Abstract
Between 2015 and the beginning of 2018 (January-March), 30 cetaceans were found stranded along the Ligurian Sea coast of Italy. Necropsies were performed in 22 cases and infectious diseases resulted the most common cause of death. Three striped dolphins, showed a severe coinfection involving the monophasic variant of Salmonella Typhimurium (Salmonella 1,4,[5],12:i:-). The isolates were characterized based on antimicrobial resistance, Multiple-Locus Variable-number tandem-repeat Analysis (MLVA) and whole-genome sequencing (WGS). All isolates demonstrated the same multidrug resistant genotype (ASSuT isolates), showed three different MLVA profiles, two of which closely related, and were identified as Sequence Type 34. Moreover, Single nucleotide polymorphisms (SNP) analysis confirmed strong correlations between two out of the three isolates. To our knowledge, S. 1,4,[5],12:i:-, one of the most common serovars in cases of human infection and food sources worldwide, has not previously been described in marine mammals, and reports of Salmonella-associated disease in free-ranging cetaceans are rare. These results highlight the role of cetaceans as sentinel species for zoonotic and terrestrial pathogens in the marine environment, suggest a potential risk for cetaceans and public health along the North Western Italian coastline and indicate cetaceans as a novel potential reservoir for one of the most widespread Salmonella serovars.
Introduction
Many marine mammal species share the coastal environment with humans and may serve as efficient sentinels for infectious agents, including those with a zoonotic potential, thus becoming important indicators for public health-related issues1.
The Mediterranean basin represents the largest enclosed sea on earth and an important marine biodiversity hotspot, being surrounded by heavily populated and industrialized coastal areas, which makes the impact of anthropic activities proportionally stronger than in any other basins. Marine mammals from such regions, moreover, show high tissue concentrations of organochlorine (OC) xenobiotics2,3, increasing their susceptibility to other anthropogenic stressors.
Between 2015 and the beginning of 2018 (January-March), 30 cetaceans, of which 23 striped dolphins (Stenella coeruleoalba), 2 bottlenose dolphins (Tursiops truncatus), 1 sperm whale (Physeter macrocephalus) and 4 undetermined specimens stranded along the Ligurian coast of Italy, in the Pelagos Sanctuary, a marine protected area for marine mammals covering about 90,000 km2 in the North Western Mediterranean basin, between Italy, Monaco and France, thereby encompassing the Northern coast of Sardinia, the Tuscan Archipelago and the entire Corsica Island (Fig. 1). Thanks to the surveillance activity of the Italian National Reference Centre for Diagnostic Activities on Stranded Marine Mammals (C.Re.Di.Ma.), necropsies were performed in 22 cases, the cause of stranding was determined with confidence in 19 cases and infectious diseases resulted the most common cause of death (18 out of 19). In half of the cases, the cetaceans under study were affected by severe coinfections involving cetacean-specific viruses, zoonotic pathogens and/or microbial agents indicative of environmental contamination.
Figure 1.
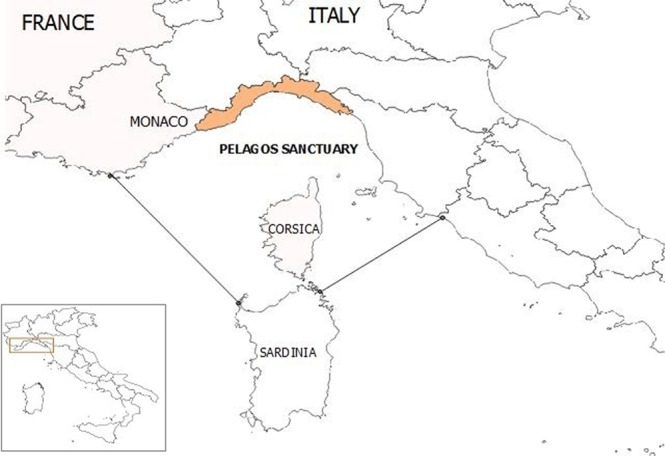
Map of the study area (Ligurian coastline) in the Pelagos Sanctuary area. The map was created by A.P. with QGIS (QGIS Development Team (2018). QGIS Geographic Information System. Open Source Geospatial Foundation Project. http://qgis.osgeo.org). Copyright © 2018 Alessandra Pautasso. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.3 or any later version published by the Free Software Foundation; with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts.
Herein we report three cases of coinfection, involving striped dolphins found stranded along the coast of the province of Savona (Fig. 2), and characterized by the detection of a monophasic variant of Salmonella 1,4,[5],12:i:- in association with cetacean-specific viruses along with pathogens indicative of environmental contamination.
Figure 2.
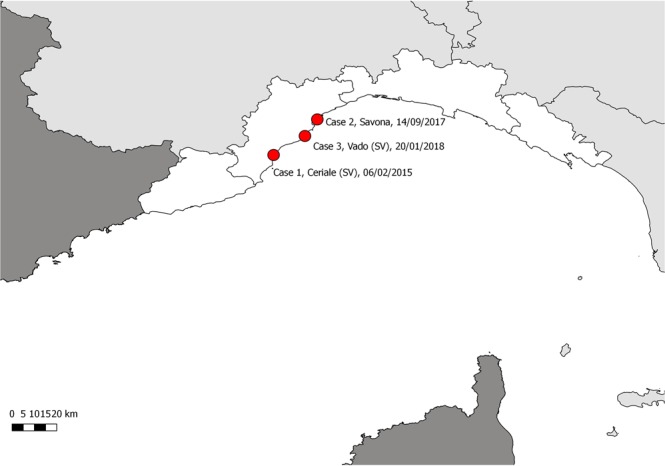
Map of the study area (Ligurian coastline), displaying the stranding locations of the 3 striped dolphins infected by Salmonella 1,4,[5],12:i:- variant (marked by red dots). The map was created by A.P. with QGIS (QGIS Development Team (2018). QGIS Geographic Information System. Open Source Geospatial Foundation Project. http://qgis.osgeo.org). Copyright © 2018 Alessandra Pautasso. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.3 or any later version published by the Free Software Foundation; with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts.
Although many marine species are known to harbor Salmonella enterica, reports of Salmonella-associated disease in free-ranging cetaceans are rare4,5, usually occurring in debilitated or stressed animals6. Clinically, severe enteritis with necrosis and/or haemorrhage are usually reported. Animals that develop septicaemia can die without showing clinical signs or, in some cases, can show complications such as bronchopneumonia, necrotizing hepatitis, splenitis, meningoencephalitis and abscessation6,7.
To our knowledge, the role of the environmental factors affecting Salmonella spp. persistence in the marine environment remains poorly understood, together with the infection’s transmission pathways to marine organisms. This faecal bacterium, in fact, is not indigenous to the marine environment, and its presence in coastal waters has been linked to heavy rain and storm-generated flows, transporting the contamination from their sources to the sea via river waters8, as well as to in situ defecation by infected marine animals9. Enteric bacteria may concentrate in sediments as well as in invertebrates and vertebrates of marine environments contaminated with faecal materials for prolonged periods. The presence of zooplankton and suspended marine particles colonized by Salmonella has been also reported, suggesting additional pathways for bacterial dissemination in marine habitats10.
Monophasic variant of Salmonella Typhimurium (Salmonella 1,4,[5],12:i:-) began to emerge in the mid-1990s and is currently one of the most common serovars in human clinical cases as well as in animal and food sources (mainly pigs and pork products) worldwide11–15, being responsible for a number of outbreaks, including those reported in Luxembourg, Germany, France and United States16.
In the last few years, in Italy, S. 1,4,[5],12:i:- has shown an increase in terms of prevalence, thereby ranking from the third most frequent Salmonella serovar isolated from clinical samples in 200817 to the second in 200918, and ultimately gaining the first position from 2011 onwards19.
This serovar exhibits the highest rate of multidrug resistance at the European level, thus representing a significant concern to public health authorities and also showing extremely high survival rates in the environment20. To the best of our knowledge, the monophasic variant of Salmonella Typhimurium has not been previously described in marine mammals.
In this study, three cases of coinfection, characterized by S. 1,4,[5],12:i:- detection, were analysed in detail, focusing on the pathogenic role of the identified agents, the association with the pathological findings and the most likely sources of exposure, considering the characteristics of the concerned coastline and the bacterial isolates’ features.
Results
A summary of the results obtained is presented here below, with reference to necropsy, histopathological and analytical data for each case considered. Microbiological, virological and immunohistochemical results are also summarized in Table 1.
Table 1.
Microbiological, virological and immunohistochemical results of investigations performed on the 3 striped dolphins under study.
| Samples | Organs/Tissues | Investigations | Results | Reference |
|---|---|---|---|---|
| Case 1 | faeces, liver | Salmonella spp. isolation | positive (S. 1,4,[5],12:i:-) | 38 |
| brain, lung, PSC ln, TB ln, liver, spleen, kidney | standard aerobic bacterial culture | Aeromonas hydrophila | — | |
| brain | Listeria spp. isolation | negative | 38 | |
| brain, PSC ln, spleen | Brucella spp. isolation | negative | 38 | |
| brain, lung, PSC ln, spleen | Brucella spp. (PCR) | negative | 50 | |
| serum | anti- Brucella spp. AB detection (S.A.R.) | negative | 52 | |
| brain, lung, PSC ln, spleen | Morbillivirus (PCR, sequence analysis) | positive (Dolphin Morbillivirus) | 27 | |
| brain, lung, spleen, kidney | Morbillivirus (IHC) | negative | 25 | |
| lung, PSC ln | Morbillivirus (isolation) | negative | 51 | |
| serum | anti-Morbillivirus AB detection (SN) | negative | 53 | |
| brain, PSC ln, liver, spleen, muscle | Toxoplasma gondii (PCR) | positive | 26 | |
| brain | Toxoplasma gondii (IHC) | negative | 25 | |
| serum | anti-T. gondii AB detection (IFAT) | positive (>1:160) | 25 | |
| brain, lung, PSC ln, spleen | Herpesvirus (PCR) | negative | 49 | |
| Case 2 | faeces, liver | Salmonella spp. isolation | positive (S. 1,4,[5],12:i:-) | 38 |
| brain, lung, PSC ln, TB ln, liver, spleen, kidney, blood | standard aerobic bacterial culture | positive (S. 1,4,[5],12:i:-) | — | |
| brain | Listeria spp. isolation | negative | 38 | |
| brain, lung, PS ln, TB ln, liver, spleen | Brucella spp. isolation | negative | 38 | |
| brain, lung, tonsils, PS ln, TB ln, liver, spleen, kidney, blood | Brucella spp. (PCR) | negative | 50 | |
| serum, HA, CSF | anti- Brucella spp. AB detection (S.A.R.) | negative | 52 | |
| brain, lung, tonsils, PS ln, TB ln, liver, spleen, kidney, bladder, blood | Morbillivirus (PCR, sequence analysis) | positive (Dolphin Morbillivirus) | 27 | |
| brain, lung, PSC ln, TB ln, spleen, kidney, bladder, pancreas | Morbillivirus (IHC) | positive | 25 | |
| brain, lung, TB ln, spleen, kidney, bladder | Morbillivirus (isolation) | negative | 51 | |
| serum, HA, CSF | anti-Morbillivirus AB detection (SN) | negative | 53 | |
| brain, PS ln, TB ln, liver, spleen, heart, muscle | Toxoplasma gondii (PCR) | positive | 26 | |
| brain, heart | Toxoplasma gondii (IHC) | positive | 25 | |
| serum, HA, CSF | anti-T. gondii AB detection (IFAT) | positive (serum,1:160; HA, 1:5120) | 25 | |
| brain, lung, PSC ln, TB ln, spleen kidney | Herpesvirus (PCR, sequence analysis) | positive (Alphaherpesvirus) | 49 | |
| Case 3 | faeces | Salmonella spp. isolation | positive (S. 1,4,[5],12:i:-) | 38 |
| brain, lung, PSC ln, PUL ln, liver, spleen, kidney | Salmonella spp. isolation (with pre-enrichment) | positive (S. 1,4,[5],12:i:-) | 38 | |
| brain | Listeria spp. isolation | negative | 38 | |
| brain | Brucella spp. isolation | negative | 38 | |
| brain, lung, PSC ln, kidney | Brucella spp. (PCR) | negative | 50 | |
| serum, HA, CSF | anti- Brucella spp. AB detection (S.A.R.) | negative | 52 | |
| brain, lung, PSC ln, PUL ln, liver, spleen, kidney, bladder | Morbillivirus (PCR, sequence analysis) | positive (Dolphin Morbillivirus) | 27 | |
| brain, lung, PSC ln, PUL ln, liver, spleen, kidney, bladder | Morbillivirus (isolation) | negative | 51 | |
| serum, HA, CSF | anti-Morbillivirus AB detection (SN) | negative | 53 | |
| brain, PSC ln, PUL ln, liver, spleen, heart, muscle | Toxoplasma gondii (PCR) | negative | 26 | |
| serum, HA, CSF | anti-T. gondii AB detection (IFAT) | negative | 25 | |
| brain, lung, PSC ln, PUL ln, liver, spleen | Herpesvirus (PCR, sequence analysis) | positive (Alphaherpesvirus) | 49 |
Legend - PSC ln: prescapular lymph node; TB ln: tracheo-bronchial lymph node; AB: antibodies; S.A.R: rapid serum agglutination; IHC: immunohistochemistry; IFAT: indirect immunofluorescence test; HA, humor acqueous; CSF: cerebrospinal fluid; PUL ln: pulmonary lymph node; SN: serum neutralization.
Tissues positive are shown in bold.
Case 1
The first animal, a 204 cm (Total Length, TL) adult female, was found stranded in Ceriale (SV) (44.09N, 8.23E) on 2nd February 2015, in a moderate nutritional status and a post-mortem condition code 3 (moderately decomposed). The estimated age was 17 years. Significant gross necropsy findings included a severe state of dehydration (related to prolonged exposure to dry and windy conditions before necropsy), a widespread internal congestion and a mild Phyllobotrium delphini and Monorygma grimaldi infection. The forestomach contained scanty material, represented by few highly digested cephalopod beaks, correlated to a non-recent meal. The cephalopod species were identified as 1 Goliteuthis armata (family Cranchiidae), typical of offshore and deep pelagic waters, 2 Loligo vulgaris (family Loginidae), which generally inhabits coastal waters and 6 Todarodes sagitattus (family Ommastrephidea), a species widely distributed both in coastal and offshore waters.
Microscopically, a non-suppurative meningoencephalitis characterized by vasculitis, gliosis, meningitis and prominent perivascular mononuclear cell cuffing was observed in all brain areas, associated with one protozoan cyst located in the frontal cortex.
Despite the poor post-mortem preservation degree, a mild broncho-interstitial pneumonia with oedema and multifocal parasitic granulomas, multifocal chronic cholangiohepatitis, lymphoid depletion and a mild mixed inflammatory infiltrate in spleen, were observed.
The diagnosis of Salmonella infection was achieved by the isolation of Salmonella 1,4,[5],12:i:- from faeces (intestine) and liver using a specific bacteriological approach based on selective broth enrichment and subculture onto selective solid media, as shown in Table 2.
Table 2.
Salmonella spp. isolation from the 3 striped dolphins (Stenella coreuleoalba)under study: methods, results and classification of Salmonella infection.
| Method | Tissues | Infection | |
|---|---|---|---|
| Case 1 | A | brain-lung-prescapular and tracheobronchial lymph nodes-liver-spleen-kidney | sub-clinic/asymptomatic |
| B | feces (intestine)-liver | ||
| Case 2 | A | brain-lung-prescapular and tracheobronchial lymph nodes-liver-spleen-kidney-blood | septicaemic |
| B | feces (intestine)-liver | ||
| Case 3 | B | feces (intestine) | disseminated |
| C | brain-lung-prescapular and pulmonary lymph nodes-liver-spleen-kidney |
Legend –
A: standard aerobic bacterial culture;
B: enrichment step in selective liquid media (Selenyte Cystine broth-SC and Mueller-Kaufmann Tetrathionate-Novobiocin –MKTTn broth) and subculture onto selective solid media (Brilliant Green Agar –BGA and Xylose Lysine Desoxycholate –XLD agar);
C: pre-enrichment step in Buffered Peptone Water (BPW) and subculture onto semisolid medium (Rappaport Vassiliadis Semi-Solid Medium Modified-MSRV agar).
Tissues positive are shown in bold.
The antimicrobial susceptibility results are reported in Table 3.
Table 3.
Antimicrobial phenotipic profiles of the 3 Salmonella 1,4,[5],12:i:- isolates recovered from the 3 striped dolphins under study (R = resistant; I = intermediate; S = susceptible).
| A | AMC | C | CAZ | CIP | CTX | G | K | KF | NAL | S | SXT | T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | R | I | S | I | S | I | S | S | I | I | R | I | R |
| Case 2 | R | I | S | S | S | S | S | S | S | S | R | R | R |
| Case 3 | R | S | S | S | S | S | S | S | S | S | R | S | R |
Abbreviations: A, ampicillin; AMC, amoxicillin/clavulanic acid; C, chloramphenicol; CZ, ceftazidime; CIP, ciprofloxacin; CTX, cefotaxime; G, gentamicin; K, kanamycin; KF, cefalotin; NAL, nalidixic acid; S, streptomycin; SXT, trimethoprim-sulfamethoxazole; T, tetracycline.
The Multiple-Locus Variable-number tandem-repeat Analysis (MLVA) profile associated to this isolate was 3-13-12-n.a.-0211 (Table 4).
Table 4.
Biomolecular typing results related to MLVA, in silico MLST and AMR genotype.
| MLVA | MLST | AMR genotype | Template | Related Phenotype | query_coverage | |
|---|---|---|---|---|---|---|
| Case 1 | 3-13-12-n.a.-0211 | ST34 | ASSuT | aph(3″)-Ib_5_AF321551 | Streptomycin-Resistance | 100.00 |
| aph(6)-Id_1_M28829 | Aminoglycoside Resistance | 100.00 | ||||
| blaTEM-1B_1_JF910132 | Beta-lactam Resistance | 100.00 | ||||
| sul2_3_HQ840942 | Sulphonamide Resistance | 100.00 | ||||
| tet(B)_4_AF326777 | Tetracycline Resistance | 100.00 | ||||
| Case 2 | 3-16-8-n.a.-0211 | ST34 | ASSuT | aph(3″)-Ib_5_AF321551 | Streptomycin-Resistance | 100.00 |
| aph(6)-Id_1_M28829 | Aminoglycoside Resistance | 100.00 | ||||
| blaTEM-1B_1_JF910132 | Beta-lactam Resistance | 100.00 | ||||
| sul2_3_HQ840942 | Sulphonamide Resistance | 100.00 | ||||
| tet(B)_4_AF326777 | Tetracycline Resistance | 100.00 | ||||
| Case 3 | 3-15-8-n.a.-0211 | ST34 | ASSuT | aph(3″)-Ib_5_AF321551 | Streptomycin-Resistance | 100.00 |
| aph(6)-Id_1_M28829 | Aminoglycoside Resistance | 100.00 | ||||
| blaTEM-1B_1_JF910132 | Beta-lactam Resistance | 100.00 | ||||
| sul2_3_HQ840942 | Sulphonamide Resistance | 100.00 | ||||
| tet(B)_4_AF326777 | Tetracycline Resistance | 100.00 |
Abbreviations: MLVA, Multiple-Locus Variable-number tandem-repeat Analysis; MLST, Multilocus Sequence Typing; AMR, Antimicrobial Resistance; ST, Sequence Type; ASSuT, Ampicillin, Streptomycin, Sulfonamide and Tetracycline resistant isolate.
The multilocus sequence typing (MLST) in silico showed that Salmonella 1,4,[5],12:i:- isolate belonged to ST34 (Table 4); the results of KmerResistance 2.2 are listed in Table 4 and the results of SPIFinder 1.0 “Pathogenic Island” are listed in Table 5.
Table 5.
Salmonella Pathogenicity Islands extracted using SPIFinder 1.0.
| Gene | Origin | %Identity | HSP/Query length | Insertion location | SPI Accession | |
|---|---|---|---|---|---|---|
| Case 1 | SPI-1 | Salmonella Typhimurium LT2 | 100.00 | 44279/44279 | fhlA-mutS | gi|16763390:3005842-3050120 |
| SPI-2 | Salmonella Typhimurium LT2 | 100.00 | 40071/40071 | tRNA-valV | gi|16763390:1461740-1501810 | |
| SPI-3 | Salmonella Typhimurium LT2 | 99.99 | 16616/16616 | tRNA-selC | gi|16763390:3948576-3965191 | |
| SPI-4 | Salmonella Choleraesuis str SC-B67 | 98.94 | 26699/26698 | ssb-soxSR | gi|62178570:4411902-4438599 | |
| SPI-5 | Salmonella Typhimurium LT2 | 100.00 | 9069/9069 | tRNA-serT | gi|16763390:1175321-1184389 | |
| SPI-13 | Salmonella Gallinarum SGD-3 | 99.41 | 338/338 | tRNA-pheV | AY956832 | |
| SPI-13 | Salmonella Gallinarum SGG-1 | 100.00 | 404/404 | tRNA-pheV | AY956833 | |
| SPI-13 | Salmonella Gallinarum SGA-10 | 100.00 | 341/341 | tRNA-pheV | AY956834 | |
| SPI-14 | Salmonella Gallinarum SGA-8 | 100.00 | 501/501 | Not_published | AY956835 | |
| SPI-14 | Salmonella Gallinarum SGC-8 | 99.55 | 441/441 | Not_published | AY956836 | |
| C63PI | Salmonella Typhimurium SL1344 | 99.88 | 21435/25252 | fhlA | AF128999 | |
| Case 2 | SPI-1 | Salmonella Typhimurium LT2 | 100.00 | 44280/44279 | fhlA-mutS | gi|16763390:3005842-3050120 |
| SPI-2 | Salmonella Typhimurium LT2 | 100.00 | 40071/40071 | tRNA-valV | gi|16763390:1461740-1501810 | |
| SPI-3 | Salmonella Typhimurium LT2 | 99.99 | 16616/16616 | tRNA-selC | gi|16763390:3948576-3965191 | |
| SPI-4 | Salmonella Choleraesuis str SC-B67 | 98.94 | 26699/26698 | ssb-soxSR | gi|62178570:4411902-4438599 | |
| SPI-5 | Salmonella Typhimurium LT2 | 100.00 | 9069/9069 | tRNA-serT | gi|16763390:1175321-1184389 | |
| SPI-13 | Salmonella Gallinarum SGG-1 | 100.00 | 404/404 | tRNA-pheV | AY956833 | |
| SPI-13 | Salmonella Gallinarum SGA-10 | 100.00 | 341/341 | tRNA-pheV | AY956834 | |
| SPI-13 | Salmonella Gallinarum SGD-3 | 99.41 | 338/338 | tRNA-pheV | AY956832 | |
| SPI-14 | Salmonella Gallinarum SGC-8 | 99.55 | 441/441 | Not_published | AY956836 | |
| SPI-14 | Salmonella Gallinarum SGA-8 | 100.00 | 501/501 | Not_published | AY956835 | |
| C63PI | Salmonella Typhimurium SL1344 | 99.91 | 21140/25252 | fhlA | AF128999 | |
| Case 3 | SPI-1 | Salmonella Typhimurium LT2 | 100.00 | 44280/44279 | fhlA-mutS | gi|16763390:3005842-3050120 |
| SPI-2 | Salmonella Typhimurium LT2 | 100.00 | 40071/40071 | tRNA-valV | gi|16763390:1461740-1501810 | |
| SPI-3 | Salmonella Typhimurium LT2 | 99.99 | 16616/16616 | tRNA-selC | gi|16763390:3948576-3965191 | |
| SPI-5 | Salmonella Typhimurium LT2 | 100.00 | 9069/9069 | tRNA-serT | gi|16763390:1175321-1184389 | |
| SPI-13 | Salmonella Gallinarum SGG-1 | 100.00 | 404/404 | tRNA-pheV | AY956833 | |
| SPI-13 | Salmonella Gallinarum SGA-10 | 100.00 | 341/341 | tRNA-pheV | AY956834 | |
| SPI-14 | Salmonella Gallinarum SGA-8 | 100.00 | 501/501 | Not_published | AY956835 | |
| SPI-14 | Salmonella Gallinarum SGC-8 | 99.55 | 441/441 | Not_published | AY956836 | |
| C63PI | Salmonella Typhimurium SL1344 | 99.91 | 25244/25252 | fhlA | AF128999 | |
| SPI-13 | Salmonella Gallinarum SGD-3 | 99.41 | 338/338 | tRNA-pheV | AY956832 |
Abbreviations: SPI, Salmonella Pathogenic Island.
Aeromonas hydrophila was additionally isolated from brain, spleen and lung by aerobic bacterial culture, while neither Listeria spp nor Brucella spp. were isolated.
No biomolecular evidence of Brucella spp. was found, with no anti-Brucella spp. antibodies being detected in the blood serum.
Dolphin Morbillivirus (DMV) infection targeting the lung and prescapular lymph node was demonstrated by means of PCR, followed by amplicon sequencing and BLAST analysis. Nevertheless, viral isolation attempts were negative, with no anti-Morbillivirus antibodies being detected in serum. Furthermore, no immunohistochemical (IHC) evidence of morbilliviral antigens was found in tissues from this dolphin.
Direct evidence of Toxoplasma gondii infection was obtained through the observation of one protozoan cyst, coupled with PCR positivity in the animal’s brain, alongside simultaneous evidence of anti-T. gondii antibodies in serum (>1:160). No IHC evidence of T. gondii-specific antigens was additionally found in tissues from this dolphin.
No biomolecular evidence of Herpesvirus was found in the examined tissues.
Case 2
The second animal, a 202 cm (TL), adult female, was found stranded dead in Savona (44.29N, 8.45E) on 14th September 2017, in a poor nutritional status and in post-mortem condition code 2 (fresh). The estimated age was 15 years. This was a lactating individual. Relevant pathological findings included haemorrhagic suffusions at blubber and myocardal levels, along with a severe Phyllobotrium delphini and Monorygma grimaldi infection, a mild pulmonary congestion, a multicentric reactive lymphadenopathy associated with splenic hypertrophy and the presence of numerous, focal ulcers in the keratinized gastric compartment. The forestomach contained scanty ingesta, mostly represented by undigested seagrass (Posidonia oceanica).
Microscopically, a non-suppurative meningoencephalitis characterized by multifocal malacic areas associated with oedema, gitter cells, mononuclear cell infiltrates, perivascular cuffs, syncytial cells and multiple protozoan cysts, some of which destroyed, were observed. Areas characterized by gliosis and neuronophagia, together with focal and moderate non-suppurative meningitis, were also apparent. One protozoan cyst, characterized by a huge diameter, without reaction in the surrounding tissue, was evident at parietal cortex level. Protozoan cysts were also detected in the heart. A severe membranous glomerular nephropathy, associated with multifocal interstitial chronic nephritis, was additionally found. Lymphoid depletion and sinus hypercellularity were observed in spleen and lymph nodes. Mild endocardiosis of the left atrio-ventricular valve, with fibrosis and lymphocytic epicarditis, were also found. Multifocal superficial erosions of the forestomach mucosa and necrosis of the intestinal crypts’ epithelium were observed. An intravascular embolus in the intestinal mesentery was also detected (Fig. 3).
Figure 3.
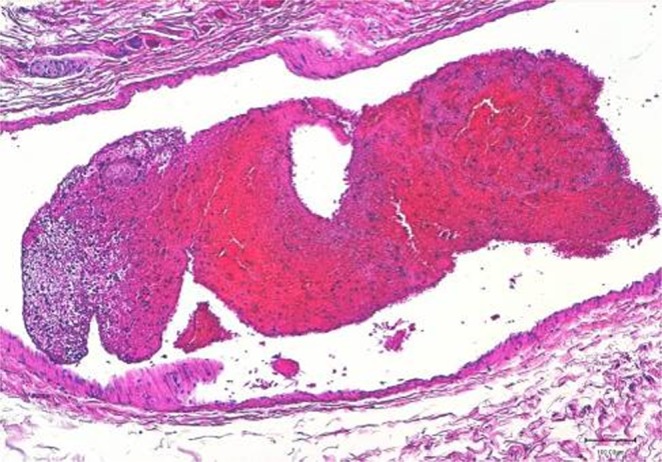
Striped dolphin (Stenella coeruleoalba). Intestinal mesentery. A large embolus is clearly shown inside the lumen of a blood vessel. 20x HE (created by K.V.).
The diagnosis of a septicaemic form of salmonellosis was achieved by the isolation of Salmonella 1,4,[5],12:i:- from samples of brain, lung, prescapular and tracheo-bronchial lymph nodes as well as from liver, spleen, kidney and blood, by standard aerobic bacterial culture, and from feces (intestine) and liver using a a specific bacteriological approach based on selective broth enrichment and subculture onto selective solid media (Table 2).
The antimicrobial susceptibility results, corresponding to a multidrug resistant phenotype (ASSuT isolate, Ampicillin, Streptomycin, Sulphonamide and Tetracycline resistant), are reported in Table 3.
The MLVA profile associated to this isolate was 3-16-8-n.a.-0211 (Table 4).
In silico MLST showed that Salmonella 1,4,[5],12:i:- isolate belonged to ST34 (Table 4); the results of KmerResistance 2.2 are listed in Table 4 and the results of SPIFinder 1.0 “Pathogenic Island” are listed in Table 5.
No other significant bacteria were isolated, including Listeria spp. and Brucella spp.
No biomolecular evidence of Brucella spp. was found, with no anti-Brucella spp. antibodies were being detected in serum, humor acqueous or cerebrospinal fluid.
A generalized DMV infection was demonstrated, by means of PCR, in brain, lung, laryngeal tonsils, tracheo-bronchial lymph node, spleen, kidney and bladder, being subsequently confirmed through amplicon sequencing and BLAST analysis. Morbillivirus-specific antigens were additionally detected in brain by IHC. Nevertheless, no anti-Morbillivirus antibodies were detected in serum, humor aqueous or cerebrospinal fluid and virus isolation attempts failed.
Direct evidence of T. gondii infection was obtained through the observation of protozoal cysts and PCR positivity in brain and heart, together with the simultaneous occurrence of anti -T. gondii antibodies in serum (1:160) as well as in humor aqueous (1:5120), IHC evidence of T. gondii - specific antigens was also found in brain.
Furthermore, PCR investigations, followed by amplicon sequencing and BLAST analysis, yelded positive results for cetacean Alphaherpesvirus at the level of prescapular lymph node.
Among the different organochlorine (OC) tissue concentrations, hexachlorobenzene (HCB) was the compound with the lowest levels (47.94 ng/g l.w.) followed by dichlorodiphenyltrichloroethane and its metabolites (DDTs) (10074.67 ng/g l.w.) and polychlorinated biphenyls (PCBs) (41059.22 ng/g l.w.). The Extracted Organic Material percent (EOM%) was 50.66, showing a relevant depletion of blubber layer. The PCB levels were greater than the estimated toxicity threshold (17 mg/kg l.w.)21,22, confirming that the specimen experienced deleterious health effects.
Case 3
The third animal, a 204 cm (TL), adult male, was found stranded dead in Vado (SV) (44.26N, 8.45E) on 20th January 2018, in good nutritional status and in post-mortem condition code 4 (decomposed). The estimated age was 18 years. Significant gross necropsy findings included a generalized post-mortem autolysis, with loss of more than 50% of the skin, associated with a mild Phyllobotrium delphini and Monorygma grimaldi infection and 2 Pholeter gastrophilus gastric nodules.
The forestomach contained scanty material, represented by few fully digested cephalopod beaks, correlated to a non-recent meal. The cephalopod species were identified as 1 Ancitroteuthis lichtensteini (family Onychiteuthidae) and 3 Todarodes sagittatus (family Ommastrephidae), both of which are normally found in coastal and pelagic waters.
Considering the advanced stage of post-mortem autolysis, a limited panel of investigations was performed and histology and immunohistochemistry were not attempted.
The diagnosis of a generalized form of salmonellosis was achieved by the isolation of Salmonella 1,4,[5],12:i:- initially just from faeces (intestine), using a specific bacteriological approach based on selective broth enrichment and subculture onto selective solid media (Table 2). This positive finding subsequently led us to carry out supplementary investigations, in order to ascertain the potential dissemination of the infection. Considering the features of the samples available, represented by brain, lung, prescapular and pulmonary lymph nodes, liver, spleen and kidney, to enhance the diagnostic sensitivity the isolation was therefore attempted using a specific isolation protocol, based on non selective pre-enrichment broth and subculturing on semi-solid media, with positive results in all tissues tested (Table 2).
The antimicrobial susceptibility results are reported in Table 3.
The MLVA profile associated to this isolate was 3-15-8-n.a.-0211 (Table 4).
In silico MLST showed that Salmonella 1,4,[5],12:i:- isolate belonged to ST34 (Table 4); the results of KmerResistance 2.2 are listed in Table 4 and the results of SPIFinder 1.0 “Pathogenic Island” are listed in Table 5.
Neither Listeria spp. nor Brucella spp. were isolated from brain.
No biomolecular evidence of Brucella spp. was found nor anti-Brucella spp. antibodies were detected.
Furthermore, biomolecular evidence of systemic DMV infection was obtained through PCR positivity in brain, lung, prescapular and pulmonary lymph nodes, liver, spleen, kidney and bladder, being subsequently and confirmed by amplicon sequencing and BLAST analysis. Nevertheless, no anti-Morbillivirus antibodies were detected in serum, humor aqueous or cerebrospinal fluid and virus isolation attempts were all negative.
No microscopic nor IHC and biomolecular (PCR) evidence of T. gondii infection was found, with no anti-T. gondii antibodies being additionally detected in this dolphin’s blood serum, humor aqueous and cerebrospinal fluid.
Finally, PCR investigations, followed by amplicon sequencing and BLAST analysis, yelded positive results for cetacean Alphaherpesvirus in the liver and prescapular lymph node.
The tissue concentrations of HCB, DDTs and PCBs in blubber were 173.84 ng/g l.w., 19809.83 ng/g l.w. and 35757.92 ng/g l.w. respectively, and also in this case the PCB levels were greater than the toxicity threshold of 17 mg/kg l.w21,22. The EOM% was very high (94.95). In this specimen the toxicological stress was evaluated also using a theoretical model23 using DDT and PCB levels expressed in ng g-1 dry weight (d.w.): the result of Canonical Variable (CAN) was −0.514, so it did not reveal the presence of hazardous levels of OC pollutants (CAN > 0.47).
Discussion
The herein presented results clearly show that cetaceans in the Pelagos Sanctuary deal with some pathogens of anthropogenic origin (e.g. Salmonella spp., Toxoplasma gondii), which may adversely affect both their individual health and their population conservation status.
Moreover, the interaction of multiple pathogens, as represented in these cases, may have significantly impacted the course and severity of the disease conditions experienced by the herein investigated dolphins, leading to their subsequent stranding and death.
Considering the infection by a monophasic variant of Salmonella Typhimurium, the first case (Case 1) was characterized by the presence of the bacterium only on faeces and liver, the latter possibly related to bacterial migration from the intestine to the liver in a moderately decomposed carcass (Code 3). In this case, the striped dolphin was supposed to carry Salmonella asymptomatically, thus harbouring the bacterium in faeces and shedding into the surrounding environment without displaying clinical symptoms.
We hypothesize a multifactorial cause of death for this individual, involving a chronic systemic DMV infection, confirmed by molecular analysis from lung and prescapular lymph node and specific lesions in spleen, which could have seriously compromised the health status, along with an additional impairment likely caused by cerebral toxoplasmosis, which may have led in turn to the animal’s disorientation and subsequent stranding.
The second case (Case 2) was instead characterized by the isolation of Salmonella 1,4,[5],12:i:- from multiple tissues and/or biological fluids, including blood samples. Salmonella was the only primary bacterial pathogen detected, in a fresh carcass (Code 2), as the cause of a septicaemic form of infection associated with microscopic lesions (intestinal necrosis, vascular embolus in intestinal mesentery). Such severe evolution of the infection likely played a role in this animal’s stranding and death and it could be related to a multifactorial impairment of this individual. Herein, we hypothesize that a subacute systemic Morbillivirus infection, associated with Herpesvirus infection and hazardous levels of pollutants, could have impacted the severity of toxoplasmosis, and that the cerebral impairment may have led to the animal’s disorientation and behavioural changes, such as debris ingestion24. The role of the ulcers in stomach, possibly related to gastric pain following the ingestion of abnormal material as a substitute of fish25–27, must not be underestimated, as they could have acted as a door of entry for Salmonella infection, due to the ingestion of abnormal, contaminated material (Posidonia oceanica, found in coastal polluted waters) in a weakened individual, incapable of catching.
In the third case (Case 3), a diagnosis of a disseminated form of salmonellosis was obtained thanks to supplementary investigations performed after the first isolation of Salmonella from faeces (intestine). The isolation from multiple tissues like brain, lung, prescapular and pulmonary lymph nodes, spleen and kidney, in addition to the liver, cannot be related to a post-mortem migration of bacteria from the intestine to contiguous and/or more distant organs; therefore, considering the lack of information for the few ancillary tests available, a septicaemic form of infection, with a likely high rate of bacterial dissemination, cannot be ruled out. The systemic DMV infection, confirmed by molecular analysis from multiple tissues, including blood, associated with Alphaherpesvirus infection, could have debilitated and predisposed the animal to a fatal systemic infection by Salmonella 1,4,[5],12:i:-.
Considering the data retrieved from molecular characterization, the Salmonella 1,4,[5],12:i:- isolates detected in the 3 herein investigated dolphins appear to be highly related to each other. In particular, all the isolates, as shown by KmerResistance 2.2 tool (Table 4), harboured the same genes for resistance to ampicillin, streptomycin, sulphonamides, tetracycline and are identifiable as ASSut, which is described as one of the most common profiles of resistance circulating in Italy and in Europe associated to monophasic variant of Salmonella Typhimurium28. Case 1 and Case 2 share the same Salmonella Pathogenic Island (SPI) genes’ profiles, with Case 3 differing only for the absence of SPI-4 (Table 5).
Furthermore, due to the results of in silico molecular typing, all isolates belonging to the same MLST type (Sequence Type, ST) and the MLVA results show a high correlation (Table 4).
Based on the query performed on the MLVA database profiles of Italian Reference Laboratory for Salmonellosis, the MLVA profile related to Case 1 (3-13-12-n.a.-0211) was found to be associated with 38 Salmonella isolated namely of human (N = 15), swine (N = 22) and shellfish (N = 1) origin, respectively (Table 6). All profiles were epidemiologically unrelated to each other.
Table 6.
MLVA profile associated to the cases described in this work, compared to database MLVA of Italian Reference Laboratory for Salmonellosis.
| MLVA Profile Case 1 3-13-12-n.a.-0211 (no of isolates) |
MLVA Profile Case 2 3-16-8-na-0211 (no of isolates) |
MLVA Profile Case 3 3-15-8-na-0211 (no of isolates) |
|
|---|---|---|---|
| Sources | human (N = 15) | swine (N = 1) | swine (N = 3) |
| swine (N = 22) | poultry (N = 1) | ||
| shellfish (N = 1) | cat (N = 1) |
Legend – MLVA: Multiple Locus Variable number tandem repeats Analysis.
These findings suggest that such specific MLVA profile is quite common in swine and human sources; however, they also highlight the need to deeply investigate the role of seawater environment in the spread of this zoonotic pathogen.
The MLVA profiles concerning Cases 2 and 3 (3-16-8-n.a.-0211 and 3-15-8-n.a.-0211, respectively) are tightly related, differing by a unique repeat unit occurring on STTR5 locus (Short Typhimurium Tandem repeat).
The MLVA profile of Case 2 was not already present into the database, differently from that of Case 3, which was found to be associated with 5 isolated of swine (N = 3), poultry(N = 1) and cat (N = 1) origin, respectively, overall collected from 2017 to 2018 (Table 6).
Moreover, Single Nucleotide Polymorphisms (SNP) analysis confirmed a high degree of correlation between Cases 2 and 3, as shown in the SNP matrix reported in Table 7, with only 2 different SNPs; when Cases 2 and 3 were compared with Case 1, they both differed from Case 1 by 38 SNPs. The genetic differences between these isolates may indicate that Cases 2 and 3 belong to the same clonal cluster, while Case 1 can be placed on the threshold, identifying a different clonal cluster, even if a threshold SNP number for Salmonella might not be identifiable29 and, to our knowledge, has not been defined yet for Salmonella 1,4,[5],12:i:-.
Table 7.
SNP matrix inferred by the mean of CSI Phylogeny 1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) to represent the phylogenetic relationships between strains.
| Case 1 | Case 2 | Case 3 | |
|---|---|---|---|
| Case 1 | 0 | 38 | 38 |
| Case 2 | 38 | 0 | 2 |
| Case 3 | 38 | 2 | 0 |
| min: 2 max: 38 |
Abbreviations: SNP, Single Nucleotide Polymorphism.
Although the source of infection by this serovar to the dolphins remains unknown, different considerations and hypotheses can be drawn about the Salmonella contamination of Pelagos Sanctuary marine waters and the transmission pathways for these striped dolphins.
Salmonella spp. is a marker for faecal contamination. The waters of highly populated coastal areas, like those of the Pelagos Sanctuary, receive large quantities of wastewater discharged from human, animal and industrial sources, treated and sometimes untreated.
Based on available data of the Regional Reference Laboratory for Salmonella typing of the Istituto Zooprofilattico (CeRTIS), retrospectively investigated, the circulation of S.1,4,[5],12:i:- at human, animal and environmental levels has increased in recent years in the Liguria Region as well as in whole Italy. Indeed, this serovar was isolated from 32 patients suffering from salmonellosis in all provinces of Liguria, in 2016, 2017 and January 2018, as well as from 6 wild boars sampled in the Genoa province, in 2014 and 2015, and from one environmental sample, in the Genoa province, at the beginning of February 2018. No positivity was found for for Salmonella spp. in seafood (fish, mussels, edible sea molluscs) submitted to official controls carried out on the coastline from 2015.
In the area surrounding the Ligurian coastal zone there is not a significant livestock production, but a consistent presence of pig farms characterizes Corsica as well as Tuscany and Northern Sardinia, the coasts of which are encompassed in the Pelagos Sanctuary. Therefore, along with human sewage, untreated wastewater discharged from pig farms might also represent a potential source of Salmonella in the concerned area.
In addition, considering the significant presence of wild boars in Liguria, Tuscany and Corsica, the risk of bacterial shedding through their faeces should not be underestimated.
Moreover, rivers, rainfall and extreme weather events may transfer enteric pathogens from distant sources to coastal waters8,30. Noteworthy, the contamination of Pelagos Sanctuary marine waters by bacteria originating from human and/or animal wastes could have been potentially increased by the severe flooding repeatedly occurred on the Ligurian and Tuscany coasts in recent years (Genova 2014, Nice 2016, Livorno 2017).
As the Mediterranean basin represents one of the busiest navigation crossroads and top tourist destinations in the world, an additional potential source of bacterial contamination in the Pelagos Sanctuary could be represented by wastewaters discharged from ships and boats31. The presence of marine birds represents an additional potential reservoir of infection, and in this regard the last tract of the Magra River, in Liguria, characterized by watersheds highly populated by water birds, should be also taken into account as a potential contamination source.
Additionally, focusing on the eating habits of Stenella coeruleoalba, the transmission of Salmonella infection could be derived from the consumption of infected or contaminated marine organisms like cephalopods or filter-feeders clupeids.
Furthermore, keeping in mind the typical respiratory patterns in cetaceans, the aerolized sea surface microlayer (SML), laden with microorganisms and contaminants, could have been inhaled deep into the tracheobronchial tree during porpoising, being subsequently deposited into alveolar spaces with more forceful exhalation and inhalation processes32.
Analysing more in depth the features of the striped dolphin stranded in 2017 (Case 2), another hypothesis could be represented by direct ingestion of contaminated water while trying to eat something or by assumption of contaminated seagrass, which does not represent a common food source for any cetacean.
The potential impact that Salmonella could have on offshore, resident, or transient populations is unknown and deserves further considerations. In order to better define the role that Salmonella plays in causing disease in cetaceans it would be appropriate to test all marine mammals found stranded for this pathogen and perform antimicrobial sensitivity tests on all Salmonella isolates. The detection of multiresistant isolates, as shown in these cases, has important implications, confirming the seepage of land-based antimicrobial compounds and/or antibiotic resistant microorganisms into the marine environment, coupled with the bioaccumulation, and possible biomagnification, into the highest marine trophic levels32,33.
The long survival of Salmonella spp in seawater, up to 17 months9, supports even more the health risks for susceptible marine animals and humans.
In conclusion, these results highlight the role of cetaceans as sentinel species for zoonotic and terrestrial pathogens in the marine environment and suggest a high level of seawater contamination in the North Western Italian coast, which may adversely impact both cetacean species and public health.
Moreover, the herein reported findings represent an additional, relevant matter of concern for the zoonotic importance of the Salmonella serovar isolated from the dolphins under investigation, which is considered as the emerging one in humans in Europe34.
The potential risk of Salmonella isolates circulating in the marine environment to public health should not be discounted9, considering the bacterial release from both carriers and infected marine mammals, along with the spread of multiresistant isolates and the medical implications for humans sharing the same habitat or working with marine mammals32.
Our observations indicate that Salmonella 1,4,[5],12:i:- is characterized by a potential pathogenic role in striped dolphins and, consequently, it should be added to the list of pathogenic bacteria causing generalized infections in cetaceans.
Moreover, these results suggest cetaceans as novel potential reservoir for one of most important Salmonella serovars.
In order to characterize the possible transmission routes, further research is needed to determine if this zoonotic pathogen poses a significant risk to cetaceans, by investigations on the infections’s prevalence as well as on its pathogenicity and epidemiology and the comparison of isolates from wild birds, wild mammals, livestock, agricultural sources and clinical samples, using not only Pulsed-field gel electrophoresis (PFGE) and MLVA35 but also Whole Genome Sequencing typing (WGS). Specific investigations for Salmonella spp. on surface river and coastal waters would be also an appropriate tool to assess the levels of contamination in the aquatic environments under consideration, while the reduction of run-offs, erosion and urban pollution in coastal areas, by an ad hoc surveillance on public waterways, would be an appropriate additional measure, preventing marine environmental contamination by Salmonella spp. as well as by other oro-faecally transmitted microorganisms.
Materials and Methods
Materials
The carcasses of three striped dolphins, stranded along the Ligurian Sea coast between 2015 and the beginning of 2018 (January-March), were submitted for necropsy to Istituto Zooprofilattico Sperimentale, Diagnostic Laboratory of Imperia, where a complete examination was performed according to standard protocols36.
The animals’ age was estimated by counting dentine growth layers (GLGs) on longitudinal section of teeth37. Three mandibular teeth collected from each specimen were mounted in epoxy resin and cut along the sagittal plane in several sections (0,3 mm thick), using a Low Spread Saw endowed with a diamond blade. The total number of GLGs in each of the tooth sections was determined in three separate sessions by three independent readers.
Gastric contents were collected during necropsies and preserved frozen for later analysis. They were then weighed, filtered and sorted, and the species were identified to the lowest taxonomic level possible. The foreign bodies were also weighed and sorted.
During necropsy, the tissue samples of all the major organs and lesions were collected and split into aliquots for subsequent analyses: one was kept frozen at −20 °C for microbiological and toxicological investigations, one at −80 °C for biomolecular and virological analyses, and the other was preserved in 10% buffered formalin for histological and immunohistochemical (IHC) investigations. Blood serum, aqueous humour and cerebrospinal fluid (CFS) were collected, when available, and kept frozen at −20 °C for serological investigations.
Microbiology
Tissue samples including brain, lung, prescapular and tracheobronchial lymph nodes, liver, spleen and kidney (Cases 1 and 2) and blood (Case 2) were processed for standard aerobic, anaerobic and microaerobic (5% CO2) bacterial culture and identification, by biochemical and/or molecular analyses. Following international recommendations38, samples from target tissues underwent specific bacteriological procedures to screen Listeria spp. and Brucella spp. (Case 1 and 2). Samples of brain (Case 3), recovered retrospectively, underwent the same specific bacteriological approach.
Salmonella isolation, identification and typing
Samples of liver and faeces (intestine) (Case 1 and 2), and of faeces (intestine) (Case 3) underwent specific bacteriological procedures to screen Salmonella spp. by means of an enrichment step in selective liquid media (Selenyte Cystine broth, Microbiol, Cagliari; MKTTn broth, Liofilchem, Roseto degli Abruzzi, TE) for 24 h at 37 °C, followed by subculture on solid media (BGA and XLD agar, Oxoid, Rodano, MI) for 24 hr at 37 °C38.
Samples of brain, lung, prescapular and pulmonary lymph nodes, liver, spleen and kidney (Case 3), recovered retrospectively, underwent specific bacteriological procedures for Salmonella spp. in order to enhance the diagnostic sensitivity, by means of a pre-enrichment step in Buffered Peptone Water (BPW, Oxoid, Rodano, MI), followed by subculture on semisolid medium (MSRV agar, Oxoid, Rodano, MI) for 48 hrs at 42 °C38.
Salmonella spp. were identified based on colony morphology on selective media and biochemical properties, according to ISO 6579-1:201739.
Serotyping was carried out according to the Kauffman-White scheme40.
Salmonella antimicrobial susceptibility testing
The Kirby–Bauer disc diffusion test was performed using Mueller-Hinton agar plates (Microbiol, Uta (CA), Italy) using the following antimicrobials and concentrations (μg):
ampicillin (A, 10), amoxicillin/clavulanic acid 2:1 (AMC, 30), chloramphenicol (C, 30), ceftazidime (CAZ, 10), cyprofloxacin (CIP, 5), cefotaxime (CTX, 5), gentamicin (G, 10), kanamycin (K, 30), cefalotin (KF, 30), nalidixic acid (NAL, 30), streptomycin (S, 10), trimethoprim–sulfamethoxazole (SXT, 1.25/23.75), and tetracycline (T, 30). Data were interpreted using CLSI guidelines (M100-S25, 2015).
Each bacterial isolate was classified as susceptible, intermediate, or resistant, depending on the growth inhibition diameter.
Salmonella sequencing and Whole Genome Sequencing (WGS) typing
WGS of the DNA extracted from the Salmonella 1,4,[5],12:i:- isolates was performed on the MiSeq platform (Illumina, San Diego, United States) using paired-end libraries which were prepared by following the Nextera™ DNA Flex Library Prep Kit (Illumina, San Diego, United States), with 150-bp read length. The reads were first subjected to the Galaxy tool “FastQC Read Quality reports”, accessed via the Galaxy public server at https://usegalaxy.org 41, to provide the quality control checks on raw sequence data and then the reads were trimmed using the Galaxy tool Trimmomatic42. We used Unicycler (ver. 0.4.1.1) via Galaxy to assemble genomes43. The assembled genomes were processed to determinate the multilocus sequence typing (MLST) in silico with MLST 1.8 (accessed via https://cge.cbs.dtu.dk/services/MLST)44, to identify antimicrobial resistance genes with KmerResistance 2.2 (accessed via https://cge.cbs.dtu.dk/services/KmerResistance/)45, thereafter being investigated for Salmonella Pathogenicity Islands with SPIFinder 1.0 (accessed via https://cge.cbs.dtu.dk/services/SPIFinder/). The fastq files of paired-nd reads were processed with CSI Phylogeny 1.4 (accessed via https://cge.cbs.dtu.dk/services/CSIPhylogeny/) to call and filter single nucleotide polymorphisms (SNPs) and infer phylogeny based on the concatenated alignment of the high quality SNPs46.
Multiple Locus Variable number tandem repeat Analysis (MLVA) was performed as previously described47. MLVA results were reported as a string of five numbers (STTR9–STTR5–STTR6–STTR10pl–STTR3) (STTR: Short Typhimurium Tandem Repeat), representing the Variable Number of Tandem Repeats (VNTR) at the corresponding locus, or as n.a. in cases where a PCR product was not obtained for a locus. The MLVA nomenclature suggested by Larsson et al.48 was adopted in this study. The MLVA profiles obtained were compared with MLVA database profiles of Italian Reference Laboratory for Salmonellosis (over a period from 2015 to 2018).
Histology and immunohistochemistry
Representative tissue samples from Case 1 (brain, lung, heart, liver, spleen, kidney, skeletal muscle, adrenal gland and reproductive system) and from Case 2 (brain, tonsils, lung, prescapular and tracheobronchial lymph nodes, heart, liver, spleen, pancreas, stomach, intestine, skeletal muscle, skin, kidney, urinary bladder, adrenal gland and reproductive system) were collected and fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 4 ± 2 µm, stained with haematoxylin and eosin (H&E) and examined under a light microscope.
Five different areas from the brain were sampled, including the telencephalon, diencephalon, mesencephalon, cerebellum and brainstem (Cases 1 and 2).
Immunohistochemistry (IHC) for Morbillivirus was performed on tissue sections from Case 1 (brain, lung, spleen and kidney) and from Case 2 (brain, lung, prescapular and tracheo-bronchial lymph nodes, spleen, kidney, bladder and pancreas), using a monoclonal anti-Canine Distemper Virus (CDV) antibody (VMRD)25.
Toxoplasma gondii IHC was carried out on samples from Cases 1 and 2 (the five aforementioned brain areas, along with the H&E-stained sections of all tissues in which coccidian parasites were identified by histopathology and/or biomolecular tests)25, using a polyclonal serum of caprine origin (VMRD).
Considering the advanced post-mortem autolysis, tissue samples from Case 3 were not investigated.
PCR, sequence analysis, virological investigations and serology
Molecular detection of Dolphin Morbillivirus (DMV), T. gondii, Herpesvirus and Brucella spp. were routinely achieved on target tissues availables from Cases 1, 2 and 3, identified, respectively, in brain, lung, tonsils, lymph nodes, liver, spleen, kidney, bladder and blood for DMV, in brain, lymph nodes, liver, spleen, heart and muscle for Toxoplasma, in brain, lung, lymph nodes, spleen, kidney for Herpesvirus (integrated by liver for Case 3), and brain, lung, tonsils, lymph nodes, liver, spleen, kidney and blood for Brucella spp26,27,49,50.
For DMV and Herpesvirus assays, amplicons were directly sequenced using PCR primers on a 3130XL Genetic Analyzer (Thermo Fisher).Sequences were aligned using the SeqMan software (Lasergene package. DNASTAR Inc.) to obtain a consensus sequence and BLAST analysis was performed.
Virus isolation was carried out for Morbillivirus and Herpesvirus from PCR-positive frozen tissues, using, respectively, confluent monolayers of Vero Dog Slam cells51 and MDBK cells (Madin-Darby Bovine Kidney). All virus isolation attempts were unsuccessful.
The presence of anti-T. gondii, anti-Brucella spp. and anti-Morbillivirus antibodies was investigated in blood serum from Case 1 and in blood serum, humor aqueous and cerebrospinal fluid (CSF) from Case 2 and 325,52,53. Specifically, anti-Brucella spp. antibodies were detected by rapid serum agglutination (S.A.R.), utilizing both B. abortus and B. melitensis as antigens, whereas anti-T. gondii antibodies were detected by an indirect immunofluorescence test (IFAT) and anti-Morbillivirus antibodies by virus neutralization assay (serum neutralization, SN), using the Onderstepoort strain of CDV25.
Toxicology
Hexachlorobenzene (HCB), dichlorodiphenyltrichloroethane (DDT) and its metabolites (dichlorodiphenyldichloroethane (DDD) and dichlorodiphenyldichloroethylene (DDE)) and polychlorinated biphenyls (PCBs), as well as Extracted Organic Material percent (EOM%), were measured in blubber (Cases 2 and 3), according to the Environmental Protection Agency method 8081/8082, with revisions54. The OC levels are expressed both in ng/g dry weight and in ng/g lipid weight basis (l.w.) since variation in lipid content among organisms can affect contaminant concentrations.
Toxicological stress was evaluated using both a theoretical model23 and the toxicity threshold value set by Jepson et al.21 and Kannan et al.22. For Case 2 blubber residue levels were measured only on a lipid basis since this female specimen was experiencing a marked starvation, leading to an intense mobilization of her subcutaneous fat reserves. For this reason, the Extracted Organic Material (EOM) percentage was very similar in all the tissues and very low in the blubber (EOM% = 50.66), proving this metabolic imbalance.
Accession codes
The Salmonella whole-genome sequences assembled have been deposited at DDBJ/ENA/GenBank under the accession no. QIBF00000000, QIBE00000000 and QIBD00000000; the version described in this paper is version 0.1 for all genomes.
Acknowledgements
The Authors are grateful to Dr. C.E. Di Francesco for her valuable support in serological analyses.
Author Contributions
C.G., F.G. and W.M. performed the necropsies of the three study cases. F.Ga. performed analyses to determine the life-history of the three cases. L.S., A.D. and S.Z. performed complementary analyses for microbiology and Salmonella isolation. S.G., A.R. and A.A.L. performed Salmonella identification and sequencing. M.G., S.P., L.M. and G.D.G. performed virological investigations. K.V., B.I., F.G., F.E.S. and C.C. did the histopathological studies of the two cases examinated. L.M. performed toxicological investigations of the two cases examinated. C.G., C.C. and A.F. conceived the study. C.G. wrote the manuscript. M.B., K.V., S.P., G.D.G., S.G., A.A.L., A.P. and L.M. provided inputs on the text. A.P. created the figures. All authors reviewed the manuscript.
Data Availability
All data generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bossart GD. Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 2011;48(3):676–90. doi: 10.1177/0300985810388525. [DOI] [PubMed] [Google Scholar]
- 2.Marsili L. Lipophilic contaminants in marine mammals: review of the results of ten years’ work at the Department of Environmental Biology, Siena University (Italy) Int. J. Environ. Pollut. 2000;13:416–452. doi: 10.1504/IJEP.2000.002329. [DOI] [Google Scholar]
- 3.Marsili, L. et al. Test tube cetaceans: from the evaluation of susceptibility to the study of genotoxic effects of different environmental contaminants using cetacean fibroblast cell cultures in New approaches to the study of marine mammals (eds Romero, A. & Keith, E. O.) 49−76 (INTECH, 2012).
- 4.Minette HP. Salmonellosis in the marine environment. A review and commentary. Int. J. Zoonoses. 1986;13(2):71–5. [PubMed] [Google Scholar]
- 5.Colegrove KM, et al. Salmonella Newport omphaloarteritis in a stranded killer whale (Orcinus orca) neonate. J. Wildl. Dis. 2010;46(4):1300–4. doi: 10.7589/0090-3558-46.4.1300. [DOI] [PubMed] [Google Scholar]
- 6.Moeller, R. B. JR. Pathology of marine mammals with special reference to infectious disease in Toxicology of Marine Mammals (eds Vos, J. G., Bossart, G. D., Fournier, M. & O’Shea, T.) 3–37 (Taylor & Francis, 2003).
- 7.Valderrama Vasquez CA, Macgregor SK, Rowcliffe JM, Jepson PD. Occurrence of a monophasic strain of Salmonella group B isolated from cetaceans in England and Wales between 1990 and 2002. Environ. Microbiol. 2008;10(9):2462–8. doi: 10.1111/j.1462-2920.2008.01651.x. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Urtaza J, et al. Influence of environmental factors and human activity on the presence of Salmonella serovars in a marine environment. Appl. Environ. Microbiol. 2004;70(4):2089–97. doi: 10.1128/AEM.70.4.2089-2097.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson, M. C., Berardi, T., Aguilar, B., Byrne, B. A. & Shapiro, K. Effects of transparent exopolymer particles and suspended particles on the survival of Salmonella enterica serovar Typhimurium in seawater. FEMS Microbiol. Ecol. 91(3), 10.1093/femsec/fiv005 (2015). [DOI] [PubMed]
- 10.Miller MA, et al. Enteric bacterial pathogen detection in southern sea otters (Enhydra lutris nereis) is associated with coastal urbanization and freshwater runoff. Vet. Res. 2010;41(1):1–13. doi: 10.1051/vetres/2009049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeita MA, Díez R, Usera MA. Distribution of Salmonella spp. serotypes isolated in Spain during a 4-year period (1993–1996) Enferm. Infect. Microbiol. Clin. 1999;17(1):9–14. [PubMed] [Google Scholar]
- 12.Hauser E, et al. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Appl. Environ. Microbiol. 2010;76(14):4601–10. doi: 10.1128/AEM.02991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins KL, et al. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro. Surveill. 2010;15:19580. [PubMed] [Google Scholar]
- 14.Laorden L, et al. Genetic evolution of the Spanish multidrug-resistant Salmonella enterica 4,5,12:i:- monophasic variant. J. Clin. Microbiol. 2010;48(12):4563–6. doi: 10.1128/JCM.00337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cito F, et al. Outbreak of unusual Salmonella enterica serovar Typhimurium monophasic variant 1,4,[5],12:i:-, Italy, June 2013 to September 2014. Euro. Surveill. 2016;21(15):30194. doi: 10.2807/1560-7917.ES.2016.21.15.30194. [DOI] [PubMed] [Google Scholar]
- 16.Arai, N. et al. Phylogenetic Characterization of Salmonella enterica Serovar Typhimurium and Its Monophasic Variant Isolated from Food Animals in Japan Revealed Replacement of Major Epidemic Clones in the Last 4 Decades. J. Clin. Microbiol. 56(5), 10.1128/JCM.01758-17 (2018). [DOI] [PMC free article] [PubMed]
- 17.Dionisi AM, et al. Molecular characterization of multidrug-resistant strains of Salmonella enterica serotype Typhimurium and Monophasic variant (S. 4,[5],12:i:-) isolated from human infections in Italy. Foodborne Pathog. Dis. 2009;6(6):711–7. doi: 10.1089/fpd.2008.0240. [DOI] [PubMed] [Google Scholar]
- 18.Istituto Superiore di Sanità (ISS). ENTERNET Salmonella. Sorveglianza delle infezioni da Salmonella. Dati Storici dal 2007 al 2009. Rome: ISS. Available from, http://www.iss.it/binary/salm/cont/4f_Dati_SALMONELLA_2007_2009.pdf (2010).
- 19.Graziani C, et al. Distribution of Salmonella enterica isolates from human cases in Italy, 1980 to 2011. Euro. Surveill. 2013;18:20519. doi: 10.2807/1560-7917.ES2013.18.7.20519. [DOI] [PubMed] [Google Scholar]
- 20.Frasson I, Bettanello S, De Canale E, Richter SN, Palù G. Serotype epidemiology and multidrug resistance patterns of Salmonella enterica infecting humans in Italy. Gut. Pathog. 2016;8:26. doi: 10.1186/s13099-016-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jepson PD, et al. Relationships between polychlorinated biphenyls and health status in harbor porpoises (Phocoena phocoena) stranded in the United Kingdom. Environ. Toxicol. Chem. 2005;24(1):238–48. doi: 10.1897/03-663.1. [DOI] [PubMed] [Google Scholar]
- 22.Kannan K, Blankenship AL, Jones PD, Giesy JP. Toxicity reference values for the toxic effects of polychlorinated biphenyls to aquatic mammals. Hum. Ecol. Risk Assess. 2000;6(1):181–20. doi: 10.1080/10807030091124491. [DOI] [Google Scholar]
- 23.Marsili L, D’Agostino A, Bucalossi D, Malatesta T, Fossi MC. Theoretical models to evaluate hazard due to organochlorine compounds (OCs) in Mediterranean striped dolphin (Stenella coeruleoalba) Chemosphere. 2004;56:791–801. doi: 10.1016/j.chemosphere.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Krzyszczyk E, et al. A report on six cases of seagrass-associated gastric impaction in bottlenose dolphins (Tursiops sp.) Mar. Mamm. Sci. 2013;29(3):548–554. doi: 10.1111/j.1748-7692.2012.00579.x. [DOI] [Google Scholar]
- 25.Di Guardo G, et al. Cerebral toxoplasmosis in striped dolphins (Stenella coeruleoalba) stranded along the Ligurian Sea coast of Italy. Vet. Pathol. 2010;47(2):245–53. doi: 10.1177/0300985809358036. [DOI] [PubMed] [Google Scholar]
- 26.Vitale M, et al. A High Sensitive Nested PCR for Toxoplasma gondii Detection in Animal and Food Samples. J. Microb. Biochem. Technol. 2013;5:039–041. doi: 10.4172/1948-5948.1000097. [DOI] [Google Scholar]
- 27.Verna F, et al. Detection of morbillivirus infection by RT-PCR RFLP analysis in cetaceans and carnivores. J. Virol. Methods. 2017;247:22–27. doi: 10.1016/j.jviromet.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Barco L, et al. Molecular characterization of Salmonella enterica serovar 4,[5],12:i:- DT193 ASSuT strains from two outbreaks in Italy. Foodborne Pathog. Dis. 2014;11(2):138–44. doi: 10.1089/fpd.2013.1626. [DOI] [PubMed] [Google Scholar]
- 29.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One. 2014;9(2):e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funari E, Manganelli M, Sinisi L. Impact of climate change on waterborne diseases. Ann Ist Super Sanita. 2012;48(4):473–487. doi: 10.4415/ANN_12_04_13. [DOI] [PubMed] [Google Scholar]
- 31.Perić T. Wastewater pollution from cruise ships in coastal sea area of the Republic of Croatia. Scientific Journal of Maritime Research. 2016;30:160–164. [Google Scholar]
- 32.Raverty SA, et al. Respiratory Microbiome of Endangered Southern Resident Killer Whales and Microbiota of Surrounding Sea Surface Microlayer in the Eastern North Pacific. Sci. Rep. 2017;7(1):394. doi: 10.1038/s41598-017-00457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer AM, Goldstein JD, Reif JS, Fair PA, Bossart GD. Antibiotic-Resistant Organisms Cultured from Atlantic Bottlenose Dolphins (Tursiops truncatus) Inhabiting Estuarine Waters of Charleston, SC and Indian River Lagoon, FL. EcoHealth. 2009;6:33–41. doi: 10.1007/s10393-009-0221-5. [DOI] [PubMed] [Google Scholar]
- 34.EFSA Scientific Opinion on monitoring and assessment of the public health risk of “Salmonella Typhimurium-like” strain. EFSA Journal. 2010;8(10):1826. doi: 10.2903/j.efsa.2010.1826. [DOI] [Google Scholar]
- 35.Baily JL, et al. Salmonella infection in grey seals (Halichoerus grypus), a marine mammal sentinel species: pathogenicity and molecular typing of Salmonella strains compared with human and livestock isolates. Environ. Microbiol. 2016;18(3):1078–87. doi: 10.1111/1462-2920.13219. [DOI] [PubMed] [Google Scholar]
- 36.Geraci, J. R. & Lounsbury, V. J. Marine mammals ashore: a field guide for strandings, 2nd edn. National Aquarium in Baltimore, Baltimore, MD (2005).
- 37.Evans, K., Kemper, C., McKenzie, J. & McIntosh, R. Age determination of marine mammals using tooth structure. Marine Mammal Ageing Facility, South Australian Museum, South Australia, publ.: 74pp (2008).
- 38.OIE (World Organisation for Animal Health). Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris (2018).
- 39.International Organisation for Standardisation, Geneva, Switzerland, 2007. EN ISO 6579-1:2017. Microbiology of food and animal feeding stuffs–Horizontal method for the detection, enumeration and serotyping of Salmonella spp.
- 40.Guibourdenche M, et al. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2010;161(1):26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wick Ryan R., Judd Louise M., Gorrie Claire L., Holt Kathryn E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Computational Biology. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larsen MV, et al. Multilocus sequence typing of total genome sequenced bacteria. J. Clin. Microbiol. 2012;50(4):1355–61. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clausen PT, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J. Antimicrob. Chemother. 2016;71(9):2484–8. doi: 10.1093/jac/dkw184. [DOI] [PubMed] [Google Scholar]
- 46.Kaas Rolf S., Leekitcharoenphon Pimlapas, Aarestrup Frank M., Lund Ole. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE. 2014;9(8):e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lettini AA, et al. Characterization of an unusual Salmonella phage type DT7a and report of a foodborne outbreak of salmonellosis. Int. J. Food. Microbiol. 2014;189:11–7. doi: 10.1016/j.ijfoodmicro.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Larsson, J. T. et al. Development of a new nomenclature for Salmonella typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill. 14(15), 10.2807/ese.14.15.19174 (2009). [PubMed]
- 49.VanDevanter DR, et al. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baily GG, Krahn JB, Drasar BS, Stoker NG. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J. Trop. Med. Hyg. 1992;95:271–275. [PubMed] [Google Scholar]
- 51.Peletto S, et al. Efficient isolation on Vero.DogSLAMtag cells and full genome characterization of Dolphin Morbillivirus (DMV) by next generation sequencing. Sci. Rep. 2018;8(1):860. doi: 10.1038/s41598-018-19269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernández-Mora G, et al. Neurobrucellosis in stranded dolphins, Costa Rica. Emerg. Infect. Dis. 2008;14:1430–1433. doi: 10.3201/eid1409.071056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Profeta F, et al. Retrospective seroepidemiological investigations against Morbillivirus, Toxoplasma gondii and Brucella spp. in cetaceans stranded along the Italian coastline (1998–2014) Res. Vet. Sci. 2015;101:89–92. doi: 10.1016/j.rvsc.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Marsili L, Focardi S. Chlorinated hydrocarbon (HCB, DDTs and PCBs) levels in cetaceans stranded along the Italian coasts: an overview. Environ. Monit. Assess. 1997;45:129–180. doi: 10.1023/A:1005786627533. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


