Summary
Bacteriocins are secreted bacterial proteins that selectively kill related strains. Lectin‐like bacteriocins are atypical bacteriocins not requiring a cognate immunity factor and have been primarily studied in Pseudomonas. These so‐called LlpAs are composed of a tandem of B‐lectin domains. One domain interacts with d‐rhamnose residues in the common polysaccharide antigen of Pseudomonas lipopolysaccharide (LPS). The other lectin domain is crucial for interference with the outer membrane protein assembly machinery by interacting with surface‐exposed loops of its central component BamA. Via genome mining, we identified a second subclass of Pseudomonas lectin‐like proteins, termed LlpB, consisting of a single B‐lectin domain. We show that these proteins also display bactericidal activity. Among LlpB‐resistant transposon mutants of an LlpB‐susceptible Pseudomonas strain, a major subset was hit in an acyltransferase gene, predicted to be involved in LPS core modification, hereby suggesting that LlpBs equally attach to LPS for surface anchoring. This indicates that LPS binding and target strain specificity are condensed in a single B‐lectin domain. The identification of this second subclass of lectin‐like bacteriocins further expands the toolbox of antibacterial warfare deployed by bacteria and holds potential for their integration in biotechnological applications.
Introduction
Bacteriocins are secreted ribosomally encoded antibacterial peptides, proteins or multi‐protein complexes that selectively kill phylogenetically related strains, thus facilitating the colonization of competitive environments. Among Gram‐negative bacteria, bacteriocins from Escherichia coli (colicins) and Pseudomonas aeruginosa (pyocins) serve as model systems for studying receptor binding, cell import mechanisms and toxin‐immunity interactions (Cascales et al., 2007; Papadakos et al., 2012; Ghequire and De Mot, 2014; Chassaing and Cascales, 2018). These compounds are potent antibacterials and their use in food and therapeutic applications is currently being investigated (Schulz et al., 2015; Paškevičius et al., 2017; Scholl, 2017; Schneider et al., 2018). Major advantages of bacteriocins include biodegradability, selective killing and eligibility (of some bacteriocins) for large‐scale production in plants (Behrens et al., 2017; Ghequire and De Mot, 2018).
To date, four main classes of Pseudomonas bacteriocins have been described, highly diverse in molecular architecture and killing mechanism: R‐ and F‐type tailocins (Ghequire and De Mot, 2015; Scholl, 2017), modular (or S‐type) bacteriocins (Jamet and Nassif, 2015), B‐type microcins (Metelev et al., 2013) and lectin‐like bacteriocins (Ghequire et al., 2018b). The latter set of bacteriocins (also called LlpAs) are composed of two B‐lectin domains followed by a short carboxy‐terminal extension and share structural similarity with lectins from monocot plants (Ghequire et al., 2013; McCaughey et al., 2014). The carboxy‐terminal lectin domain of these antibacterial proteins binds to d‐rhamnose (McCaughey et al., 2014), the major constituent of the common polysaccharide antigen in the lipopolysaccharide (LPS) layer (Lam et al., 2011), in contrast to B‐lectins from plants which show a much higher affinity for d‐mannose (Barre et al., 1996). The amino‐terminal lectin domain selectively interacts with the essential outer membrane protein BamA (Ghequire et al., 2018b). The latter protein acts as an insertase responsible for the integration of new proteins in the outer membrane (Noinaj et al., 2015). It remains unclear how LlpA interacts with the surface‐exposed loops of BamA and how cellular killing is achieved. Given the lack of a distinct toxin domain and cognate immunity factor as found in modular bacteriocins (Sharp et al., 2017), LlpA killing is likely initiated upon contact with the outer membrane. This way no subsequent bacteriocin import, as is the case for modular bacteriocins, would be required (White et al., 2017). Several other hypothetical prokaryotic proteins in which a B‐lectin domain is combined with (an)other domain(s) have been identified (Ghequire et al., 2012b). For a protein with an amino‐terminal B‐lectin domain fused to a putative peptidase domain, bacteriocin activity has been described: albusin B from ruminal bacterium Ruminococcus albus 7 kills Ruminococcus flavefaciens (Chen et al., 2004). However, how these domains contribute to bacteriocin activity has not been studied. Homologues of this bacteriocin gene are present in some other strains of this Firmicutes species (Azevedo et al., 2015). In Mycobacterium smegmatis MC2155, a protein consisting of a B‐lectin and a LysM domain has been described (Patra et al., 2011), though it remains unclear whether this compound serves a role in bacterial antagonism.
In this paper, we report on the bacteriocin activity of a distinct type of Pseudomonas lectin‐like protein, termed LlpB, consisting of a single B‐lectin domain and a short carboxy‐terminal extension. Characterization of transposon mutants resistant to an LlpB from a Pseudomonas fluorescens strain indicates that target recognition involves LPS of susceptible cells.
Results and discussion
LlpB: a distinct type of lectin‐like protein in Pseudomonas
Using proteobacterial B‐lectin modules (Pfam PF01453) of Pseudomonas LlpAs as search queries, BlastP homology searches previously revealed a second group of lectin‐like proteins in pseudomonads (Ghequire et al., 2012b; Loper et al., 2012; Ghequire and De Mot, 2014). These proteins (~19.8 kDa) consist of a single B‐lectin domain and a carboxy‐terminal extension of ~32 AA. The latter stretch is poorly conserved but typified by a number of basic and aromatic residues (Fig. S1), similarly to Pseudomonas LlpAs (Ghequire et al., 2013). Phylogenetic analysis shows that the predicted lectin modules of these proteins, further called LlpBs, cluster with the amino‐terminal domains of LlpAs, acting as target selectivity determinants in these bacteriocins (Ghequire et al., 2013). The LlpB sequences fall apart in two distinct branches, of which the smaller one is exclusively populated by representatives belonging to the P. fluorescens species group (Fig. 1). As seen for LlpAs, the putative sugar‐binding motifs in LlpBs display strongly differing degrees of sequence conservation, with the first and last of the tree pockets being well conserved (Fig. S1).
Figure 1.
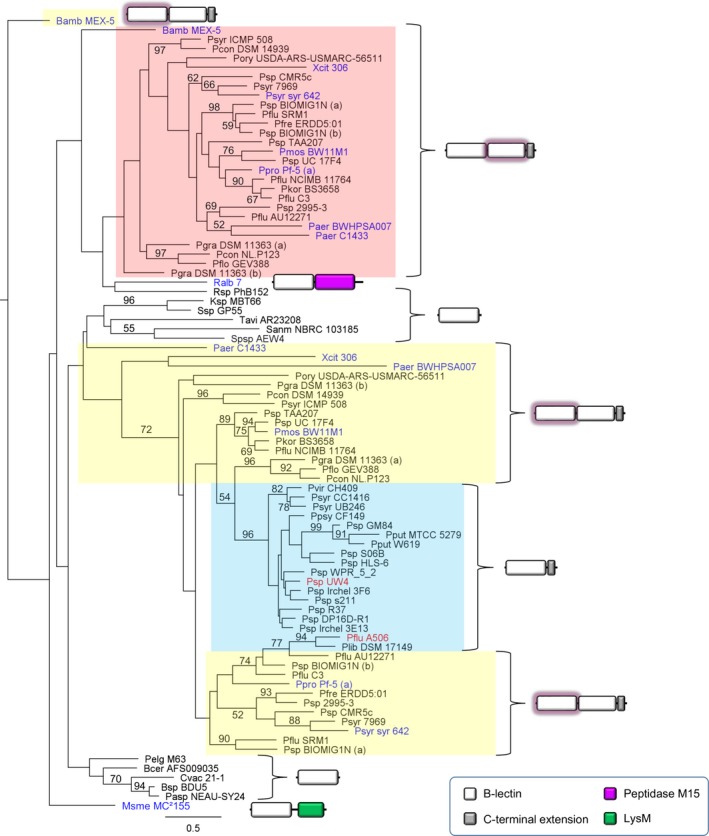
Maximum likelihood phylogenetic tree of B‐lectin domains from LlpA and LlpB proteins in Pseudomonas, characterized LlpA and B‐lectin domain‐containing proteins retrieved in other bacteria, and select B‐lectin mono‐domain proteins in other bacteria. The domain architecture is specified by a schematic representation, and domains are coloured according to function (see colour legend in box). Amino‐terminal and carboxy‐terminal lectin domains from LlpAs and lectin domains from LlpBs are shown on a yellow, red and blue background, respectively. B‐lectin domains from LlpBs cluster with the amino‐terminal domain of LlpAs. In the case of LlpAs, the B‐lectin domain shown in the respective cluster is highlighted by a glowing background. Highly similar sequences (> 75% pairwise amino acid sequence identity for full length LlpA/LlpB proteins) are represented by one sequence only. Previously characterized proteins with a B‐lectin domain are labelled in blue, and LlpBs characterized in this study in red. Multiple LlpA/LlpB bacteriocins in a particular strain are specified by extensions (a) and (b). Phylogenetic analysis was performed with PhyML, using the JTT substitution model. Bootstrap values (percentages of 1000 replicates) higher than 50 are shown at the branches. The tree is rooted to the amino‐terminal B‐lectin domain of the LlpA from Burkholderia ambifaria MEX‐5. Scale bar represents 0.5 substitutions per site. Bamb, Burkholderia ambifaria; Bcer, Bacillus cereus; Bsp, Burkholderia sp.; Cvac, Chromobacterium vaccinii; Ksp, Kitasatospora sp.; Msme, Mycobacterium smegmatis; Paer, Pseudomonas aeruginosa; Pasp, Paraburkholderia sp.; Pcon, Pseudomonas congelans; Pelg, Paenibacillus elgii; Pflo, Pseudomonas floridensis; Pflu, Pseudomonas fluorescens; Pfre, Pseudomonas frederiksbergensis; Pgra, Pseudomonas graminis; Pkor, Pseudomonas koreensis; Plib, Pseudomonas libanensis; Pmos, Pseudomonas mosselii; Pory, Pseudomonas oryzihabitans; Ppro, Pseudomonas protegens; Ppsy, Pseudomonas psychrophila; Pput, Pseudomonas putida; Psp, Pseudomonas sp.; Psyr (syr), Pseudomonas syringae (pathovar syringae); Pvir, Pseudomonas viridiflava; Ralb, Ruminococcus albus; Rsp, Rathayibacter sp.; Sanm, Streptacidiphilus anmyonensis; Spsp, Sphingobium sp.; Ssp, Streptomyces sp.; Tavi, Tumebacillus avium; Xcit, Xanthomonas citri.
llpB genes mainly occur in plant‐ and soil‐associated Pseudomonas isolates, but are lacking from P. aeruginosa genomes. They are rather rare overall (~3.4% of assembled Pseudomonas genomes, excluding the P. aeruginosa species group), but appear relatively more frequent in Pseudomonas syringae (~7.5% of strains belonging to the P. syringae species group). Interestingly, llpB genes do not co‐occur with llpA genes within a single strain. Furthermore, quite some bacteria encode LlpB‐like proteins lacking a carboxy‐terminal extension (Fig. 1). Such mono‐B‐lectin domain proteins, often preceded by a (predicted) SecA secretion signal sequence (http://www.compgen.org/tools/PRED-TAT) , are for example found in Actinobacteria (e.g. Kitasatospora, Streptacidiphilus and Streptomyces) and Firmicutes (e.g. Brevibacillus and Paenibacillus). As seen for LlpA‐encoding Pseudomonas strains (Parret et al., 2005; Ghequire et al., 2018b), isolates may host (up to) two llpB genes, for example Pseudomonas sp. FW104‐R4. If so, llpB genes are organized in tandem, whereas llpA genes in strains carrying two representatives usually appear at distant loci (Ghequire and De Mot, 2014). As noted for other (mid‐sized) bacteriocins (Ghequire et al., 2015, 2017a,2017b; Dingemans et al., 2016; Sharp et al., 2017), llpB genes are typified by a lower GC content than the genomic average (~47% versus ~60%), pointing towards foreign origin. Yet another similarity with llpA genes is that some of these llpB genes arise in prophage/tailocin clusters (Ghequire et al., 2015), for example in a Rp3 tailocin gene cluster of Pseudomonas libanensis DSM 17149. Such association is confined to the minor branch of llpB‐carrying isolates (Fig. 1). In contrast, in the large clade they mainly occur at two other loci: downstream of sulphate adenylyltransferase cysN or downstream of a flavin monoamine oxidase gene (data not shown). In some strains, a modular bacteriocin‐immunity gene tandem is integrated between cysN and llpB, for example in P. putida MTCC 5279 (putative HNH DNase toxin), underlining the plasticity of the locus. Taken together, the striking parallels of LlpBs with other bacteriocins suggest that these proteins may also exert an antibacterial function. To explore this further, representative and divergent LlpBs (Fig. 1) from biocontrol strain P. fluorescens A506 (Loper et al., 2012) and plant growth‐promoting rhizobacterium Pseudomonas sp. UW4 (Duan et al., 2013) were selected for further characterization.
Bacteriocin activity of LlpBs
llpB genes from strains A506 (locus_tag PflA506_2041) and UW4 (locus_tag PPUTUW4_RS25815, codon‐optimized) were PCR‐amplified, digested and cloned in pET28a to encode an amino‐terminal His6‐tagged protein (primers in Table S1), resulting in pCMPG6205 and pCMPG6207, respectively. Sequence‐verified plasmids (GATC Biotech, Constance, Germany) were transformed to E. coli BL21(DE3). Cells grown in 500‐ml LB erlenmeyers were induced with isopropyl‐β‐d‐thiogalactopyranoside and incubated overnight, as described earlier (Ghequire et al., 2012b). After, cells were harvested, dissolved and sonicated, and soluble proteins isolated via centrifugation. His‐tagged proteins were purified via affinity chromatography on Ni‐NTA agarose. The presence of recombinant protein in the imidazole‐eluted fractions was confirmed via SDS‐PAGE, and samples were further polished by gel filtration. The calculated molecular weights of His6‐tagged LlpBs (20.6 kDa LlpBPfluA506; 21.4 kDa LlpBPspUW4) match well with the apparent sizes of the recombinant proteins as estimated by SDS‐PAGE (Fig. 2).
Figure 2.
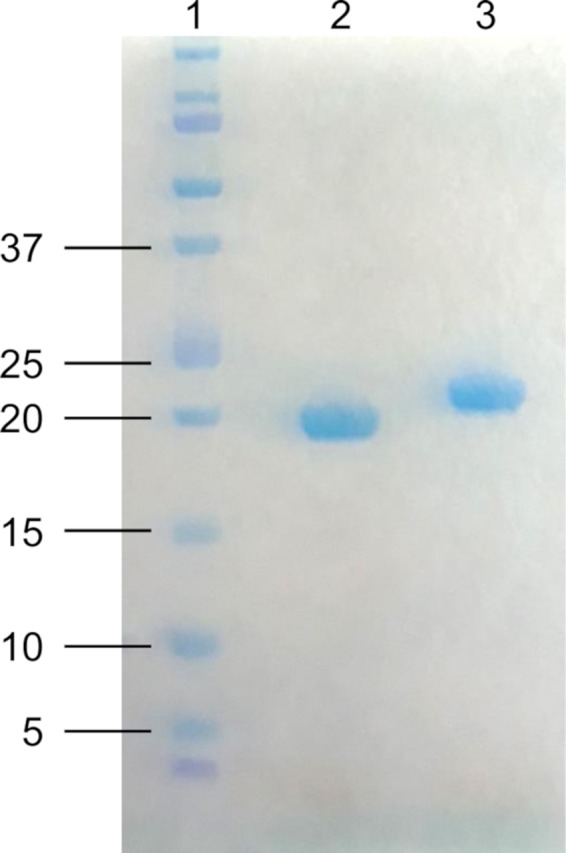
SDS‐PAGE electrophoresis of purified recombinant LlpB proteins from strains P. fluorescens A506 and Pseudomonas sp. UW4. Lane 1, Precision Plus Dual Xtra size marker (kDa); lane 2, LlpBP fluA506 (~19 kDa, predicted size 20.6 kDa); lane 3, LlpBP sp UW 4 (~21 kDa, predicted size 21.4 kDa).
Antagonistic activity of the LlpBs was evaluated via spot assay against a panel of pseudomonads, including several Pseudomonas reference strains. Ten‐μl drops of recombinant protein (concentration 1 mg ml−1) were applied onto bacterial cell lawns, incubated overnight, and scored for the presence of zones of growth inhibition the following day (Hockett and Baltrus, 2017). For both LlpBs, eight out of 49 strains in the Pseudomonas test panel proved susceptible (Table 1), confirming the bactericidal function of these proteins. Five strains were killed by both LlpBs, despite their low sequence identity (~34%). As seen for other Pseudomonas bacteriocins (LlpAs and other (non‐P. aeruginosa) bacteriocins) (Ghequire et al., 2012a, 2015), LlpB activity surpasses species boundaries: the bacteriocins from P. fluorescens A506 and Pseudomonas sp. UW4 [P. jesseni group (Gomila et al., 2015; Garrido‐Sanz et al., 2016)] both also kill strains from the P. stutzeri and P. syringae groups.
Table 1.
Antibacterial activity of purified recombinant LlpBs against Pseudomonas isolates
| Indicator strain | Growth inhibitiona by | |
|---|---|---|
| LlpBPfluA506 | LlpBPspUW4 | |
| P. aeruginosa group | ||
| P. aeruginosa LMG 1242 | − | − |
| P. aeruginosa ATCC27853 | − | − |
| P. aeruginosa PAO1 | − | − |
| P. aeruginosa UCBPP‐PA14 | − | − |
| P. resinovorans LMG 2274 | − | − |
| P. fluorescens complex | ||
| P. chlororaphis subsp. aureofaciens LMG 1245 | − | − |
| P. chlororaphis subsp. chlororaphis LMG 5004 | − | − |
| P. fluorescens 2‐79 | − | − |
| P. fluorescens 13‐79 | + | − |
| P. fluorescens A1‐B | − | − |
| P. fluorescens CC‐848406‐E | − | − |
| P. fluorescens F113 | − | − |
| P. fluorescens LMG 1794 | + | − |
| P. fluorescens LMG 2210 | − | − |
| P. fluorescens OE 39.4 | − | − |
| P. fluorescens OE 48.2 | − | − |
| P. fluorescens Pf0‐1 | − | − |
| P. fluorescens PGSB 7705 | + | T |
| P. fluorescens PGSB 7716 | − | − |
| P. fluorescens PGSB 7947 | − | − |
| P. fluorescens PGSB 8301 | − | + |
| P. fluorescens PGSB 8472 | − | − |
| P. fluorescens SBW25 | − | − |
| P. fluorescens WCS141 | − | − |
| P. fluorescens WCS365 | − | T |
| P. protegens CHA0b | − | − |
| P. tolaasii CH36 | − | − |
| P. tolaasii LMG 2342 | − | − |
| P. tolaasii LMG 2344 | − | − |
| P. putida group | ||
| P. putida KT2440 | − | − |
| P. putida LMG 2257 | − | − |
| P. putida OE 53.2 | − | − |
| P. putida WCS358 | − | − |
| P. stutzeri group | ||
| P. stutzeri LMG 11199 | − | − |
| P. stutzeri LMG 1228 | + | + |
| P. syringae group | ||
| P. cichorii LMG 2162 | − | T |
| P. savastanoi LMG 2209 | − | − |
| P. savastanoi LMG 5154 | − | − |
| P. savastanoi LMG 5485 | + | − |
| P. savastanoi LMG 6768 | − | − |
| P. savastanoi LMG 17581 | − | − |
| P. syringae GR12‐2R3 | + | + |
| P. syringae pv. glycinea LMG 5066 | + | + |
| P. syringae pv. syringae LMG 1247 | − | − |
| P. syringae pv. tabaci LMG 5192 | − | − |
| P. syringae pv. tomato DC3000 | − | − |
| P. viridiflava LMG 2352 | + | + |
| Other Pseudomonas spp. | ||
| P. agarici LMG 2112 | − | − |
| P. mendocina LMG 1223 | − | − |
a. Growth inhibition due to LlpB bacteriocin activity was scored as follows: +, clear halo; T, turbid halo; −, no zone of growth inhibition. Running buffer was used as a negative control.
b. Of the strains used in the test panel (and for which genome sequence information is available), only P. protegens CHA0 carries an llpA gene in its genome. No strain contains an llpB gene.
Genes affected in LlpB‐resistant mutants indicate a key role of LPS in target cell susceptibility
The first and last of the three sugar‐binding motifs in LlpBs show sequence similarity with the consensus motif accounting for d‐mannose binding in plant lectins, QxDxNxVxY (Ghequire et al., 2012b). Given the role assigned to d‐rhamnose as a ligand for LlpAs, we hypothesized that one or both of these lectin motifs in LlpBs may bind to carbohydrates from lipopolysaccharides as well, enabling target cell attachment in a similar way.
In search for susceptibility determinants of LlpB killing, a mutant library was created in P. fluorescens LMG 1794T (sequenced as NCTC10038T) using transposon delivery vector pRL27 (Larsen et al., 2002), via triparental conjugation. Transposon mutants were pooled, supplemented with concentrated LlpBPfluA506 (~5 mg ml−1), and subsequently plated. Following day, colonies were selected, verified for bacteriocin resistance and transposon insertion sites determined, as described earlier (Ghequire et al., 2017b). Interestingly, of the 34 (independent) LlpB‐resistant mutants isolated, 24 were hit in an acyltransferase gene oatA (NCTC10038_05872) (Fig. 3). The encoded protein shares 27% amino acid identity with oafA, previously studied in Salmonella Typhimurium and shown to function as an O‐antigen acetylase (Slauch et al., 1996). Gene synteny and significant sequence similarity (48% pairwise amino acid identity) can be noted for PA5238 from Pseudomonas aeruginosa PAO1. Lipopolysaccharide acetylation activity has been proposed for the latter enzyme (King et al., 2009), but remains to be verified. The repeating units constituting the O‐specific polysaccharide chains of LPS in P. fluorescens LMG 1794 have been determined and consist of l‐rhamnose and N‐acetyl‐d‐fucose (Veremecheĭnko et al., 2005). Given that PA5238 was suggested to play a role in O‐acetylation of the LPS core and not of the repeating units (King et al., 2009), we thus do not expect these two carbohydrate residues to interact with LlpBPfluA506. It remains to be assessed whether other LlpBs equally depend for killing on the activity of this acyltransferase gene in target cells, which would be expected if these lectin‐like bacteriocins share a common LPS moiety as receptor. It should be emphasized that polar effects on the two genes downstream of NCTC10038_05872 cannot be excluded a priori, though the multiple transposon insertions independently hitting oatA argue against this. When evaluating our strain panel for the presence of oatA and oatA‐like genes, we found that the majority of the strains (for which a full or draft genome is available, 23/30) encodes such an acyltransferase, including all the strains killed by one or both of the LlpBs.
Figure 3.
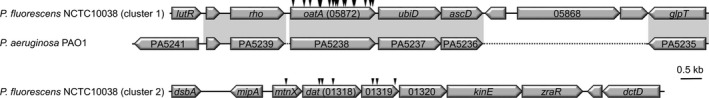
Schematic gene representation of two genomic regions in Pseudomonas fluorescens LMG 1794T (NCTC10038T) susceptible to bacteriocin LlpBP fluA506. Genes are shown as arrows and insert locations of transposon pRL27 are indicated with black triangles. Gene synteny of the locus of oatA, target of the large majority of the LlpB‐resistant mutants, with the corresponding region in reference strain P. aeruginosa PAO1 is shown by grey shading. Dotted lines indicate the lack of an equivalent region.
A second set of seven transposon mutants were hit in an operon that is possibly involved in LPS biogenesis as well (Fig. 3). This cluster is conserved in Pseudomonas species, but apparently lacks from P. aeruginosa genomes. In LPS of P. fluorescens NCTC10038, different amino sugars have been detected (Wilkinson, 1972), which may require dat aminotransferase activity. Whether this second cluster plays a role in LPS biosynthesis remains speculative however. In the nearby future, chemical characterization of LPS constituents of different mutants obtained in this study will shed further light on the carbohydrates interacting with LlpB. Whether BamA or (an)other outer membrane protein(s) contribute to LlpB killing also remains to be investigated.
Conflict of interest
None declared.
Supporting information
Fig. S1. Multiple sequence alignment of LlpBs included in Figure 1.
Table S1. Primers used in this study.
Acknowledgements
MGKG is the recipient of a FWO postdoctoral fellowship (12M4618N). This work was supported by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Grants 1523116N, 1525618N and 1529718N to MGKG).
Microbial Biotechnology (2019) 12(3), 567–573
Funding Information
MGKG is the recipient of a FWO postdoctoral fellowship (12M4618N). This work was supported by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Grants 1523116N, 1525618N and 1529718N to MGKG).
References
- Azevedo, A.C. , Bento, C.B. , Ruiz, J.C. , Queiroz, M.V. , and Mantovani, H.C. (2015) Distribution and genetic diversity of bacteriocin gene clusters in rumen microbial genomes. Appl Environ Microbiol 81: 7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre, A. , Van Damme, E.J. , Peumans, W.J. , and Rougé, P. (1996) Structure‐function relationship of monocot mannose‐binding lectins. Plant Physiol 112: 1531–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, H.M. , Six, A. , Walker, D. , and Kleanthous, C. (2017) The therapeutic potential of bacteriocins as protein antibiotics. Emerg Top Life Sci 1: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales, E. , Buchanan, S.K. , Duché, D. , Kleanthous, C. , Lloubès, R. , Postle, K. , et al (2007) Colicin biology. Microbiol Mol Biol Rev 71: 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing, B. , and Cascales, E. (2018) Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol 26: 329–338. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Stevenson, D.M. , and Weimer, P.J. (2004) Albusin B, a bacteriocin from the ruminal bacterium Ruminococcus albus 7 that inhibits growth of Ruminococcus flavefaciens . Appl Environ Microbiol 70: 3167–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemans, J. , Ghequire, M.G. , Craggs, M. , De Mot, R. , and Cornelis, P. (2016) Identification and functional analysis of a bacteriocin, pyocin S6, with ribonuclease activity from a Pseudomonas aeruginosa cystic fibrosis clinical isolate. Microbiologyopen 5: 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, J. , Jiang, W. , Cheng, Z. , Heikkila, J.J. and Glick, B.R. (2013) The complete genome sequence of the plant growth‐promoting bacterium Pseudomonas sp. UW4. PLoS ONE 8, e58640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido‐Sanz, D. , Meier‐Kolthoff, J.P. , Göker, M. , Martín, M. , Rivilla, R. , and Redondo‐Nieto, M. (2016) Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 11: e0150183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G. , and De Mot, R. (2014) Ribosomally encoded antibacterial proteins and peptides from Pseudomonas . FEMS Microbiol Rev 38: 523–568. [DOI] [PubMed] [Google Scholar]
- Ghequire, M.G. , and De Mot, R. (2015) The tailocin tale: peeling off phage tails. Trends Microbiol 23: 587–590. [DOI] [PubMed] [Google Scholar]
- Ghequire, M.G.K. , and De Mot, R. (2018) Turning over a new leaf: bacteriocins going green. Trends Microbiol 26: 567–2. [DOI] [PubMed] [Google Scholar]
- Ghequire, M.G. , Li, W. , Proost, P. , Loris, R. , and De Mot, R. (2012a) Plant lectin‐like antibacterial proteins from phytopathogens Pseudomonas syringae and Xanthomonas citri . Environ Microbiol Rep 4: 373–380. [DOI] [PubMed] [Google Scholar]
- Ghequire, M.G. , Loris, R. , and De Mot, R. (2012b) MMBL proteins: from lectin to bacteriocin. Biochem Soc Trans 40: 1553–1559. [DOI] [PubMed] [Google Scholar]
- Ghequire, M.G. , Garcia‐Pino, A. , Lebbe, E.K. , Spaepen, S. , Loris, R. , and De Mot, R. (2013) Structural determinants for activity and specificity of the bacterial toxin LlpA. PLoS Pathog 9: e1003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G. , Dillen, Y. , Lambrichts, I. , Proost, P. , Wattiez, R. , and De Mot, R. (2015) Different ancestries of R tailocins in rhizospheric Pseudomonas isolates. Genome Biol Evol 7: 2810–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G. , Kemland, L. , and De Mot, R. (2017a) Novel immunity proteins associated with colicin M‐like bacteriocins exhibit promiscuous protection in Pseudomonas . Front Microbiol 8: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G.K. , Kemland, L. , Anoz‐Carbonell, E. , Buchanan, S.K. , and De Mot, R. (2017b) A natural chimeric Pseudomonas bacteriocin with novel pore‐forming activity parasitizes the ferrichrome transporter. MBio 8: e01961‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M. , Swings, T. , Michiels, J. , Buchanan, S. , and De Mot, R. (2018a) Hitting with a BAM: selective killing by lectin‐like bacteriocins. MBio 9: e02138‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M.G.K. , Öztürk, B. , and De Mot, R. (2018b) Lectin‐like bacteriocins. Front Microbiol 9: 2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila, M. , Peña, A. , Mulet, M. , Lalucat, J. , and García‐Valdés, E. (2015) Phylogenomics and systematics in Pseudomonas . Front Microbiol 6: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett, K.L. , and Baltrus, D.A. (2017) Use of the soft‐agar overlay technique to screen for bacterially produced inhibitory compounds. J Vis Exp 119: e55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet, A. , and Nassif, X. (2015) New players in the toxin field: polymorphic toxin systems in bacteria. MBio 6: e00285‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.D. , Kocíncová, D. , Westman, E.L. , and Lam, J.S. (2009) Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa . Innate Immun 15: 261–312. [DOI] [PubMed] [Google Scholar]
- Lam, J.S. , Taylor, V.L. , Islam, S.T. , Hao, Y. , and Kocíncová, D. (2011) Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, R.A. , Wilson, M.M. , Guss, A.M. , and Metcalf, W.W. (2002) Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178: 193–201. [DOI] [PubMed] [Google Scholar]
- Loper, J.E. , Hassan, K.A. , Mavrodi, D.V. , Davis, E.W. 2nd , Lim, C.K. , Shaffer, B.T. , et al (2012) Comparative genomics of plant‐associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8, e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey, L.C. , Grinter, R. , Josts, I. , Roszak, A.W. , Waløen, K.I. , Cogdell, R.J. , et al (2014) Lectin‐like bacteriocins from Pseudomonas spp. utilise D‐rhamnose containing lipopolysaccharide as a cellular receptor. PLoS Pathog 10, e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metelev, M. , Serebryakova, M. , Ghilarov, D. , Zhao, Y. , and Severinov, K. (2013) Structure of microcin B‐like compounds produced by Pseudomonas syringae and species specificity of their antibacterial action. J Bacteriol 195: 4129–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj, N. , Rollauer, S.E. , and Buchanan, S.K. (2015) The β‐barrel membrane protein insertase machinery from Gram‐negative bacteria. Curr Opin Struct Biol 31: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakos, G. , Wojdyla, J.A. , and Kleanthous, C. (2012) Nuclease colicins and their immunity proteins. Q Rev Biophys 45: 57–103. [DOI] [PubMed] [Google Scholar]
- Parret, A.H. , Temmerman, K. , and De Mot, R. (2005) Novel lectin‐like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf‐5. Appl Environ Microbiol 71: 5197–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paškevičius, S. , Starkevič, U. , Misiūnas, A. , Vitkauskien≐, A. , Gleba, Y. , and Ražanskien≐, A. (2017) Plant‐expressed pyocins for control of Pseudomonas aeruginosa . PLoS ONE 12: e0185782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra, D. , Sharma, A. , Chandran, D. , and Vijayan, M. (2011) Cloning, expression, purification, crystallization and preliminary X‐ray studies of the mannose‐binding lectin domain of MSMEG_3662 from Mycobacterium smegmatis . Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, T. , Hahn‐Löbmann, S. , Stephan, A. , Schulz, S. , Giritch, A. , Naumann, M. , et al (2018) Plant‐made Salmonella bacteriocins salmocins for control of Salmonella pathovars. Sci Rep 8: 4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl, D. (2017) Phage tail‐like bacteriocins. Annu Rev Virol 4: 453–467. [DOI] [PubMed] [Google Scholar]
- Schulz, S. , Stephan, A. , Hahn, S. , Bortesi, L. , Jarczowski, F. , Bettmann, U. , et al (2015) Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant‐produced colicins. Proc Natl Acad Sci USA 112: E5454–E5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, C. , Bray, J. , Housden, N.G. , Maiden, M.C.J. , and Kleanthous, C. (2017) Diversity and distribution of nuclease bacteriocins in bacterial genomes revealed using Hidden Markov Models. PLoS Comput Biol 13: e1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch, J.M. , Lee, A.A. , Mahan, M.J. , and Mekalanos, J.J. (1996) Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O‐antigen: oafA is a member of a family of integral membrane trans‐acylases. J Bacteriol 178: 5904–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veremecheĭnko, S.N. , Vodianik, M.A. , and Zdorovenko, G.M. (2005) Structural characteristics and biological properties of Pseudomonas fluorescens lipopolysaccharides. Prikl Biokhim Mikrobiol 41: 414–421. [PubMed] [Google Scholar]
- White, P. , Joshi, A. , Rassam, P. , Housden, N.G. , Kaminska, R. , Goult, J.D. , et al (2017) Exploitation of an iron transporter for bacterial protein antibiotic import. Proc Natl Acad Sci USA 114: 12051–12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, S.G. (1972) Amino sugars in the wall of Pseudomonas fluorescens . J Gen Microbiol 70: 365–369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Multiple sequence alignment of LlpBs included in Figure 1.
Table S1. Primers used in this study.


