–antibiotic combinations has been revisited, this time with clinical success. Cefiderocol (a catechol-substituted siderophore–cephalosporin combination) has excellent in vitro [109] and in vivo [110] efficacy against a range of Gram-negative multidrug-resistant organisms and is currently undergoing Phase 3 clinical trials [111].
The Gram-negative bacterial OM is not just an intrinsic resistance mechanism, but organisms can also acquire resistance through mutations in the porins through which antibiotics gain access over the OM. Hence, antibiotic adjuvants that permeabilise the OM or therapeutics designed to be transported over the OM are excellent ways to breach both these intrinsic and acquired resistance mechanisms of the OM and is gaining track as valuable treatment options to reverse resistance in multidrug-resistant Gram-negative bacteria.
(iv) Gene silencing technologies
Gene silencing technologies for the reversal of resistance is still in its infancy with many hurdles, notably on-target delivery of these technologies, still to be overcome. The expression of resistance genes could be suppressed by either antisense oligomers [24] or by utilising the bacterial CRISPR-Cas immune system [25].
Antisense oligomers targeted at AMR resistance genes
Antisense oligomers are short, single-stranded oligomers that mimic the structure of DNA or RNA. Based on the chemistry of the sugar-phosphate backbone the antisense oligomers can be divided into RNase H-incompetent or RNase H-competent. RNase H-incompetent antisense oligomers bind to the target RNA and prevents binding of the 30s Ribosome, thereby preventing transcription of the mRNA while binding of RNase H-competent antisense oligomers leads to activation of RNase H and degradation of the target mRNA. The latter approach has the distinct advantage that RNase H-dependent oligonucleotides can inhibit protein expression when targeted to virtually any region of the mRNA while the RNase H-incompetent oligonucleotides are efficient only when targeted to the 5′- or AUG initiation codon region [112]. For this reason the majority of antisense drugs investigated for clinical use are function via the RNase H-dependent mechanism. Fomivirsin, the first FDA-approved antisense therapeutic that targets a microorganism (cytomegalovirus) is also based on an RNase H-dependent mechanisms [113].
The New Delhi metallo-β-lactamase (NDM-1) is a plasmid-associated metallo β-lactamase that confers resistance to carbapenem antibiotics. Sully et al. [114] developed a phosphorodiamidate morpholino antisense oligomer targeted to the blaNDM-1 gene for the NDM-1 carbapemase. The antisense oligomer was conjugated to an arginine-rich peptide, which improves penetration of the oligomers into bacteria [114]. This peptide-conjugated antisense oligomer restored bacterial susceptibility to carbapenems and protected mice in a lethal model of sepsis when co-administered with meropenem.
CRISPR-Cas to selectively remove AMR genes
CRISPR-Cas is a bacterial immune system that protects bacteria against invading nucleic acids. This system is widely used for genome editing and has great potential to be utilised to selectively remove AMR genes from bacterial populations. RNA-guided nucleases target and remove specific DNA sequences and hence the system could be programmed to remove genes coding for resistance determinants. The β-lactamase coding genes blaSHV-18 and blaNDM-1 has been targeted by designed RNA-guided nucleases [115]. Similarly, Bikard et al. managed to selectively target the mecA gene which codes for an alternative penicillin binding protein and is the main resistance determinant in methicillin-resistant Staphylococcus aureus (MRSA). Using a phagemid delivery system, the authors were able to drastically reduce the level of MRSA in a mixed population of bacteria [116]. The efficacy of this system was also demonstrated in vivo with a mouse skin colonisation model [116].
The biggest obstacle facing gene silencing technologies is high-efficiency delivery of the genetic constructs to the bacterial cells. Antisense and CRISP-Cas has yet to reach the clinic however, provided that the issues with delivery can be overcome, these techniques hold great promise for future therapies to target resistance mechanisms in bacteria.
New developments – antisense oligonucleotides restores sensitivity to a last line antibiotic
Colistin is a last line antibiotic used to treat carbapenem-resistant Gram-negative infections. Worryingly, the mobile colistin resistance gene (mcr-1) which was first identified in a pig in China [117] has now spread world-wide and colistin resistance is on the rise. Peptide-conjugated phosphorodiamidate morpholino oligomers targeted to mcr-1 mRNA were developed and could effectively resensitise mcr-1-positive E. coli strains to polymyxins. Moreover, addition of the peptide-conjugated antisense oligomers in combination with colistin significantly reduced the bacterial count and morbidity in a mouse model of septicaemia when compared with the effect of colistin alone [118].
Discussion
Antimicrobial resistance is now a worldwide therapeutic problem with MDR Gram-negative bacteria, which are untreatable with any current antibiotic, fast becoming a reality in healthcare settings. With most big Pharma lacking the financial incentive to address this problem, it is up to research laboratories to provide solutions. One way of accelerating antimicrobial drug discovery and development is to reverse resistance to our currently used antibiotics by co-administering resistance breakers with these antibiotics. Huge success has already been reached by the use of β-lactams in combination with β-lactamase inhibitors. However, there is ample scope for increasing the use of our current arsenal of antibiotics even more.
Inhibition of drug efflux pumps would resensitise cells to antibiotics to which it have developed resistance (e.g. efflux-mediated resistance to carbapenems and fluoroquinolines). In addition, EPIs could also render antibiotic such as linezolid that is used to treat highly resistant Gram-positives but lack efficacy against Gram-negatives due to efflux, as new treatment options for MDR Gram-negative infections.
The arsenal of available antibiotics will be greatly expanded if compounds that are currently only active against Gram-positive bacteria but share a common target with Gram-negative bacteria could be delivered to their target site. Additionally, several last-line antibiotics that are used to treat resistant Gram-positives e.g. vancomycin and the vancomycin analogues telavancin, oritavancin and dalbavancin, the glycopeptide antibiotic teicoplanin or mupirocin [95] could potentially be rendered active against Gram-negatives by using chemosensitisers to breach the OM permeability barrier. Similarly teixobactin, the only new class of drug discovered in the last 33 years [119], has high efficacy against Gram-positive bacteria but lacks any Gram-negative antibacterial activity. The target for teixobactin is lipid II [120,121] an essential precursor for both Gram-positive and Gram-negative cell wall synthesis. Hence, the Gram-positive selective activity of teixobactin could most probably be attributed to inability to reach its target in Gram-negatives; this issue could potentially be solved by co-administration of an OM permeabiliser.
Three out of the eleven antibiotic treatments currently in Phase III trials are combinations of antibiotics with molecules designed to overcome resistance (data from Pew charitable trust, www.pewtrusts.org/en/research-and-analysis/data-visualizations/2014/antibiotics-currently-in-clinical-development). As drug discovery and development are not able to keep up with the development of resistance, efforts should be made to speed up this process. In our opinion cutting the discovery time by revitalising antibiotics to which resistance have developed or to which intrinsic resistance mechanisms exist is the most sensible way of reducing the antimicrobial drug discovery and development timeline; this would be imperative for addressing the treatment void for the organisms in the WHO’s most critical priority for antibacterial drug development.
Abbreviations
- AMR
antimicrobial-resistant
- EDTA
ethylenediaminetetraacetic acid
- EPI
efflux pump inhibitor
- IM
inner membrane
- IMP
inner-membrane protein
- MRSA
methicillin-resistant Staphylococcus aureus
- NDM-1
New Delhi metallo-β-lactamase
- OM
outer membrane
- OMP
outer-membrane protein
- PAβN
phenylarginyl-β-naphthylamide
- PAP
periplasmic adapter protein
- RND
resistance nodulation cell division
Competing interests
The author declares that there are no competing interests associated with the manuscript.
Funding
This work was supported by the NHMRC [grant number GN1147538].
References
- 1.Willyard C. (2017) The drug-resistant bacteria that pose the greatest health threats. Nature 543, 15 10.1038/nature.2017.21550 [DOI] [PubMed] [Google Scholar]
- 2.Arzanlou M., Chai W.C. and Venter H. (2017) Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 61, 49–59 10.1042/EBC20160063 [DOI] [PubMed] [Google Scholar]
- 3.Nicolas-Chanoine M.H., Mayer N., Guyot K., Dumont E. and Pages J.M. (2018) Interplay between membrane permeability and enzymatic barrier leads to antibiotic-dependent resistance in Klebsiella pneumoniae. Front. Microbiol. 9, 1422 10.3389/fmicb.2018.01422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zgurskaya H.I., Lopez C.A. and Gnanakaran S. (2015) Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 1, 512–522 10.1021/acsinfecdis.5b00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aires J.R., Kohler T., Nikaido H. and Plesiat P. (1999) Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43, 2624–2628 10.1128/AAC.43.11.2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X., Yu T., Xu P., Xu X., Ji S., Gao W.. et al. (2018) Role of efflux pumps in the in vitro development of ciprofloxacin resistance in listeria monocytogenes. Front. Microbiol. 9, 2350 10.3389/fmicb.2018.02350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhodes K.A. and Schweizer H.P. (2016) Antibiotic resistance in Burkholderia species. Drug Resist. Updat. 28, 82–90 10.1016/j.drup.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher A., Trittler R., Bohnert J.A., Kummerer K., Pages J.M. and Kern W.V. (2007) Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. J. Antimicrob. Chemother. 59, 1261–1264 10.1093/jac/dkl380 [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Garcia A., Rocha-Gracia R.D.C., Bello-Lopez E., Juarez-Zelocualtecalt C., Saenz Y., Castaneda-Lucio M.. et al. (2018) Characterization of antimicrobial resistance mechanisms in carbapenem-resistant Pseudomonas aeruginosa carrying IMP variants recovered from a Mexican Hospital. Infect. Drug Resist. 11, 1523–1536 10.2147/IDR.S173455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheesman M.J., Ilanko A., Blonk B. and Cock I.E. (2017) Developing new antimicrobial therapies: are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn. Rev. 11, 57–72 10.4103/phrev.phrev_21_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole S.T. (2014) Who will develop new antibacterial agents? Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130430 10.1098/rstb.2013.0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper M.A. and Shlaes D. (2011) Fix the antibiotics pipeline. Nature 472, 32 10.1038/472032a [DOI] [PubMed] [Google Scholar]
- 13.Bettiol E., Wetherington J.D., Schmitt N., Harbarth S. and Consortium C. (2015) Challenges and solutions for clinical development of new antibacterial agents: results of a survey among pharmaceutical industry professionals. Antimicrob. Agents Chemother. 59, 3695–3699 10.1128/AAC.00638-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes P. and Martens E. (2017) Antibiotics in late clinical development. Biochem. Pharmacol. 133, 152–163 10.1016/j.bcp.2016.09.025 [DOI] [PubMed] [Google Scholar]
- 15.Payne D.J., Miller L.F., Findlay D., Anderson J. and Marks L. (2015) Time for a change: addressing R&D and commercialization challenges for antibacterials. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne D.J., Gwynn M.N., Holmes D.J. and Pompliano D.L. (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]
- 17.Butler M.S., Blaskovich M.A. and Cooper M.A. (2017) Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. (Tokyo) 70, 3–24 10.1038/ja.2016.72 [DOI] [PubMed] [Google Scholar]
- 18.(2015) BEAM to fight antimicrobial resistance. Nat. Biotechnol. 33, 889 10.1038/nbt0915-889b [DOI] [PubMed] [Google Scholar]
- 19.Blaskovich M.A., Butler M.S. and Cooper M.A. (2017) Polishing the tarnished silver bullet: the quest for new antibiotics. Essays Biochem. 61, 103–114 10.1042/EBC20160077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desselle M.R., Neale R., Hansford K.A., Zuegg J., Elliott A.G., Cooper M.A.. et al. (2017) Institutional profile: community for open antimicrobial drug discovery – crowdsourcing new antibiotics and antifungals. Future Sci. OA 3, FSO171 10.4155/fsoa-2016-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolla J.M., Alibert-Franco S., Handzlik J., Chevalier J., Mahamoud A., Boyer G.. et al. (2011) Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria. FEBS Lett. 585, 1682–1690 10.1016/j.febslet.2011.04.054 [DOI] [PubMed] [Google Scholar]
- 22.McCusker M.P., Ferreira D.A., Cooney D., Alves B.M., Fanning S., Pages J.M.. et al. (2018) Modulation of antibiotic resistance in clinical isolates of Enterobacter aerogenes – a strategy combining antibiotics and chemosensitisers. J. Global Antimicrob. Resist. [DOI] [PubMed] [Google Scholar]
- 23.Wright G.D. (2016) Antibiotic adjuvants: rescuing antibiotics from resistance: (Trends in Microbiology 24, 862–871; October 17, 2016). Trends Microbiol. 24, 928 10.1016/j.tim.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 24.Sully E.K. and Geller B.L. (2016) Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 33, 47–55 10.1016/j.mib.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pursey E., Sunderhauf D., Gaze W.H., Westra E.R. and van Houte S. (2018) CRISPR-Cas antimicrobials: challenges and future prospects. PLoS Pathog. 14, e1006990 10.1371/journal.ppat.1006990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins R.R., Papp-Wallace K.M., Drawz S.M. and Bonomo R.A. (2013) Novel beta-lactamase inhibitors: a therapeutic hope against the scourge of multidrug resistance. Front. Microbiol. 4, 392 10.3389/fmicb.2013.00392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Docquier J.D. and Mangani S. (2018) An update on beta-lactamase inhibitor discovery and development. Drug Resist. Updat. 36, 13–29 10.1016/j.drup.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 28.Queenan A.M. and Bush K. (2007) Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamma P.D., Han J.H., Rock C., Harris A.D., Lautenbach E., Hsu A.J.. et al. (2015) Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum beta-lactamase bacteremia. Clin. Infect. Dis. 60, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambrose P.G., Lomovskaya O., Griffith D.C., Dudley M.N. and VanScoy B. (2017) beta-Lactamase inhibitors: what you really need to know. Curr. Opin. Pharmacol. 36, 86–93 10.1016/j.coph.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 31.Drawz S.M. and Bonomo R.A. (2010) Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201 10.1128/CMR.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ju L.C., Cheng Z., Fast W., Bonomo R.A. and Crowder M.W. (2018) The continuing challenge of metallo-beta-lactamase inhibition: mechanism matters. Trends Pharmacol. Sci. 39, 635–647 10.1016/j.tips.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papp-Wallace K.M., Barnes M.D., Alsop J., Taracila M.A., Bethel C.R., Becka S.A.. et al. (2018) Relebactam is a potent inhibitor of the KPC-2 beta-lactamase and restores imipenem susceptibility in KPC-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 62, 10.1128/AAC.00174-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papp-Wallace K.M., Nguyen N.Q., Jacobs M.R., Bethel C.R., Barnes M.D., Kumar V.. et al. (2018) Strategic approaches to overcome resistance against Gram-negative pathogens using beta-lactamase inhibitors and beta-lactam enhancers: activity of three novel diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234. J. Med. Chem. 61, 4067–4086 10.1021/acs.jmedchem.8b00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hecker S.J., Reddy K.R., Totrov M., Hirst G.C., Lomovskaya O., Griffith D.C.. et al. (2015) Discovery of a cyclic boronic acid beta-lactamase inhibitor (RPX7009) with utility vs class a serine carbapenemases. J. Med. Chem. 58, 3682–3692 10.1021/acs.jmedchem.5b00127 [DOI] [PubMed] [Google Scholar]
- 36.Hackel M.A., Lomovskaya O., Dudley M.N., Karlowsky J.A. and Sahm D.F. (2018) In vitro activity of meropenem-vaborbactam against clinical isolates of KPC-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomovskaya O., Sun D., Rubio-Aparicio D., Nelson K., Tsivkovski R., Griffith D.C.. et al. (2017) Vaborbactam: spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob. Agents Chemother. 61, 10.1128/AAC.01443-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blair J.M., Richmond G.E. and Piddock L.J. (2014) Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 9, 1165–1177 10.2217/fmb.14.66 [DOI] [PubMed] [Google Scholar]
- 39.Chitsaz M. and Brown M.H. (2017) The role played by drug efflux pumps in bacterial multidrug resistance. Essays Biochem. 61, 127–139 10.1042/EBC20160064 [DOI] [PubMed] [Google Scholar]
- 40.Du D., Wang-Kan X., Neuberger A., van Veen H.W., Pos K.M., Piddock L.J.V.. et al. (2018) Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539 10.1038/s41579-018-0048-6 [DOI] [PubMed] [Google Scholar]
- 41.Li X.Z., Plesiat P. and Nikaido H. (2015) The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole K. (2005) Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56, 20–51 10.1093/jac/dki171 [DOI] [PubMed] [Google Scholar]
- 43.Venter H., Mowla R., Ohene-Agyei T. and Ma S. (2015) RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6, 377 10.3389/fmicb.2015.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alibert S., N’Gompaza Diarra J., Hernandez J., Stutzmann A., Fouad M., Boyer G.. et al. (2017) Multidrug efflux pumps and their role in antibiotic and antiseptic resistance: a pharmacodynamic perspective. Expert Opin. Drug Metab. Toxicol. 13, 301–309 [DOI] [PubMed] [Google Scholar]
- 45.Cheng Y.H., Lin T.L., Lin Y.T. and Wang J.T. (2018) A putative RND-type efflux pump, H239_3064, contributes to colistin resistance through CrrB in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1509–1516 10.1093/jac/dky054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuanchuen R., Karkhoff-Schweizer R.R. and Schweizer H.P. (2003) High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control 31, 124–127 10.1067/mic.2003.11 [DOI] [PubMed] [Google Scholar]
- 47.Hassan K.A., Liu Q., Elbourne L.D.H., Ahmad I., Sharples D., Naidu V.. et al. (2018) Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 169, 450–454 10.1016/j.resmic.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machado D., Antunes J., Simoes A., Perdigao J., Couto I., McCusker M.. et al. (2018) Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J. Med. Microbiol. 67, 740–749 10.1099/jmm.0.000741 [DOI] [PubMed] [Google Scholar]
- 49.Ni W., Li Y., Guan J., Zhao J., Cui J., Wang R.. et al. (2016) Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 60, 3215–3218 10.1128/AAC.00248-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole K. (2002) Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 92, 55S–64S 10.1046/j.1365-2672.92.5s1.8.x [DOI] [PubMed] [Google Scholar]
- 51.Jin M., Lu J., Chen Z., Nguyen S.H., Mao L., Li J.. et al. (2018) Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ. Int. 120, 421–430 10.1016/j.envint.2018.07.046 [DOI] [PubMed] [Google Scholar]
- 52.Kurenbach B., Gibson P.S., Hill A.M., Bitzer A.S., Silby M.W., Godsoe W.. et al. (2017) Herbicide ingredients change Salmonella enterica sv. Typhimurium and Escherichia coli antibiotic responses. Microbiology 163, 1791–1802 10.1099/mic.0.000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J., Jin M., Nguyen S.H., Mao L., Li J., Coin L.J.M.. et al. (2018) Non-antibiotic antimicrobial triclosan induces multiple antibiotic resistance through genetic mutation. Environ. Int. 118, 257–265 10.1016/j.envint.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 54.Ricci V., Tzakas P., Buckley A. and Piddock L.J. (2006) Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 50, 38–42 10.1128/AAC.50.1.38-42.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pu Y., Ke Y. and Bai F. (2017) Active efflux in dormant bacterial cells – new insights into antibiotic persistence. Drug Resist. Updat. 30, 7–14 10.1016/j.drup.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 56.Wang Y., Venter H. and Ma S. (2016) Efflux pump inhibitors: a novel approach to combat efflux-mediated drug resistance in bacteria. Curr. Drug Targets 17, 702–719 10.2174/1389450116666151001103948 [DOI] [PubMed] [Google Scholar]
- 57.Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T.. et al. (2014) Structure of the AcrAB-TolC multidrug efflux pump. Nature 509, 512–515 10.1038/nature13205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neuberger A., Du D. and Luisi B.F. (2018) Structure and mechanism of bacterial tripartite efflux pumps. Res. Microbiol. 169, 401–413 10.1016/j.resmic.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 59.Ruggerone P., Murakami S., Pos K.M. and Vargiu A.V. (2013) RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 13, 3079–3100 10.2174/15680266113136660220 [DOI] [PubMed] [Google Scholar]
- 60.Poole K. (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3, 255–264 [PubMed] [Google Scholar]
- 61.Poole K. (2001) Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4, 500–508 10.1016/S1369-5274(00)00242-3 [DOI] [PubMed] [Google Scholar]
- 62.Zhang L., Li X.Z. and Poole K. (2001) SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45, 3497–3503 10.1128/AAC.45.12.3497-3503.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lomovskaya O. and Bostian K.A. (2006) Practical applications and feasibility of efflux pump inhibitors in the clinic – a vision for applied use. Biochem. Pharmacol. 71, 910–918 10.1016/j.bcp.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 64.Aron Z. and Opperman T.J. (2016) Optimization of a novel series of pyranopyridine RND efflux pump inhibitors. Curr. Opin. Microbiol. 33, 1–6 10.1016/j.mib.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mowla R., Wang Y., Ma S. and Venter H. (2018) Kinetic analysis of the inhibition of the drug efflux protein AcrB using surface plasmon resonance. Biochim. Biophys. Acta Biomembr. 1860, 878–886 10.1016/j.bbamem.2017.08.024 [DOI] [PubMed] [Google Scholar]
- 66.Sjuts H., Vargiu A.V., Kwasny S.M., Nguyen S.T., Kim H.S., Ding X.. et al. (2016) Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA 113, 3509–3514 10.1073/pnas.1602472113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y., Mowla R., Ji S., Guo L., De Barros Lopes M.A., Jin C.. et al. (2018) Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 143, 699–709 10.1016/j.ejmech.2017.11.102 [DOI] [PubMed] [Google Scholar]
- 68.Wang Y., Mowla R., Guo L., Ogunniyi A.D., Rahman T., De Barros Lopes M.A.. et al. (2017) Evaluation of a series of 2-napthamide derivatives as inhibitors of the drug efflux pump AcrB for the reversal of antimicrobial resistance. Bioorg. Med. Chem. Lett. 27, 733–739 [DOI] [PubMed] [Google Scholar]
- 69.Daury L., Orange F., Taveau J.C., Verchere A., Monlezun L., Gounou C.. et al. (2016) Tripartite assembly of RND multidrug efflux pumps. Nat. Commun. 7, 10731 10.1038/ncomms10731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Picard M., Tikhonova E.B., Broutin I., Lu S., Verchere A. and Zgurskaya H.I. (2018) Biochemical reconstitution and characterization of multicomponent drug efflux transporters. Methods Mol. Biol. 1700, 113–145 10.1007/978-1-4939-7454-2_8 [DOI] [PubMed] [Google Scholar]
- 71.Verchere A., Dezi M., Adrien V., Broutin I. and Picard M. (2015) In vitro transport activity of the fully assembled MexAB-OprM efflux pump from Pseudomonas aeruginosa. Nat. Commun. 6, 6890 10.1038/ncomms7890 [DOI] [PubMed] [Google Scholar]
- 72.Atzori A., Malviya V.N., Malloci G., Dreier J., Pos K.M., Vargiu A.V.. et al. (2019) Identification and characterization of carbapenem binding sites within the RND-transporter AcrB. Biochim. Biophys. Acta Biomembr. 1861, 62–74 10.1016/j.bbamem.2018.10.012 [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi N., Tamura N., van Veen H.W., Yamaguchi A. and Murakami S. (2014) beta-Lactam selectivity of multidrug transporters AcrB and AcrD resides in the proximal binding pocket. J. Biol. Chem. 289, 10680–10690 10.1074/jbc.M114.547794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakashima R., Sakurai K., Yamasaki S., Hayashi K., Nagata C., Hoshino K.. et al. (2013) Structural basis for the inhibition of bacterial multidrug exporters. Nature 500, 102–106 10.1038/nature12300 [DOI] [PubMed] [Google Scholar]
- 75.Gupta S., Tyagi S., Almeida D.V., Maiga M.C., Ammerman N.C. and Bishai W.R. (2013) Acceleration of tuberculosis treatment by adjunctive therapy with verapamil as an efflux inhibitor. Am. J. Respir. Crit. Care Med. 188, 600–607 10.1164/rccm.201304-0650OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delcour A.H. (2009) Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794, 808–816 10.1016/j.bbapap.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Acosta-Gutierrez S., Ferrara L., Pathania M., Masi M., Wang J., Bodrenko I.. et al. (2018) Getting drugs into Gram-negative bacteria: rational rules for permeation through general porins. ACS Infect. Dis. 4, 1487–1498 10.1021/acsinfecdis.8b00108 [DOI] [PubMed] [Google Scholar]
- 79.Krishnamoorthy G., Leus I.V., Weeks J.W., Wolloscheck D., Rybenkov V.V. and Zgurskaya H.I. (2017) Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. mBio 8, 10.1128/mBio.01172-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krishnamoorthy G., Wolloscheck D., Weeks J.W., Croft C., Rybenkov V.V. and Zgurskaya H.I. (2016) Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob. Agents Chemother. 60, 7372–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lome V., Brunel J.M., Pages J.M. and Bolla J.M. (2018) Multiparametric profiling for identification of chemosensitizers against Gram-negative bacteria. Front. Microbiol. 9, 204 10.3389/fmicb.2018.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Masi M., Dumont E., Vergalli J., Pajovic J., Refregiers M. and Pages J.M. (2018) Fluorescence enlightens RND pump activity and the intrabacterial concentration of antibiotics. Res. Microbiol. 169, 432–441 10.1016/j.resmic.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 83.Vergalli J., Dumont E., Cinquin B., Maigre L., Pajovic J., Bacque E.. et al. (2017) Fluoroquinolone structure and translocation flux across bacterial membrane. Sci. Rep. 7, 9821 10.1038/s41598-017-08775-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vergalli J., Dumont E., Pajovic J., Cinquin B., Maigre L., Masi M.. et al. (2018) Spectrofluorimetric quantification of antibiotic drug concentration in bacterial cells for the characterization of translocation across bacterial membranes. Nat. Protoc. 13, 1348–1361 10.1038/nprot.2018.036 [DOI] [PubMed] [Google Scholar]
- 85.Bajaj H., Acosta Gutierrez S., Bodrenko I., Malloci G., Scorciapino M.A., Winterhalter M.. et al. (2017) Bacterial outer membrane porins as electrostatic nanosieves: exploring transport rules of small polar molecules. ACS Nano 11, 5465–5473 10.1021/acsnano.6b08613 [DOI] [PubMed] [Google Scholar]
- 86.Mamelli L., Petit S., Chevalier J., Giglione C., Lieutaud A., Meinnel T.. et al. (2009) New antibiotic molecules: bypassing the membrane barrier of gram negative bacteria increases the activity of peptide deformylase inhibitors. PLoS One 4, e6443 10.1371/journal.pone.0006443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morones-Ramirez J.R., Winkler J.A., Spina C.S. and Collins J.J. (2013) Silver enhances antibiotic activity against gram-negative bacteria. Sci. Transl. Med. 5, 190ra81 10.1126/scitranslmed.3006276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van der Heijden J., Reynolds L.A., Deng W., Mills A., Scholz R., Imami K.. et al. (2016) Salmonella rapidly regulates membrane permeability to survive oxidative stress. mBio 7, 10.1128/mBio.01238-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cinquin B., Maigre L., Pinet E., Chevalier J., Stavenger R.A., Mills S.. et al. (2015) Microspectrometric insights on the uptake of antibiotics at the single bacterial cell level. Sci. Rep. 5, 17968 10.1038/srep17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vaara M. (1992) Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56, 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lewinson O., Adler J., Poelarends G.J., Mazurkiewicz P., Driessen A.J. and Bibi E. (2003) The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl. Acad. Sci. USA 100, 1667–1672 10.1073/pnas.0435544100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ogunniyi A.D., Khazandi M., Stevens A.J., Sims S.K., Page S.W., Garg S.. et al. (2017) Evaluation of robenidine analog NCL195 as a novel broad-spectrum antibacterial agent. PLoS One 12, e0183457 10.1371/journal.pone.0183457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flora S.J. and Pachauri V. (2010) Chelation in metal intoxication. Int. J. Environ. Res. Public Health 7, 2745–2788 10.3390/ijerph7072745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Escolar E., Lamas G.A., Mark D.B., Boineau R., Goertz C., Rosenberg Y.. et al. (2014) The effect of an EDTA-based chelation regimen on patients with diabetes mellitus and prior myocardial infarction in the Trial to Assess Chelation Therapy (TACT). Circ. Cardiovasc. Qual. Outcomes 7, 15–24 10.1161/CIRCOUTCOMES.113.000663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vaara M. (1992) The outer membrane as the penetration barrier against mupirocin in gram-negative enteric bacteria. J. Antimicrob. Chemother. 29, 221–222 10.1093/jac/29.2.221 [DOI] [PubMed] [Google Scholar]
- 96.Landman D., Georgescu C., Martin D.A. and Quale J. (2008) Polymyxins revisited. Clin. Microbiol. Rev. 21, 449–465 10.1128/CMR.00006-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Velkov T., Dai C., Ciccotosto G.D., Cappai R., Hoyer D. and Li J. (2018) Polymyxins for CNS infections: pharmacology and neurotoxicity. Pharmacol. Ther. 181, 85–90 [DOI] [PubMed] [Google Scholar]
- 98.Allam A., Maigre L., Vergalli J., Dumont E., Cinquin B., Alves de Sousa R.. et al. (2017) Microspectrofluorimetry to dissect the permeation of ceftazidime in Gram-negative bacteria. Sci. Rep. 7, 986 10.1038/s41598-017-00945-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneider E.K., Reyes-Ortega F., Velkov T. and Li J. (2017) Antibiotic-non-antibiotic combinations for combating extremely drug-resistant Gram-negative ‘superbugs’. Essays Biochem. 61, 115–125 10.1042/EBC20160058 [DOI] [PubMed] [Google Scholar]
- 100.Blaskovich M.A.T., Pitt M.E., Elliott A.G. and Cooper M.A. (2018) Can octapeptin antibiotics combat extensively drug-resistant (XDR) bacteria? Expert Rev. Anti Infect. Ther. 16, 485–499 10.1080/14787210.2018.1483240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Han M.L., Shen H.H., Hansford K.A., Schneider E.K., Sivanesan S., Roberts K.D.. et al. (2017) Investigating the interaction of octapeptin A3 with model bacterial membranes. ACS Infect. Dis. 3, 606–619 10.1021/acsinfecdis.7b00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pletzer D., Mansour S.C. and Hancock R.E.W. (2018) Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 14, e1007084 10.1371/journal.ppat.1007084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muheim C., Gotzke H., Eriksson A.U., Lindberg S., Lauritsen I., Norholm M.H.H.. et al. (2017) Increasing the permeability of Escherichia coli using MAC13243. Sci. Rep. 7, 17629 10.1038/s41598-017-17772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Braun V., Pramanik A., Gwinner T., Koberle M. and Bohn E. (2009) Sideromycins: tools and antibiotics. Biometals 22, 3–13 10.1007/s10534-008-9199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mollmann U., Heinisch L., Bauernfeind A., Kohler T. and Ankel-Fuchs D. (2009) Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals 22, 615–624 10.1007/s10534-009-9219-2 [DOI] [PubMed] [Google Scholar]
- 106.Page M.G., Dantier C. and Desarbre E. (2010) In vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant gram-negative bacilli. Antimicrob. Agents Chemother. 54, 2291–2302 10.1128/AAC.01525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim A., Kutschke A., Ehmann D.E., Patey S.A., Crandon J.L., Gorseth E.. et al. (2015) Pharmacodynamic profiling of a siderophore-conjugated monocarbam in Pseudomonas aeruginosa: assessing the risk for resistance and attenuated efficacy. Antimicrob. Agents Chemother. 59, 7743–7752 10.1128/AAC.00831-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tomaras A.P., Crandon J.L., McPherson C.J., Banevicius M.A., Finegan S.M., Irvine R.L.. et al. (2013) Adaptation-based resistance to siderophore-conjugated antibacterial agents by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 4197–4207 10.1128/AAC.00629-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kohira N., West J., Ito A., Ito-Horiyama T., Nakamura R., Sato T.. et al. (2016) In vitro antimicrobial activity of a siderophore cephalosporin, S-649266, against Enterobacteriaceae clinical isolates, including carbapenem-resistant strains. Antimicrob. Agents Chemother. 60, 729–734 10.1128/AAC.01695-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakamura R., Toba S., Tsuji M., Yamano Y. and Shimada J. (2014) A novel siderophore cephalosporin: IV. In vivo efficacy in various murine infection models, abstr F-1558. Abstr. 54th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 111.Saisho Y., Katsube T., White S., Fukase H. and Shimada J. (2018) Pharmacokinetics, safety, and tolerability of cefiderocol, a novel siderophore cephalosporin for Gram-negative bacteria, in healthy subjects. Antimicrob. Agents Chemother. 62, 10.1128/AAC.02163-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dias N. and Stein C.A. (2002) Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 1, 347–355 [PubMed] [Google Scholar]
- 113.Orr R.M. (2001) Technology evaluation: fomivirsen, Isis Pharmaceuticals Inc/CIBA vision. Curr. Opin. Mol. Ther. 3, 288–294 [PubMed] [Google Scholar]
- 114.Sully E.K., Geller B.L., Li L., Moody C.M., Bailey S.M., Moore A.L.. et al. (2017) Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo. J. Antimicrob. Chemother. 72, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Citorik R.J., Mimee M. and Lu T.K. (2014) Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145 10.1038/nbt.3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bikard D., Euler C.W., Jiang W., Nussenzweig P.M., Goldberg G.W., Duportet X.. et al. (2014) Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150 10.1038/nbt.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Venter H., Henningsen M.L. and Begg S.L. (2017) Antimicrobial resistance in healthcare, agriculture and the environment: the biochemistry behind the headlines. Essays Biochem. 61, 1–10 10.1042/EBC20160053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Daly S.M., Sturge C.R., Felder-Scott C.F., Geller B.L. and Greenberg D.E. (2017) MCR-1 inhibition with peptide-conjugated phosphorodiamidate morpholino oligomers restores sensitivity to polymyxin in Escherichia coli. mBio 8, 10.1128/mBio.01315-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P.. et al. (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Homma T., Nuxoll A., Gandt A.B., Ebner P., Engels I., Schneider T.. et al. (2016) Dual targeting of cell wall precursors by teixobactin leads to cell lysis. Antimicrob. Agents Chemother. 60, 6510–6517 10.1128/AAC.01050-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wen P.C., Vanegas J.M., Rempe S.B. and Tajkhorshid E. (2018) Probing key elements of teixobactin-lipid II interactions in membranes. Chem. Sci. 9, 6997–7008 10.1039/C8SC02616E [DOI] [PMC free article] [PubMed] [Google Scholar]