Abstract
PfEMP1 is a family of adhesive proteins expressed on the surface of Plasmodium falciparum-infected erythrocytes (IEs), where they mediate adhesion of IEs to a range of host receptors. Efficient PfEMP1-dependent IE sequestration often depends on soluble serum proteins, including IgM. Here, we report a comprehensive investigation of which of the about 60 var gene-encoded PfEMP1 variants per parasite genome can bind IgM via the Fc part of the antibody molecule, and which of the constituent domains of those PfEMP1 are involved. We erased the epigenetic memory of var gene expression in three distinct P. falciparum clones, 3D7, HB3, and IT4/FCR3 by promoter titration, and then isolated individual IEs binding IgM from malaria-unexposed individuals by fluorescence-activated single-cell sorting. The var gene transcription profiles of sub-clones measured by real-time qPCR were used to identify potential IgM-binding PfEMP1 variants. Recombinant DBL and CIDR domains corresponding to those variants were tested by ELISA and protein arrays to confirm their IgM-binding capacity. Selected DBL domains were used to raise specific rat anti-sera to select IEs with uniform expression of candidate PfEMP1 proteins. Our data document that IgM-binding PfEMP1 proteins are common in each of the three clones studied, and that the binding epitopes are mainly found in DBLε and DBLζ domains near the C-terminus.
Introduction
Malaria is caused by protozoan parasites of the genus Plasmodium. There are an estimated 219 million malaria cases annually, including about 435,000 fatal episodes, mainly caused by infection with P. falciparum1. The particular virulence of this species is related to its multiple strategies to evade acquired host immunity. Important among them is the capacity of P. falciparum-infected erythrocytes (IEs) to sequester in various tissues to avoid immune clearance in the spleen2. The P. falciparum erythrocyte membrane protein 1 (PfEMP1) family is essential for the adhesion of IEs to host cell membrane receptors (on endothelial cells or erythrocytes), which in some cases requires soluble host proteins3–7.
The acquisition of PfEMP1-specific protective immunity to prevent IE sequestration is frustrated by the parasite’s ability to express a single PfEMP1 variant at the IE surface at a time, and to switch among the ~60 PfEMP1 proteins encoded by the var gene family8. The var genes can be divided into several groups based on genomic location, direction of transcription, and structural features9,10. Expression of PfEMP1 variants encoded by particular var gene groups (and their sub-groups) has been associated with discrete clinical presentations and IE adhesion to specific host receptors. Thus, PfEMP1 expression in parasites isolated from severe malaria patients with little or no pre-acquired immunity is often dominated by variants encoded by the relatively conserved Group A genes. The more diverse Group B and Group C genes are more commonly found among uncomplicated and asymptomatic infections, while the Group E genes that encode VAR2CSA-type PfEMP1 variants are responsible for the pathogenesis of placental malaria11. These different subsets of PfEMP1 proteins contain specific Duffy binding-like (DBL) domains and cysteine-rich inter-domain regions (CIDR). Both DBL and CIDR domains can be divided into structurally related classes (α, β, γ, δ, ε, ζ, and α, β, γ, respectively)12. DBL and CIDR domains mediate IE adhesion to various host receptors, such as endothelial protein C receptor (EPCR), intercellular adhesion molecule 1 (ICAM-1), CD36, and oncofetal chondroitin sulfate13–18.
A handful of PfEMP1 variants has been reported to bind IgM via the Fcµ region of the antibody rather than by the hypervariable, antigen-specific Fab fragment19 (we will refer to this type of IgM-binding as “non-immune”, as it is specific in the sense that it depends on Fcµ but is independent of the antigen-specificity of the IgM molecules involved). We recently reported the existence of four additional non-immune IgM-binding PfEMP1 variants in P. falciparum 3D7 sub-clones. Relatively few sub-clones and PfEMP1 variants were tested, and the study thus probably underestimated the total number of non-immune IgM binders5. Furthermore, the study did not assess potential inter-clonal variation in the capacity for Fc-mediated binding of IgM to PfEMP1. To overcome these limitations, we report here results from single-cell sorting of parasite populations with highly heterogeneous var gene transcription to obtain a comprehensive mapping of non-immune IgM-binding PfEMP1 variants in the three distinct P. falciparum clones 3D7, HB3, and IT4/FCR3 (subsequently referred to as IT4). We also probed a multiplex array of PfEMP1 domains from P. falciparum 3D7 with non-immune IgM. Together with recombinant PfEMP1 proteins and specific rat anti-sera the study allowed us to identify several new PfEMP1 variants and constituent domains involved in non-immune IgM binding to IEs.
Results
Erasure of epigenetic memory by pVBH transfection
At the outset, our P. falciparum clones 3D7, HB3, and IT4 dominantly transcribed one var gene each (pfd1005c, hb3var06, and it4var60, respectively) (Fig. S1, left panels). To obtain P. falciparum populations with as heterogeneous var gene transcription as possible, we transfected each of the three P. falciparum clones with the pVBH plasmid, which contains a blasticidin S deaminase (bsd) gene under the control of a var promoter20,21. For each of the clones, selection by blasticidin pressure for high copy numbers of this plasmid markedly reduced transcription of the endogenous var genes (Fig. S1, center panels). Two weeks after release from the drug pressure, bsd transcription had decreased and the parasites transcribed a diverse set of endogenous var genes, without dominance of the initially most abundant var gene transcript (Fig. S1, right panels). This indicates efficient erasure of epigenetic memory, as previously described20,21.
Identification of candidate IgM-binding PfEMP1 proteins by single-cell sorting of infected erythrocytes
Erythrocytes infected with late-stage parasites, in which the var gene epigenetic memory had been erased by bsd transfection and drug selection, were labeled with non-immune IgM and subjected to fluorescence-activated single-cell sorting to isolate individual IgM+ IEs. After in vitro expansion of the sorted IEs for 3–6 weeks, we obtained 66 IgM-reactive sub-clones with varying proportions of IgM+ IEs (Fig. 1). Thirty-four sub-clones transcribed a dominant (≥35%) var gene (Figs 2, S2, Tables S1–S3). The relative level of dominant var gene transcripts generally reflected the IE reactivity with non-immune IgM. This correspondence, and analysis of the most dominant transcripts, were used to identify genes likely to encode IgM-binding PfEMP1 antigens in the three parasite clones. Apart from the Group E genes encoding VAR2CSA-type PfEMP1 in the 3D7, HB3, and IT4 (pfl0030c, hb3var2csaA and hb3var2csaB, and it4var04, respectively), all the identified genes belong to Group B or Group C. Many did not encode any DBLε and DBLζ domains, which have previously been associated with non-immune IgM binding to PfEMP14–6,22–24 (Figs 2, S2). Instead, they encoded a DBLδ1 domain near the C-terminus, which thus appeared to represent a hitherto unidentified IgM-binding domain type (Figs 2, S2). A few of the sub-clones showed low (≤15%) IgM+ IEs despite transcribing a single dominant var gene (Tables S1–S3), probably because they had largely switched away from the var gene encoding the IgM-binding PfEMP1 detected at the time of the single-cell sorting. Alternatively, these sub-clones were false positives not expressing an IgM-binding PfEMP1 at the time of sorting. Regardless of the reason for the IgM non-reactivity, this information was taken as an indication of unlikely candidates for genes encoding IgM-binding PfEMP1 variants. Examples include PFL13_0001, PFL2665c, and PF08_0107 (Table S1), HB3VAR27, HB3VAR28, HB3VAR29 (Table S2), and IT4VAR15 (Table S3).
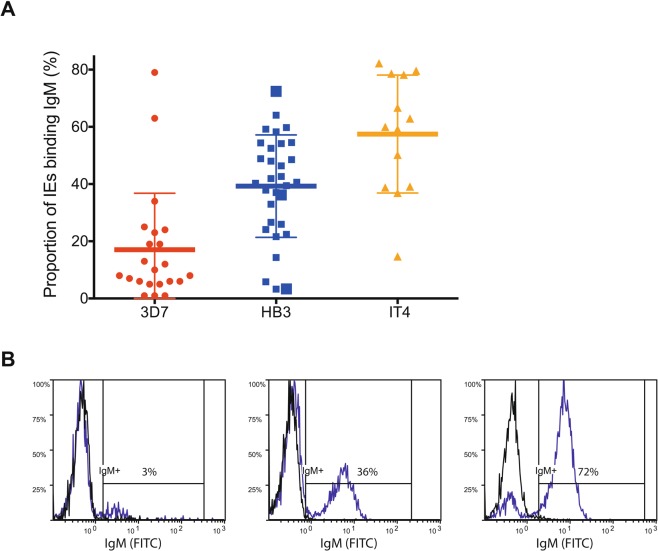
Figure 1.
Non-immune IgM Binding to P. falciparum-infected erythrocytes. Percentages of IgM-binding late-stage IEs in individual IgM+ sub-clones of 3D7 (red circles, n = 22), HB3 (blue squares, n = 31), and IT4 (orange triangles, n = 13). (A) Means and standard deviations are shown. Representative flow cytometry histograms of three HB3 sub-clones with high, medium, and low percentages of IgM+ IEs (identified in A by larger symbols) in the presence (blue) and absence of non-immune IgM (black) (B).
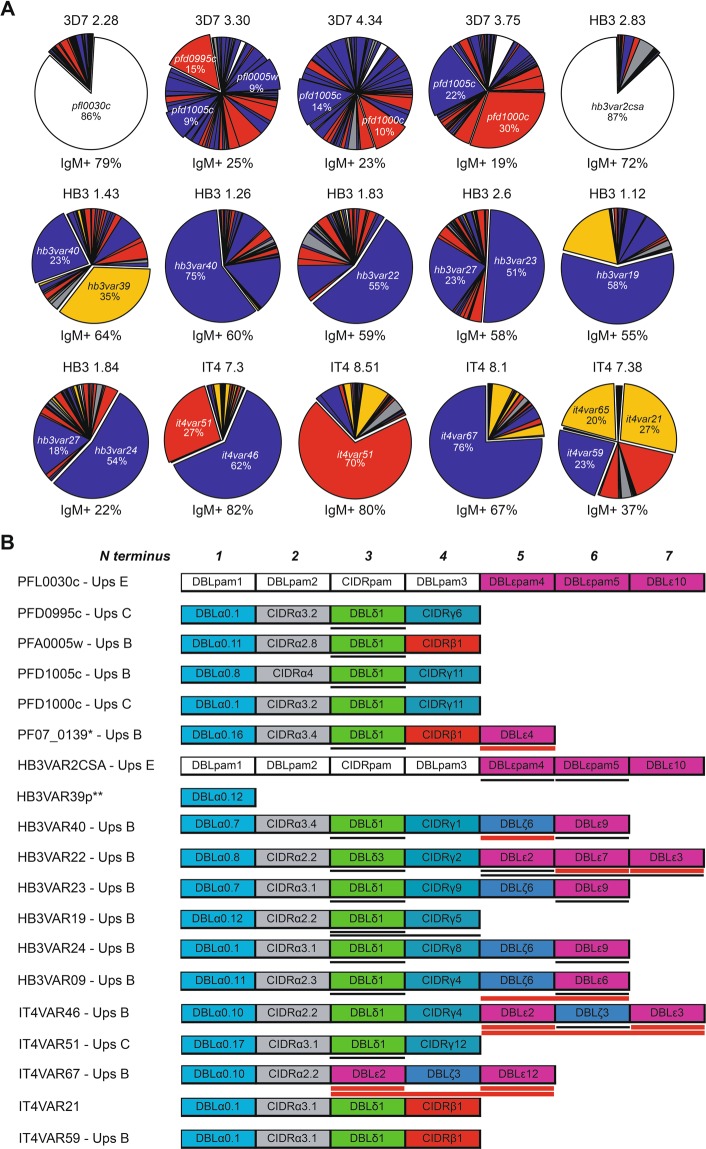
Figure 2.
Var gene transcription in P. falciparum IgM-selected sub-clones. Relative proportions of var gene transcripts in a selected subset of IgM-binding sub-clones of 3D7, HB3, and IT4 (additional data in Fig. S2 and Tables S1–S3). (A) Transcripts are color coded according to var Group (A: gray; B: blue, C: red, E: white, uncharacterized: yellow)5. Major transcripts are identified by name, and all candidate genes selected are shown as “exploded pie slices”. The percentage of corresponding IEs binding IgM is indicated below each pie diagram. Domain structures of the PfEMP1 proteins encoded by the named major transcripts in (A), color-codes as before5. (B) The orientation and domain numbers are indicated in italics along the top of the panel. Recombinant PfEMP1 constructs used in the present study are indicated by underlining, with those confirmed by ELISA to bind non-immune IgM identified by heavy red lines. *Identified as a candidate IgM-binder by Jeppesen et al.5. **Only the sequence of the DBLα_D1 domain of the PfEMP1 is known (http://www.cbs.dtu.dk/services/VarDom/).
Identification of non-immune IgM-binding domains in candidate recombinant PfEMP1 proteins
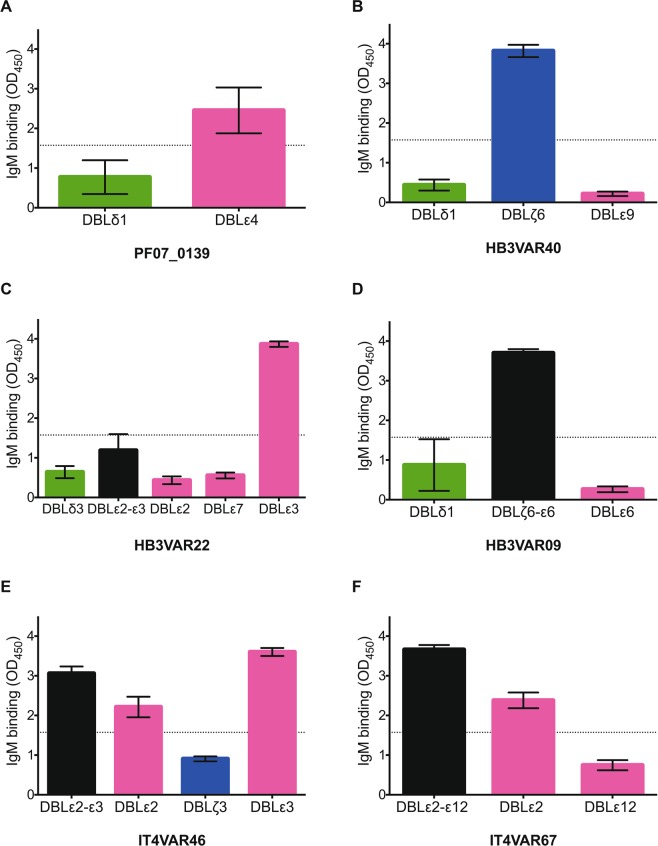
To examine further the IgM-binding properties of the candidate PfEMP1 proteins, we generated 38 recombinant domains belonging to 20 different PfEMP1 variants in 3D7 (n = 7), HB3 (n = 10), and IT4 (n = 3) (Figs 2, S2). Domains with substantial IgM-binding capacity were identified by ELISA near the C-terminus of PF07_0139 (DBLε4), HB3VAR40 (DBLζ6), HB3VAR22 (DBLε3), HB3VAR09 (DBLζ6- DBLε6), IT4VAR46 (DBLε2 and DBLε3), and IT4VAR67 (DBLε2) (Fig. 3), whereas the DBLδ1-type or DBLε9-type candidate domains generally showed limited affinity for non-immune IgM in ELISA (Figs 3 and S3). Results obtained with a “reversed” ELISA, where the plates were coated with IgM rather than the recombinant proteins, generally yielded similar results, except for HB3VAR22 and IT4VAR67 where binding to IgM was detected for additional domains (DBLε7 and DBLε12, respectively) (Fig. S4).
Figure 3.
Non-immune IgM-binding to recombinant PfEMP1 domains in ELISA. Binding of non-immune IgM to immobilized recombinant proteins representing candidate IgM-binding domains in PF07_0139 (A), HB3VAR40 (B), HB3VAR22 (C), HB3VAR09 (D), IT4VAR46 (E), and IT4VAR67 (F). Means and standard deviations of data from three independent experiments are shown. Dotted lines represent the cut-off above which binding to non-immune IgM was considered as positive. Domain types are color-coded as in Fig. 2B.
In a complementary approach, we tested a BioPlex bead array composed of 157 recombinant proteins representing almost all DBL and CIDR domains in the PfEMP1 proteins of P. falciparum 3D7 for binding by non-immune IgM, in a manner similarly to that used previously in tests of PfEMP1 binding to the endothelial receptors ICAM-1, CD36, and αVβ3 and αVβ6 integrins25–27. By this method, evidence of non-immune IgM binding was found in constructs from 13 PfEMP1 variants in P. falciparum 3D7 (Fig. S5). The data corroborated the IgM-binding capacity of domains in MAL6P1.4 (DBLε3), PFL0020w (DBLε4), and PF07_0139 (DBLε4) detected by cell-sorting and ELISA previously5 or here (Fig. S5A). With this approach, we also obtained preliminary evidence of additional IgM-binding domains in MAL6P1.4 (DBLγ13) and PFL0020w (DBLβ5), and in the head structure of MAL6P1.316 (DBLα2-CIDRα1.8) (Fig. S5A,C). MAL6P1.316 has previously been implicated in non-immune IgM binding, but the domain involved was not identified5. Furthermore, DBLα2 has been implicated in rosetting28, and CIDRα1.8 in IE adhesion to EPCR29. The BioPlex array also pointed to several PfEMP1 variants and domains not previously associated with non-immune IgM-binding. These included C-terminal domains in PF11_0008 (DBLδ5-CIDRβ4 and DBLβ9), PF08_0141 (DBLζ5), PF08_0103 (DBLδ1-CIDRβ1), and PFC0005w (DBLδ1-CIDRβ1) (Fig. S5B,C). Expression of PF08_0103 was recently associated with sporozoite-stage P. falciparum parasites, and PF08_0103-specific antisera blocked sporozoite invasion of hepatocytes30. However, it is unknown if these and other observations31,32 on these particular PfEMP1 variants are of any significance to the present findings. Finally, the array revealed non-immune IgM-binding in the N-terminal head structures of several additional PfEMP1 variants (PFE0005w, PFD0020c, PF11_0007, PF07_0048, and PF07_0051 (Fig. S5B). A weak IgM response to PF07_0051-DBLα1 was previously noted in an experimentally infected volunteer33. Overall, the BioPlex findings suggest that the capacity for Fc-mediated IgM-binding may be even more widespread than the single-IE sorting and PfEMP1-specific antibody selection experiments suggest. As such, they provide leads for future studies, ideally employing IEs expressing the corresponding native proteins.
Verification of the capacity of native candidate PfEMP1 proteins to bind non-immune IgM
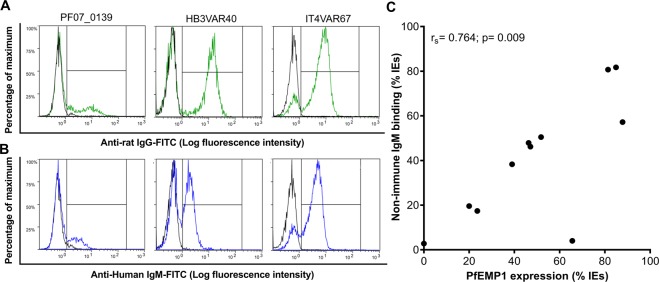
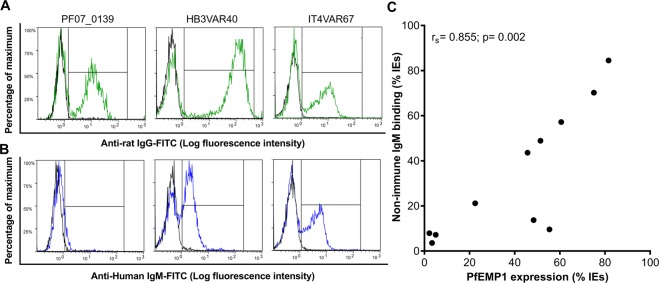
To further strengthen the link between IE surface-expression of particular native PfEMP1 variants and non-immune IgM binding to the IEs, we selected 12 recombinant domains (from 11 PfEMP1 variants) binding or not binding non-immune IgM in ELISA (Table S4). The selected domains were used to generate specific rat anti-sera and purified domain-specific IgG. In most cases, labeling of IEs from the single cell-sorted sub-clones with the domain-specific IgG (Figs 4A; S6A) and non-immune IgM (Figs 4B; S6B) demonstrated strong positive correlation between the percentage of IEs expressing candidate non-immune IgM-binding native PfEMP1 variants and the percentage of IEs that indeed bound non-immune IgM (Fig. 4C). The outlier (Fig. 4C) is HB3VAR19, where we could not verify IgM-binding to the candidate domains by ELISA (Fig. S3). The ability of this PfEMP1 protein to bind non-immune IgM must thus requires further investigation. Furthermore, repeated selection of pVBH-transfected parasites (after two weeks of culture without blasticidin pressure) with domain-specific rat IgG generally led to increased IE surface expression of the cognate native PfEMP1 variant (Figs 5A; S7A) and of non-immune IgM binding (Figs 5B; S7B), with a strong positive correlation between the two variables (Fig. 5C). The outliers (Fig. 5C) are HB3VAR19 (see above) and PF07_0139, which was found to bind IgM by ELISA (Fig. 3A) and Bioplex (Fig. S5A). However, we previously reported that the IgM-binding to native PF07_0139 on IEs surface is very low5, and the present data support that conclusion. Taken together, the data in sub-clones and selected parasites support the capacity of native PF07_0139, HB3VAR09, HB3VAR22, HB3VAR23, HB3VAR24, HB3VAR40, IT4VAR46, IT4VAR51, and IT4VAR67, to bind non-immune IgM.
Figure 4.
Non-immune IgM-binding and specific PfEMP1 expression on the surface of erythrocytes infected by P. falciparum sub-clones. Representative sub-clones generated by single-cell sorting and labeled with rat IgG specific for the PfEMP1 corresponding to the dominantly transcribed var gene (green) (A), or non-immune IgM (B). Background labeling indicated in black. Correlation (rs) of non-immune IgM-binding and labeling by PfEMP1-specific IgG for all sub-clones (C).
Figure 5.
Non-immune IgM-binding and specific PfEMP1 expression on the surface of infected erythrocytes selected by rat anti-PfEMP1 domain-specific IgG. P. falciparum 3D7 (left), HB3 (center), and IT4 (right) with heterogeneous var gene transcription (Fig. S1) after several rounds of selection for IE reactivity with rat anti-PF07_0139, anti-HB3VAR40, and anti-IT4VAR67, respectively, and labeled with the antibody used for selection (A) or non-immune IgM (B). Background labeling indicated in black. Correlation (rs) of non-immune IgM-binding and labeling by PfEMP1-specific IgG for all selected parasites expressed as percentage of IEs (C).
Overall, our data strongly support that binding of non-immune IgM to the surface of IEs is common (probably > 10% of all PfEMP1), mediated by particular PfEMP1 variants, and can be found in genetically diverse P. falciparum clones.
Discussion
P. falciparum causes the most severe form of malaria in humans. This is not least due to the evolution in this parasite species of multiple ways to evade host innate and acquired immune responses34. These include IE misappropriation of host plasma proteins7,35,36, endothelial sequestration of IEs37,38, and clonal antigenic variation39,40. The PfEMP1 family of adhesive proteins, expressed on the IE surface, are involved in all these processes4,41,42. In previous work, others and we have shown that Fcμ-mediated (i.e., non-immune) binding of IgM to particular PfEMP1 variants can function to shield IEs from recognition by PfEMP1-specific IgG43, and may also serve to augment IE adhesion to endothelial receptors6,44. Whatever function PfEMP1-bound IgM serves, it appears to be of clinical significance. The phenotype has repeatedly been associated with severe malaria in children, caused by parasites expressing Group A PfEMP1 variants3,6,28 and with malaria in pregnant women, caused by parasites expressing Group E (VAR2CSA-type) PfEMP124,43,45,46. Furthermore, it has been proposed that IgM-binding to several Group B PfEMP1 variants5, which are otherwise generally associated with uncomplicated malaria and asymptomatic infections, serves to increase the adhesive strength between IEs and endothelial receptors6,7,44. This idea supports the importance of the non-immune IgM-binding phenotype.
Both DBL and CIDR domains in the N-terminal head structure22,46 and C-terminal DBL domains4–6,46,47 have previously been implicated as the domains mediating non-immune binding of IgM to PfEMP1. In the single previous study assessing how many of the about 60 PfEMP1 variants encoded by a specific P. falciparum genome have the capacity to bind IgM that way, five were identified using the 3D7 clone5. In four of those, IgM bound to a C-terminal ε- or ζ-type DBL domain, whereas the PfEMP1 domain involved in the low-intensity IgM binding to PF07_0139 was not identified5. However, only a limited number of IgM-binding sub-clones were studied and the involvement of N-terminal domains was not systematically assessed. It is thus likely that additional IgM-binding PfEMP1 variants exist. Moreover, the study included only a single parasite clone.
To overcome these limitations, the current study was undertaken to provide an exhaustive analysis of non-immune IgM-binding PfEMP1 variants in the three P. falciparum clones 3D748, HB349, and IT440,50. The three parasite clones are representative of isolates from Africa, Central America, and Asia, respectively51. Furthermore, their var gene repertoires are highly divergent, with only two conserved genes (var1csa and var2csa) in common52.
We used two complementary approaches to identify PfEMP1 variants capable of binding non-immune IgM via Fcµ and to pinpoint the constituent domains involved in candidate proteins. In one approach, we used fluorescence-activated single-cell sorting to sub-clone individual IgM+ IEs from each of the three parasite clones (Fig. 1), followed by analysis of var gene transcription profiles and production of recombinant proteins representing specific domains in candidate PfEMP1 variants5 (Figs 2 and S2). The recombinant proteins were subsequently used to test their IgM-binding capacity directly (Figs 3 and S3). We also used them to generate antisera for detection of PfEMP1 expression on IgM-binding sub-clones (Figs 4 and S6). Finally, we used the PfEMP1-specific rat IgG to select erythrocytes infected by parasites with diverse var gene transcription (Fig. S1) for IE surface expression of the corresponding native protein and test their IgM-binding capacity (Figs 5 and S7). In the other approach, we made use of a protein array containing constructs representing almost all PfEMP1 domains or domain tandems (DBL-CIDR) in P. falciparum 3D7, immobilized on BioPlex beads26,27 (Fig. S5). Using these two approaches, we confirm the IgM-binding capacity of the PfEMP1 proteins and C-terminal DBLε and DBLζ domains implicated in our previous studies5,6. Several candidate IgM-binding PfEMP1 proteins also contained C-terminal DBLδ domains, but their capacity to bind IgM was lower (Figs 3, S3, S4). In addition, we document for the first time the IgM-binding capacity of Group E PfEMP1 in HB3 (HB3VAR2CSA) (Table S2) and of several C-terminal DBLε and DBLζ domains (PF07_0139, HB3VAR09, HB3VAR22, HB3VAR40, IT4VAR46, and IT4VAR67) (Table S1–S3). Finally, we provide preliminary evidence of IgM-binding DBLβ and DBLγ domains nearer the N-terminus (MAL6P1.4, PFL0020w) and C-terminus (PF11_0008), as well as fragments containing DBLα-CIDRα and DBLδ-CIDRβ domains in a number of PfEMP1 variants (Fig. S5). Most of those domain classes have not previously been implicated in Fcµ-mediated binding of IgM.
In other PfEMP1 variants implicated by sub-cloning of IgM+ IEs, it was not possible for us to confirm the IgM-binding capacity of selected candidate domains by ELISA (e.g., HB3VAR23_DBLε and HB3VAR24_DBLε) (Fig. S3). However, specific antisera against those domains could be used to select IEs that clearly bound non-immune IgM (Fig. S7), suggesting that the IgM-binding domains in those variants are different from those selected as candidates by us.
Overall, our study provides the first comprehensive overview of PfEMP1 proteins and domain classes that can bind IgM via its Fcµ domain. We document that the capacity for PfEMP1-dependent binding of IgM to the IE surface is prevalent in several distinct P. falciparum genomes, and show that some PfEMP1 variants (e.g., MAL6P1.4, HB3VAR22, IT4VAR46, and IT4VAR67) appear to possess several IgM-binding domains. Finally, our investigation indicates that non-immune IgM-binding may also occur among DBLα-CIDRα, DBLδ1, DBLβ, and DBLγ-type domains. However their ability to bind IgM appears less pronounced than that of DBLε- and DBLζ-type domains, and the biological relevance of this part of the study should be assessed further. While we did not investigate the functional significance of non-immune IgM-binding in the present study, previous reports have indicated roles in immune-evasion as well as pathogenesis (enabling rosetting and enhancing endothelial adhesion)6,43,44. Together, these findings support the idea that non-immune IgM binding confers survival advantages on parasites expressing PfEMP1 variants with IgM-binding domains, thereby rendering this phenotype of likely clinical importance.
Methods
Parasite in vitro culture and transfection
The P. falciparum clones 3D7, HB3, and IT4 were maintained in vitro in blood group O erythrocytes and serum-free medium, essentially as described before53. The 3D7 clone, transfected with the plasmid pVBH to erase epigenetic memory and achieve heterogeneous var gene transcription54, was generated and kindly provided by Ron Dzikowski21. Corresponding HB3 and IT4 parasites were generated by us in a similar way, except that the blasticidin concentration was increased to 10 µg/mL to recover parasites carrying high plasmid copy numbers. The genotypic identity of the parasites and the absence of Mycoplasma infection in the cultures were regularly verified.
Fluorescence-activated single-cell sorting of IgM-positive infected erythrocytes
Erythrocytes infected with pVBH-transfected late-stage parasites, obtained 14 days after release from blasticidin drug pressure, were purified by magnetic-activated cell sorting (MACS) and labeled with non-immune human IgM (10 nM; Sigma). After thorough washes in sterile phosphate-buffered saline supplemented with fetal bovine serum (2%), IEs were stained with phycoerythrin-conjugated anti-human IgM (1:200; Jackson ImmunoResearch) and washed as above. A BD FACSJazz cell sorter (BD Biosciences) was used to sort individual IgM-binding IEs into round-bottom 96-well plates (Thermo Fisher Scientific) containing serum-free culture medium (100 µL) and uninfected erythrocytes (1 µL). The parasites were maintained in the plates for 2-4 weeks as described above, changing medium twice weekly, alternating between medium alone and medium with erythrocytes as above. Parasite growth was monitored regularly by microscopy, testing small aliquots from 10 random wells per plate. Wells with microscopic evidence of parasite multiplication were transferred to culture flasks and maintained until stably growing parasite sub-clones were established.
var gene transcription profiling
Total RNA was prepared from synchronous ring-stage IEs using TRIzol (Ambion). Genomic DNA was removed by DNase I treatment (Invitrogen) and cDNA was generated using SuperScript II reverse transcriptase and random primers (Invitrogen) according to the manufacturer’s instructions. The cDNA, QuantiTec SYBR Green PCR Master Mix (Qiagen), and three validated sets of var gene-specific primers were used to assess var gene transcription profiles by real-time quantitative PCR on a Rotorgene RG-3000 thermal cycler (Corbett Research), as described in detail elsewhere55–57. The transcription levels for each var gene was calculated using the 2−ΔCT method, relative to the levels of the housekeeping gene seryl-tRNA synthetase.
Recombinant protein expression
Recombinant proteins with N-terminal his-tags and representing various PfEMP1 domains were produced in Escherichia coli Shuffle C3030 cells (New England Biolabs). Constructs were amplified from parasite genomic DNA, using specific primer sets (Table S5) and cloned into the pET15b modified plasmid58. All expressed proteins were purified by immobilized metal ion affinity chromatography using His-Trap High Performance columns (GE Healthcare). The purity of each protein was analyzed by sodium-dodecyl-sulfate polyacrylamide gel electrophoresis, followed by Coomassie staining with InstantBlue (Expedeon) following the manufacturer’s instructions. His-tags were detected by western blot using HRP-coupled anti-his antibodies (Qiagen) (Fig. S8).
Production of PfEMP1-specific anti-sera
Three male Wistar rats per domain were immunized by subcutaneous injection four times at two-weeks intervals with 30 μg (first injection) and 15 µg (subsequent injections) of selected recombinant PfEMP1 domains (Table S4) emulsified in AddaVax adjuvant (InvivoGen). Anti-sera were collected one week after the fourth immunization. All animal experiments were approved by the Danish Animal Procedures Committee “Dyreforsøgstilsynet” (permit number 2013-15-2934-00902), and were performed in accordance to the Danish acts LBK 1306 (23/11/2007) and BEK 1273 (12/12/2005).
Detection of non-immune IgM-binding to recombinant and native PfEMP1 proteins
Non-immune IgM-binding to recombinant PfEMP1 domains was assessed by ELISA as previously described6. In brief, flat-bottomed 96-well plates (Thermo Fisher Scientific) were coated overnight (4 °C) with recombinant protein (18 nM) or non-immune IgM (10 nM, for “reverse” ELISA experiments) in Tris saline magnesium buffer. After blocking and washing, plates were incubated (2 h, room temperature) with non-immune human IgM (10 nM) or recombinant protein (18 nM). Bound IgM or protein was detected using HRP-coupled anti-human IgM antibody (1:1,000; DAKO) and anti-His antibody (1:3,000; Qiagen), respectively, followed by TMB PLUS2 ready-to-use substrate (Kem-En-Tec Diagnostics) following the manufacturer’s instructions. Absorbance was read at 450 nm after stopping the colorimetric reaction by the addition of 0.2 M H2SO4. Binding was considered positive if the measured absorbance was higher than the average absorbance plus three standard deviations for the DBLγ13 (PFD1235w) expressed in an identical way as the constructs assessed in this study. This domain was chosen as a negative control, because erythrocytes infected by parasites expressing that particular variant do not bind non-immune IgM (our unpublished data).
Non-immune IgM-binding to recombinant PfEMP1 domains from P. falciparum 3D7 was also assessed by recombinant protein micro-array as previously described26,27. In brief, single- or multi-domain proteins were expressed in COS7 cells, purified and immobilized on BioPlex beads as described before25. In each experiment, a mixture of 53–55 bead regions with immobilized constructs was used. Beads with immobilized HisAdEx construct25,59 were used as negative control. The control contained all the same parts as recombinant domain constructs but short irrelevant 37-mer peptides instead of PfEMP1 domains. Some of the constructs, which showed consistent binding in these high throughput experiments (n = 4–7), were tested then in smaller sets of beads to confirm binding. PCR primers for constructs and protein domain boundaries are shown in Table S6. Uniform coating of the beads was verified using biotinylated anti-GFP antibody (Abcam), followed by incubation with streptavidin-PE (1:250; Jackson Immunoresearch). Non-immune IgM (10 nM; Sigma) binding to bead-bound PfEMP1 constructs (1,000 to 2,000 beads in the reaction) was measured in duplicates on BioPlex 200 machine (BioRad) as described before25,27. Binding of non-immune IgM to the constructs were determined by incubation with PE-conjugated anti-human IgM (1:250; Jackson Immunoresearch). The fluorescence signal (MFI, median fluorescence intensity) measured for each control construct was subtracted from the signal of each bead-bound construct, in each of duplicate well, and averaged. Experiments to test IgM binding to bead-immobilized domains were repeated at least three times with qualitatively similar results. Results of representative experiments are shown.
Non-immune IgM binding to native PfEMP1 expressed on the IE surface was detected by flow cytometry as previously described43. Briefly, late-stage IEs were purified by MACS and labeled with non-immune human IgM (10 nM; Sigma). IE-bound IgM was measured with a FITC-conjugated anti-human IgM (1:150; Sigma) and ethidium bromide (2 µg/mL) using a Beckman Coulter FC500 flow cytometer for data acquisition. Analysis was performed using FlowLogic software (Inivai Technologies, Australia).
Selection of infected erythrocytes for surface expression of specific PfEMP1 variants
Antigen-specific IgG was purified from rat sera after immunization with recombinant PfEMP1 domains using HiTrap Protein G HP and NHS-activated HP (coupled to the antigen used for immunization) columns (GE Healthcare) following manufacturer’s instructions. IEs were selected for surface expression of particular IgM-binding PfEMP1 candidates by repeated immune-magnetic selection using PfEMP1-specific rat anti-sera or affinity-purified rat IgG coupled to protein G-Dynabeads (Invitrogen) as described before60. Monospecific PfEMP1 expression on IEs was verified by flow cytometry as previously described43. In brief, late-stage IEs were labeled with affinity-purified IgG (10 μg/mL) followed by FITC-conjugated goat anti-rat IgG (1:150; Vector Laboratories). Parasite nuclei were stained with ethidium bromide (2 µg/mL) and antibody labeling of IEs quantified by flow cytometry as above.
Statistical analysis
Data were analyzed by using GraphPad Prism version 8.0.1 (GraphPad Software, San Diego, CA, USA). Two-tailed one-sample t-test relative to construct control was used to analyze the Bioplex results. Spearman’s rank correlation (rs) was used to assess the correlation between non-immune IgM-binding and labeling by PfEMP1-specific IgG. P-values < 0.05 were considered significant.
Supplementary information
Acknowledgements
We thank Ron Dzikowski (The Hebrew University of Jerusalem, Israel) for providing the pVBH-transfected 3D7 clone. Kirk Deitsch (Weill Medical College of Cornell University, NY) is thanked for providing the pVBH plasmid. Rebecca W. Olsen and Anja T.R. Jensen (University of Copenhagen, Denmark) are thanked for providing the DBLγ13 (PFD1235w) recombinant protein. Valentina Voronkova, Isaac T Frye, and Tracy Saveria are thanked for cloning some of the PfEMP1 domains used in the bead array. Justin Gullingsrud and Olga Chesnokov are thanked for the initial characterization of the PfEMP1 bead array functional activity. Maiken Visti and Nexhibe Beciri are thanked for excellent technical assistance. This work was funded by the Danish Council for Independent Research (DFF 4183-00539), Novo Nordisk Foundation (NNF15OC0017654), Rigshospitalet (R102-A4174), Sven Andersen Research Foundation, and Lundbeck Foundation (R250-2017-1289). Work related to the bead protein array was partly funded by the Florida Atlantic University start-up and by the National Institutes of Health (R01AI092120). GEM and SBD were supported by grants from the Consultative Committee for Development Research (DFC-12-081RH and 17-02-KU). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
M.d.P.Q., L.H. and M.L.P. conceived and designed the study. M.d.P.Q., G.E.M., S.O.T., S.B.D. and M.L.P. performed the experiments. A.V.O. contributed with protein array data. M.d.P.Q., A.V.O., L.H. and M.L.P. analyzed the data. M.d.P.Q., L.H. and M.L.P. wrote the manuscript. All authors read and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42585-0.
References
- 1.World Health Organization. World malaria report 2018 (2018).
- 2.David PH, Handunnetti SM, Leech JH, Gamage P, Mendis KN. Rosetting: a new cytoadherence property of malaria-infected erythrocytes. Am. J. Trop. Med. Hyg. 1988;38:289–297. doi: 10.4269/ajtmh.1988.38.289. [DOI] [PubMed] [Google Scholar]
- 3.Rowe JA, Shafi J, Kai OK, Marsh K, Raza A. Nonimmune IgM, but not IgG binds to the surface of Plasmodium falciparum-infected erythrocytes and correlates with rosetting and severe malaria. Am. J. Trop. Med. Hyg. 2002;66:692–699. doi: 10.4269/ajtmh.2002.66.692. [DOI] [PubMed] [Google Scholar]
- 4.Ghumra A, et al. Identification of residues in the Cm4 domain of polymeric IgM essential for interaction with Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) J. Immunol. 2008;181:1988–2000. doi: 10.4049/jimmunol.181.3.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeppesen A, et al. Multiple Plasmodium falciparum erythrocyte membrane protein 1 variants per genome can bind IgM via its Fc fragment Fcm. Infect. Immun. 2015;83:3972–3981. doi: 10.1128/IAI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson L, et al. Investigating the function of Fc-specific binding of IgM to Plasmodium falciparum erythrocyte membrane protein 1 mediating erythrocyte rosetting. Cell. Microbiol. 2015;17:819–831. doi: 10.1111/cmi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson L, et al. a2-macroglobulin can crosslink multiple Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) molecules and may facilitate adhesion of parasitized erythrocytes. PLoS Pathog. 2015;11:e1005022. doi: 10.1371/journal.ppat.1005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- 9.Lavstsen T, Salanti A, Jensen ATR, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar. J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol. Microbiol. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 11.Hviid L, Jensen AT. PfEMP1 - A parasite protein family of key importance in Plasmodium falciparum malaria immunity and pathogenesis. Adv. Parasitol. 2015;88:51–84. doi: 10.1016/bs.apar.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Smith JD, et al. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proceedings of the National Academy of Sciences, USA. 2000;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berendt AR, Simmons DL, Tansey J, Newbold CI, Marsh K. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature. 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- 14.Ockenhouse CF, Tandon NN, Magowan C, Jamieson GA, Chulay JD. Identification of a platelet membrane glycoprotein as a falciparum malaria sequestration receptor. Science. 1989;243:1469–1471. doi: 10.1126/science.2467377. [DOI] [PubMed] [Google Scholar]
- 15.Oquendo P, Hundt E, Lawler J, Seed B. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell. 1989;58:95–101. doi: 10.1016/0092-8674(89)90406-6. [DOI] [PubMed] [Google Scholar]
- 16.Salanti A, et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner L, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennartz F, et al. Structure-guided identification of a family of dual receptor-binding PfEMP1 that is associated with cerebral malaria. Cell Host Microbe. 2017;21:403–414. doi: 10.1016/j.chom.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pleass RJ, Moore SC, Stevenson L, Hviid L. Immunoglobulin M (IgM): restrainer of inflammation and mediator of immune evasion by Plasmodium falciparum malaria. Trends Parasitol. 2016;32:108–119. doi: 10.1016/j.pt.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Dzikowski R, Deitsch KW. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J. Mol. Biol. 2008;382:288–297. doi: 10.1016/j.jmb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fastman Y, Noble R, Recker M, Dzikowski R. Erasing the epigenetic memory and beginning to switch - the onset of antigenic switching of var genes in Plasmodium falciparum. PLoS One. 2012;7:e34168. doi: 10.1371/journal.pone.0034168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, et al. The semiconserved head structure of Plasmodium falciparum erythrocyte membrane protein 1 mediates binding to multiple independent host receptors. J. Exp. Med. 2000;192:1–10. doi: 10.1084/jem.192.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flick K, et al. Role of non-immune IgG bound to PfEMP1 in placental malaria. Science. 2001;293:2098–2100. doi: 10.1126/science.1062891. [DOI] [PubMed] [Google Scholar]
- 24.Semblat JP, Raza A, Kyes SA, Rowe JA. Identification of Plasmodium falciparum var1CSA and var2CSA domains that bind IgM natural antibodies. Mol. Biochem. Parasitol. 2006;146:192–197. doi: 10.1016/j.molbiopara.2005.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oleinikov AV, et al. High throughput functional assays of the variant antigen PfEMP1 reveal a single domain in the 3D7 Plasmodium falciparum genome that binds ICAM1 with high affinity and is targeted by naturally acquired neutralizing antibodies. PLoS Pathog. 2009;5:e1000386. doi: 10.1371/journal.ppat.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oleinikov AV, et al. A plasma survey using 38 PfEMP1 domains reveals frequent recognition of the Plasmodium falciparum antigen VAR2CSA among young Tanzanian children. PLoS One. 2012;7:e31011. doi: 10.1371/journal.pone.0031011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chesnokov O, Merritt J, Tcherniuk SO, Milman N, Oleinikov AV. Plasmodium falciparum infected erythrocytes can bind to host receptors integrins aVb3 and aVb6 through DBLd1_D4 domain of PFL2665c PfEMP1 protein. Sci. Rep. 2018;8:17871. doi: 10.1038/s41598-018-36071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghumra A, et al. Induction of strain-transcending antibodies against Group A PfEMP1 surface antigens from virulent malaria parasites. PLoS Pathog. 2012;8:e1002665. doi: 10.1371/journal.ppat.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau CK, et al. Structural conservation despite huge sequence diversity allows EPCR binding by the malaria PfEMP1 family. Cell Host Microb. 2015;17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanghi G, et al. A specific PfEMP1 is expressed in P. falciparum sporozoites and plays a role in hepatocyte infection. Cell Rep. 2018;22:2951–2963. doi: 10.1016/j.celrep.2018.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavstsen T, et al. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar. J. 2005;4:21. doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magistrado P, et al. Immunoglobulin G antibody reactivity to a Group A Plasmodium falciparum erythrocyte membrane protein 1 and protection from P. falciparum malaria. Infect. Immun. 2007;75:2415–2420. doi: 10.1128/IAI.00951-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krause DR, et al. Characterization of the antibody response against Plasmodium falciparum erythrocyte membrane protein 1 in human volunteers. Infect. Immun. 2007;75:5967–5973. doi: 10.1128/IAI.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yam XY, Preiser PR. Host immune evasion strategies of malaria blood stage parasite. Mol. Biosyst. 2017;13:2498–2508. doi: 10.1039/c7mb00502d. [DOI] [PubMed] [Google Scholar]
- 35.Scholander C, Carlson J, Kremsner PG, Wahlgren M. Extensive immunoglobulin binding of Plasmodium falciparum-infected erythrocytes in a group of children with moderate anemia. Infect. Immun. 1998;66:361–363. doi: 10.1128/iai.66.1.361-363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosa TF, et al. The Plasmodium falciparum blood stages acquire factor H family proteins to evade destruction by human complement. Cell. Microbiol. 2016;18:573–590. doi: 10.1111/cmi.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller LH. Distribution of mature trophozoites and schizonts of Plasmodium falciparum in the organs of Aoutus trivirgatus, the night monkey. Am. J. Trop. Med. Hyg. 1969;18:860–865. doi: 10.4269/ajtmh.1969.18.860. [DOI] [PubMed] [Google Scholar]
- 38.MacPherson GG, Warrell MJ, White NJ, Looaresuwan S, Warrell DA. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. American Journal of Pathology. 1985;119:385–401. [PMC free article] [PubMed] [Google Scholar]
- 39.Biggs B-A, et al. Antigenic variation in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1991;88:9171–9174. doi: 10.1073/pnas.88.20.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts DJ, et al. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su X, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 42.Smith JD, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barfod L, et al. Evasion of immunity to Plasmodium falciparum malaria by IgM masking of protective IgG epitopes in infected erythrocyte surface-exposed PfEMP1. Proc. Natl. Acad. Sci. USA. 2011;108:12485–12490. doi: 10.1073/pnas.1103708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akhouri RR, Goel S, Furusho H, Skoglund U, Wahlgren M. Architecture of Human IgM in Complex with P. falciparum Erythrocyte Membrane Protein 1. Cell Rep. 2016;14:723–736. doi: 10.1016/j.celrep.2015.12.067. [DOI] [PubMed] [Google Scholar]
- 45.Creasey A, Staalsoe T, Raza A, Arnot D, Rowe JA. Nonspecific Immunoglobulin M binding and chondroitin sulfate A binding are linked phenotypes of Plasmodium falciparum isolates implicated in malaria during pregnancy. Infect. Immun. 2003;71:4767–4771. doi: 10.1128/IAI.71.8.4767-4771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasti N, et al. Nonimmune immunoglobulin binding and multiple adhesion characterize Plasmodium falciparum-infected erythrocytes of placental origin. Proc. Natl. Acad. Sci. USA. 2006;103:13795–13800. doi: 10.1073/pnas.0601519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semblat JP, et al. Identification of the minimal binding region of a Plasmodium falciparum IgM binding PfEMP1 domain. Mol. Biochem. Parasitol. 2015;201:76–82. doi: 10.1016/j.molbiopara.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walliker D, et al. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 49.Bhasin VK, Trager W. Gametocyte-forming and non-gametocyte-forming clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 1984;33:534–537. doi: 10.4269/ajtmh.1984.33.534. [DOI] [PubMed] [Google Scholar]
- 50.Jensen JB, Trager W. Plasmodium falciparum in culture: establishment of additional strains. Am. J. Trop. Med. Hyg. 1978;27:743–746. doi: 10.4269/ajtmh.1978.27.743. [DOI] [PubMed] [Google Scholar]
- 51.Mu J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kraemer SM, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cranmer SL, Magowan C, Liang J, Coppel RL, Cooke BM. An alternative to serum for cultivation of Plasmodium falciparum in vitro. Trans. R. Soc. Trop. Med. Hyg. 1997;91:363–365. doi: 10.1016/S0035-9203(97)90110-3. [DOI] [PubMed] [Google Scholar]
- 54.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salanti A, et al. Selective upregulation of a single distinctly structured var gene in CSA-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 56.Dahlbäck M, et al. Changes in var gene mRNA levels during erythrocytic development in two phenotypically distinct Plasmodium falciparum parasites. Malar. J. 2007;6:78. doi: 10.1186/1475-2875-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soerli J, et al. Human monoclonal IgG selection of Plasmodium falciparum for the expression of placental malaria-specific variant surface antigens. Parasite Immunol. 2009;31:341–346. doi: 10.1111/j.1365-3024.2009.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins MK. The structure of a chondroitin sulfate-binding domain important in placental malaria. J. Biol. Chem. 2008;283:21842–21846. doi: 10.1074/jbc.C800086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oleinikov AV, et al. Effects of sex, parity, and sequence variation on seroreactivity to candidate pregnancy malaria vaccine antigens. J. Infect. Dis. 2007;196:155–164. doi: 10.1086/518513. [DOI] [PubMed] [Google Scholar]
- 60.Staalsoe T, et al. In vitro selection of Plasmodium falciparum 3D7 for expression of variant surface antigens associated with severe malaria in African children. Parasite Immunol. 2003;25:421–427. doi: 10.1111/j.1365-3024.2003.00652.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.