Abstract
Despite azithromycin being used in some countries to treat infections caused by Gram-negative pathogens, no resistance breakpoint for Escherichia coli exists. The aim of this study was to analyse the levels and mechanisms of azithromycin resistance in E. coli. The presence of chromosomal (rplD, rplV and 23S rRNA) mutations, 10 macrolide resistance genes (MRGs) and efflux pump overexpression was determined in 343 E. coli isolates. Overall, 89 (25.9%) isolates had MICs ≥ 32 mg/L to azithromycin, decreasing to 42 (12.2%) when assayed in the presence of Phe-Arg-β-Napthylamide, with 35 of these 42 possessing at least one MRG. Efflux pumps played a role in azithromycin resistance affecting the Minimal Inhibitory Concentration (MIC) levels of 91.2% isolates whereas chromosomal alterations seem to have a minimal role. At least one MRG was found in 22.7% of the isolates with mph(A) being the most commonly found gene. The mph(A) gene plays the main role in the development of azithromycin resistance and 93% of the mph(A)-carrying isolates showed a MIC of 32 mg/L. In the absence of a specific resistance breakpoint our results suggest a MIC of 32 mg/L to be considered in order to detect isolates carrying mechanisms able to confer azithromycin resistance.
Introduction
Infantile diarrhoea is a serious problem in developing countries and remains the second most common cause of death among children under five worldwide. In fact, it causes >800,000 deaths globally per year representing around 10–11% of the annual global child deaths1,2. Escherichia coli play a relevant role in the death of children by diarrhoea, being involved in more than 120,000 deaths annually of children under 5 years old1.
The treatment approach to diarrhoea often does not require the use of antibacterial agents being frequently limited to the replacement of lost liquids and salts by means of Oral Rehydration Salts solutions in order to fight the dehydration risks3. However, according to the patient’s nutritional status, the presence of comorbidities, the specific pathogen, illness severity and symptom duration, the use of antimicrobial agents may be required. Ampicillin and cotrimoxazole are the usual first line treatments in most low and middle-income countries4,5. Unfortunately, antimicrobial resistance has increased over time, and in different areas these antimicrobial agents are losing their usefulness as a treatment of diarrhoea4–7. Since antibiotic resistance is a severe health problem worldwide which can lead to inefficiency of antimicrobial agents and therapeutic failure8, surveillance of the development of antimicrobial resistance should be performed, establishing molecular mechanisms of resistance to thereby design alternative treatments.
Azithromycin and other macrolides have been largely used to treat Gram-positive infections and also possess good activity against different Gram-negative microorganisms, such as Bartonella spp., Campylobacter spp., Haemophilus influenzae, or Neisseria gonorrhoeae9,10. Classically, macrolides present low levels of activity against Enterobacteriaceae which have been related to the poor membrane penetration of these antimicrobial agents, preventing their use to treat Enterobacteriaceae9. Nonetheless, in comparison with other macrolides, azithromycin has a higher basic character9. Thus, while low permeability prevents the action of most of macrolide agents against Enterobacteriaceae9, this basic character confers to azithromycin a true role in the treatment of diarrhoeal infections related to different Enterobacteriaceae11,12. Thus, azithromycin is a promising alternative because of its excellent activity against most common diarrhoeagenic pathogens such as diarrhoeagenic E. coli, Shigella spp., Salmonella spp. or Campylobacter spp.9,10, and has been included in the considered armamentarium to fight against specific Enterobacteriaceae13,14.
Nonetheless, despite ranking amongst the most frequent etiological causes of diarrhoea15,16, and the association of some specific diarrhoeagenic pathotypes with high levels of children mortality16, at present no clinical breakpoint for resistance in E. coli has been established. However, a Minimal Inhibitory Concentration (MIC) ≥ 32 mg/L or a halo diameter ≤ 12 mm have been proposed as the azithromycin resistance breakpoints in some Enterobacteriaceae17,18. Furthermore, a series of questions on the use of azithromycin in the treatment of diarrhoeagenic Enterobacteriaceae remain to be fully answered. These include questions such as specific azithromycin resistance rates, azithromycin resistance mechanisms in circulation, as well as a more relevant question, such as the effect of different alterations on the final azithromycin MIC.
Chromosomal efflux pumps are bacterial systems involved in the extrusion of molecules from bacteria to the environment, including bacterial products such as siderophores as well as toxics and antibiotics19. In this line chromosomal efflux pumps are involved in intrinsic and acquired azithromycin resistance9,20. Additionally, target amino acid substitutions in the L4 (rplD) and L22 (rplV) ribosomal proteins and in 23S rRNA (rrlH) have also been involved in macrolide resistance9.
Nonetheless, the most relevant mechanisms of azithromycin resistance in Enterobacteriaceae are those encoded in mobile elements9. Different Macrolide Resistance Genes (MRGs) have been described, leading to resistance through different pathways such as target modifications produced by rRNA methylases encoded in erm genes or macrolides-inactivation, mediated by esterases such as those encoded by ere(A) or ere(B) genes or by phosphorylases such as those encoded in the mph(A) and mph(B) genes. Additionally, transferable genes such as msr(A), mef(A) or mef(B) have been reported to encode macrolide-efflux pumps9.
This study aimed to evaluate the levels and the mechanisms of resistance to azithromycin in a collection of samples of E. coli from children with and without diarrhoea. In the absence of a specific azithromycin breakpoint for E. coli, we analyse the relationship between specific mechanisms of resistance and MIC levels.
Results
Antibiotic susceptibility levels
The MICs of azithromycin ranged between 0.06 mg/L and >256 mg/L, with a MIC50 of 8 mg/L and MIC90 of 128 mg/L (Table 1).
Table 1.
Analysis of azithromycin resistance by E. coli categories.
| PAβN | Com. (84) | Diarrhoeagenic E. coli (259) | ||||||
|---|---|---|---|---|---|---|---|---|
| EPEC (120) | ETEC (41) | EAEC (78) | DAEC (20) | Total DEC | Overall (343) | |||
| MIC Range | N | 2–>256 | 0.06–>256 | 2–256 | 2–>256 | 1–>256 | 0.06–>256 | 0.06–>256 |
| Y | 0.06–>256 | 0.06–256 | 0.25–64 | 0.5–>256 | 0.25–128 | 0.06–>256 | 0.06–>256 | |
| MIC50 | N | 16 | 8 | 4 | 16 | 16 | 8 | 8 |
| Y | 2 | 1 | 1 | 2 | 4 | 1 | 1 | |
| MIC90 | N | 128 | 16 | 64 | >256 | 128 | 128 | 128 |
| Y | 32 | 2 | 4 | 64 | 32 | 32 | 32 | |
| R (No./%) | N | 23 (27.4) | 12 (10.0) | 7 (17.1) | 38 (48.7)a | 9 (45.0)b | 66 (25.5) | 89 (36.6) |
| Y | 13 (15.5) | 4 (3.3) | 1 (2.4) | 21 (26.9) | 3 (15.0) | 29 (10.8) | 42 (12.2) | |
| P | 0.0897 | 0.0671 | 0.0571 | 0.0080 | 0.0824 | <0.0001 | <0.0001 | |
PAβN: Phe-Arg-β-Napthylamide; Com: Commensal, EPEC: Enteropathogenic; ETEC: Enterotoxigenic; EAEC: Enteroaggregative; DAEC: Diffussely Adherent, DEC: Diarrhoeagenic; MIC: Minimal Inhibitory Concentration (expressed in mg/L); R: Resistance (considering MIC ≥ 32 mg/L); N: Without PAβN; Y: With PAβN; P. Differences between resistance levels in the absence and presence of PAβN (highlighted in bold the significant differences found).
aEAEC isolates were significantly more resistant than commensal (P: 0.006), EPEC (P < 0.0001) and ETEC isolates (P = 0.0007).
bDAEC isolates were significantly more resistant than EPEC (P = 0.0004) and ETEC (P = 0.0302).
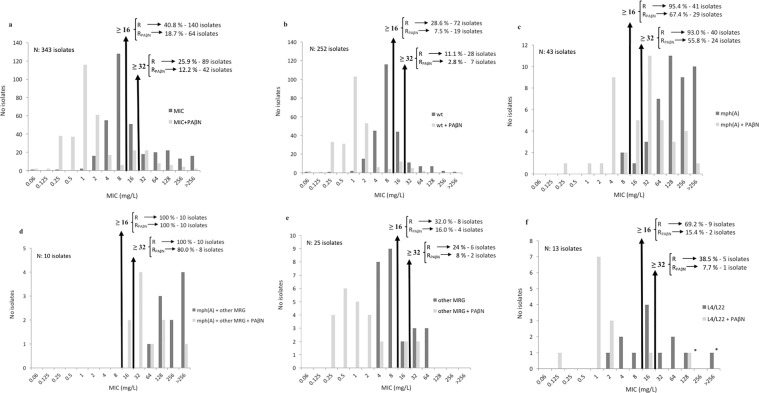
Overall, 140 (40.8%) and 89 isolates (25.9%) had a MIC ≥ 16 and ≥32 mg/L respectively, while only 18.7% and 11.9% (P < 0.0001 in both cases) remained with a MIC ≥ 16 and ≥ 32 mg/L respectively when Phe-Arg-β-Napthylamide (PAβN) was added (Table 1, Figs 1, 2 and 3). When the analysis was made comparing diarrhoeagenic and commensal E. coli no differences were observed. Nonetheless, when analysing the isolates by pathotypes the levels of resistance of enteroaggregative (EAEC) (48.7%) and diffuse-adhering (DAEC) (45%) were significantly higher than those of enterotoxigenic (ETEC) (17.1%) and enteropathogenic (EPEC) (10%). Moreover, the resistance levels of EAEC isolates were also significantly higher than those of commensal isolates (P = 0.0060) (Table 1).
Figure 1.
Analysis of Minimal Inhibitory Concentration (MIC) of 16 and 32 mg/L to detect the presence of specific macrolide-resistance mechanisms. R: Resistance; RPAβN: resistance in presence of PAβN. (a) Overall. (b) Isolates in which no sought mechanisms of resistance was found. (c) Isolates carrying the mph(A) gene alone or with a target mutation. (d) Isolates carrying the mph(A) gene together other MRG. (e) Isolates carrying a MRG different that mph(A). (f) Isolates carrying only L4 and/or L22 amino acid changes. *The single isolate (isolate 3491) which remains resistant after PAβN addition possesses an unidentified MRG.
Figure 2.
Minimal Inhibitory Concentration (MIC) distribution. MRG: Macrolide resistance gene (other than mph(A)); wt: wild type. Any MIC category with ≥5% of the isolates is highlighted in dark grey. If a strain had a L4 and/or L22 mutation(s) and a MRG, then the isolates are included in either the mph(A) or MRG category. 1One isolate (isolate 3491) in which an unidentified MRG was detected by conjugation.
Figure 3.
Minimal Inhibitory Concentration (MIC) cumulative curves (a) MIC cumulative curve in standard clinical conditions (MIC evaluation in absence of PAβN). (b) MIC cumulative curve in presence of 20 mg/L of PAβN. Horizontal lines marks the 50 and 90% of isolates inhibition.
In the presence of PAβN all groups showed decreased levels of resistance, which was significant (P = 0.0080) amongst EAEC isolates (Table 1).
Effect of PAβN
In all cases the isolates were able to grow in the presence of PAβN. As mentioned above the addition of PAβN affected the azithromycin susceptibility levels (Tables 1, 2 and 3, Figs 1, 2 and 3). Overall, when the MIC was established in the presence of PAβN (MICPAβN) the effect of PAβN on the MIC levels was observed in 91.2% of the isolates, independently of the initial MIC (MICI) of azithromycin, with 256 being the maximum MICI/MICPAβN quotient (from MICI of 64 mg/L to MICPAβN of 0.25 mg/L) (Table 2). In 47 out of 89 (52.8%) azithromycin-resistant isolates, the addition of PAβN resulted in a MIC within the range of susceptibility (Table 1, Fig. 1). On the other hand, 35 out of these 47 isolates (74.5%) possessed at least 1 MRG (unidentified in one case - see conjugation results below).
Table 2.
Analysis of MICI/MICPAβN quotient.
| E. coli | MICI/MICPAβN (N/%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MICI/MICPAβN ≤ 2 | MICI/MICPAβN > 2 | ND* | ||||||||
| 0 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | ||
| Com (84) | 2 | 24 | 29 | 16 | 7 | 2 | 1 | 3 | ||
| DEC (259) | 5 (1.9) | 17 (6.6) | 60 (23.2) | 103 (39.8) | 34 (13.1) | 24 (9.3) | 2 (0.8) | 1 | 13 (5.0) | |
| EPEC (120) | 3 | 2 | 22 | 59 | 14 | 17 | 1 | 2 | ||
| ETEC (41) | 3 | 9 | 16 | 9 | 2 | 1 | 1 | |||
| EAEC (78) | 2 | 9 | 23 | 23 | 7 | 5 | 9 | |||
| DAEC (20) | 3 | 6 | 5 | 4 | 2 | |||||
| Overall (343) | 5 | 19 | 84 | 132 | 50 | 31 | 4 | 1 | 1 | 16 |
MICI/MICPAβN = 12 (mean effect).
Com: Commensal; DEC: Diarrhoeagenic; EPEC: Enteropathogenic; ETEC: Enterotoxigenic; EAEC: Enteroaggregative; DAEC: Diffussely Adherent; PAβN: Phe-Arg-β-Napthylamide; MIC: Minimal Inhibitory Concentration (expressed in mg/L); MICI: MIC determined in the absence of PAβN; MICPAβN: MIC determined in the presence of PAβN.
ND: MICI > 256 mg/L. Note that in 10 out of these 16 cases the MICPAβN was <256 mg/L, and therefore the MICI/MICPAβN quotient was >2 (e.g.: the quotient >256/128 is at least ≥4), meaning that addition of PAβN affected the final MIC levels. In the remaining 6 cases the possible effect of PAβN was not evaluated because we only were able to determine that the MICI/MICPAβN quotient was at least 2 (i.e.: the quotient >256/256 is at least ≥2, but not necessarily >2).
Table 3.
Analysis of azithromycin resistance in the presence and absence of macrolide resistance genes.
| PAβN | Macrolide Resistant Genes (MRGs) | ||||
|---|---|---|---|---|---|
| Absence (265) | Presence | ||||
| 1 MRG (68) | 2 MRGs | ||||
| mph(A) (43) | Other (25) | Overall (10)a | |||
| MIC Range | N | 0.06–>256 | 8–>256 | 4–64 | 64–>256 |
| Y | 0.06–128 | 0.25–>256 | 0.25–32 | 16–>256 | |
| MIC50 | N | 8 | 128 | 8 | 256 |
| Y | 1 | 32 | 1 | 32 | |
| MIC90 | N | 32 | >256 | 64 | >256 |
| Y | 4 | 256 | 16 | 128 | |
| R (N/%) | N | 81/30.6% | 41/95.4%a,b | 8/32% | 10/100% |
| Y | 21/7.9% | 29/67.4% | 4/16% | 10/100% | |
| P | <0.0001 | 0.003 | 0.3209 | 1.000 | |
PAβN: Phe-Arg-β-Naphtylamyde; MRG: Macrolide resistance gene; N: Absence of PAβN; Y: Presence of PAβN. MIC: Minimal Inhibitory Concentration (expressed in mg/L). R: Azithromycin resistance (considering MIC ≥ 32 mg/L). P: Differences in azithromycin resistance levels related to the absence or presence of PAβN, being significant differences highlighted in bold.
aIn all cases the mph(A) gene was present together with: erm(A) − 4 cases; erm(B) − 3 cases; mef(A) − 2 cases; ere(A) − 1 case.
bThe isolates presenting the mph(A) were significantly more resistant than those without MRG or presenting other MRGs (P < 0.0001).
Two commensal and 4 diarrhoeagenic isolates presented a MICI > 256 mg/L and a MICPAβN ≥ 256 mg/L, thereby not allowing the effect of PAβN to be accurately established.
As a general rule the MICI/MICPAβN quotient ranged from 4 to 16 (267 isolates, 77.8% of total isolates). The MICI/MICPAβN mode was 8 (overall, and among commensal and diarrhoeagenic groups), while the mean effect was 12 (Table 2). When the diarrhoeagenic group was subdivided into pathotypes, only DAEC and EAEC showed slight differences (Tables 1 and 2).
Analysing the effect of PAβN in 255 diarrhoeagenic and 82 commensal isolates, a non-significant trend of a higher number of affected commensal isolates was observed (P = 0.0810). Thus, the effect of PAβN was not observed in 8.6% and 2.4% diarrheogenic and commensal isolates respectively. Despite the significant effect of PAβN on the MIC of EAEC isolates, 11 (14.7%) were not affected by PAβN. Interestingly, 10 out of these 11 isolates presented MICI of 64–32 mg/L and MICPAβN of 32–16 mg/L, with MRGs being detected in only 2 cases. In addition, 3 DAEC isolates (15%) were also not affected by PAβN presenting borderline significant differences with commensal isolates.
Target mutations
Only 17 out of 263 isolates analysed (6.5%) presented mutations in the rplD or rplV genes. Thus, 6 isolates had mutations in the rplD gene and 7 in the rplV gene, while 4 isolates presented amino acid codon alterations concomitantly in both genes. Thirteen of these had a MICI ≥ 32 mg/L (including 3 presenting mutations in both of the targets analysed), but only one (isolate 3491), in which an unidentified MRG was detected by conjugation (see below), remained resistant when the MICPAβN was established. In 4 cases were detected concomitant MRGs (Table 4). None of the isolates analysed had mutations in the 23S rRNA gene.
Table 4.
L4 (rplD) and L22 (rplV) amino acid substitutions.
| E. coli | L4 | L22 | MRG | MIC ± PAβN | |
|---|---|---|---|---|---|
| N | Y | ||||
| Commensal | V52I | I4L + L6Q + T72A | — | 16 | 0.125 |
| Commensal | A37S + V52L | wt | — | 16 | 1 |
| Commensal | wt | I4L + K6Q + T72A | — | 16 | 1 |
| Commensal | V52I + D91E + T173N | wt | — | 16 | 2 |
| Commensal | wt | S101T + I103L | — | 64 | 2 |
| DAEC | wt | K83N + D94H + K98N | mph(A) | 64 | 4 |
| EAEC | wt | V17I | — | 2 | 1 |
| EAEC | A37T + K74T | wt | — | 4 | 1 |
| EAEC | V120I | wt | — | 4 | 1 |
| EAEC | wt | L46Q | mph(A) | 64 | 16 |
| EAEC | K123S | I4L + K6Q | —a | >256 | 128 |
| EPEC | A190V | wt | msr(A) | 8 | 0.5 |
| EPEC | D154E | wt | — | 8 | 1 |
| EPEC | V52I + T173N | I4L + K6Q + T72A | mph(B) | 16 | 4 |
| EPEC | K123S | I4L + K6Q + T72A | — | 32 | 1 |
| ETEC | wt | L46Q | — | 64 | 16 |
| ETEC | wt | L46Q | — | 128 | 2 |
PAβN: Phe-Arg-β-Naphtylamyde; MRG: Macrolide resistance gene; wt: wild type. N: Absence of PAβN; Y: Presence of PAβN.
aA non-identified conjugative mechanism of resistance was detected (isolate 3491).
Macrolide resistance genes
Seventy-eight isolates (22.7%) possessed at least one MRG (Table 5). The MRG most frequently found was mph(A), which was present in 53 isolates (67.9% of isolates possessing MRG) belonging to all the groups analysed. In 43 cases no other MRG was detected, while in the remaining 10 cases mph(A) was detected together with the erm(A) gene in 4 cases, the erm(B) gene in 3 cases and the mef(A) and ere(A) gene in 2 and 1 cases, respectively. When more than one MRG was identified within the same isolate the mph(A) gene was always present.
Table 5.
Macrolide resistance genes.
| E. coli | N | Phosphotransferases | Methylases | Esterases | Efflux Pumps | Overall | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | Genes | |||||||||||||
| mph(A) | mph(B) | erm(A) | erm(B) | erm(C) | ere(A) | mef(A) | mef(B) | msr(A) | msr(D) | N | % | N | ||
| EAEC | 78 | 21 | 0 | 5a | 3b | 1 | 3b | 3c | 0 | 0 | 1 | 29d | 39.8 | 37 |
| EPEC | 120 | 6 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 2 | 13 | 10.8 | 13 |
| ETEC | 41 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 6 | 14.6 | 6 |
| DAEC | 20 | 10 | 0 | 0 | 1b | 0 | 0 | 0 | 0 | 0 | 0 | 10e | 50.0 | 11 |
| DEC | 259 | 39 | 1 | 5 | 4 | 4 | 5 | 3 | 1 | 2 | 3 | 58 | 23.2 | 67 |
| Comm. | 84 | 14 | 0 | 1 | 2b | 0 | 2 | 0 | 1 | 1 | 0 | 20f | 23.8 | 21 |
| Overall | 343 | 53 | 1 | 6 | 6 | 4 | 7 | 3 | 2 | 3 | 3 | 78 | 23.3 | 89 |
EAEC: Enteroaggregative; EPEC: Enteropathogenic; ETEC: Enterotoxigenic; DAEC: Diffussely Adherent; DEC: Diarrhoeagenic; Com: Commensal.
a4 of them concomitantly with mph(A); b1 of them concomitantly with mph(A); c2 of them concomitantly with mph(A).
dOverall the EAEC isolates possess more MRGs than EPEC (P < 0.0001) and ETEC (P = 0.0113).
eOverall the DAEC isolates possess more MRGs than EPEC (P < 0.0001), ETEC (P = 0.0053) and commensal (P = 0.0283).
fOverall the commensal isolates possess more MRGs than EPEC (P = 0.019).
MRG were significantly more frequent among EAEC and DAEC isolates than among the remaining groups analysed, except when EAEC were compared with commensals. In addition, significant differences were also observed in the presence of MRGs among commensal and EPEC isolates (P = 0.0195) (Table 5).
The presence of the mph(A) gene was correlated with higher MIC levels (Table 3, Figs 1, 2 and 3), while the presence of other MRGs alone seemed to have a lesser effect. In fact, 40 out of 43 isolates presenting the mph(A) gene as a single MRG had MICs ≥ 32 mg/L. Interestingly, those isolates presenting the mph(A) gene together with another MRG exhibited slightly higher MIC values than those possessing only the mph(A) gene (Fig. 1). The effect of PAβN on the 25 isolates carrying any other MRG was significantly higher (P < 0.0001) than in those isolates with the mph(A) gene. Thus, only 2 erm(B), 1 ere(A) and 1 erm(A) carrying isolates were classified as non-wt when PAβN was added.
Conjugation assay
Transconjugants with MICs ≥ 32 mg/L were observed in 16 (24.2%) out of 66 isolates analysed. The mph(A) gene was transferred in 14 cases and the erm(B) gene in 3 cases (2 together with mph(A)). Finally, 1 transconjugant was obtained from a parental isolate (strain 125: MICI > 256 mg/L; MICPAβN = 128 mg/L, carrying amino acid changes in L4 [K123S] and L22 [I4L, K6Q]) in which no MRG was previously detected.
Wt/non-wt phenotypes and MIC levels
Overall, 22 out of 78 (28.2%) isolates carrying at least one MRG presented MIC levels < 32 mg/L. Of these, 3 isolates harbouring the mph(A) gene alone (7% of isolates carrying the mph(A) gene alone; 3.8% of isolates carrying MRG) and 19 carrying MRGs other than mph(A) alone (76% of isolates carrying other MRGs; 86.4% of isolates with MIC < 32 carrying any MRG) having a MIC < 32 mg/L. No isolates possessing more than one MRG presented a MIC < 32 mg/L (Figs 1 and 2). The cumulative MIC curves of wt isolates and those presenting a MRG other than mph(A) were similar. The cumulative MIC curves of the isolates possessing target mutations, mph(A) and mph(A) plus other MRG were sequentially displaced towards higher MIC levels. When the cumulative MICs were established in presence of PAβN the results showed that those belonging to wt isolates, and those presenting MRG or L4/L22 amino acid substitutions were close similar, with only a spurious displacement towards high MIC levels of those non-wt, while isolates possessing mph(A) and mph(A) plus other MRG were sequentially displaced towards higher MIC levels in a clear manner (Fig. 3).
Discussion
Diarrhoea-related deaths in children remain among the most relevant health challenges worldwide, being of special concern in low- and middle-income countries1,2. In these countries, antibiotic therapy when needed may be crucial to achieve a successful outcome21,22. However, antibiotic resistance to commonly used antibacterial agents is dramatically increasing requiring new alternatives.
Regarding the feasibility to considered azithromycin as an alternative to treat diarrhoeagenic E. coli in the studied areas, the present study showed moderate azithromycin resistance levels highlighting some concerns about its usefulness as treatment in the absence of antibiotic susceptibility data, especially when EAEC or DAEC isolates are present.
In accordance with what has been previously described20,23, the relevant role of PAβN-inhibitible efflux pumps in azithromycin resistance has been demonstrated once more. However, differences related to the specific bacteria groups were observed. The presence of a series of EAEC isolates in which no PAβN-effect was observed opens the door to different options, including the presence of alterations in the outer membrane composition which results in a possible azithromycin impaired permeability leading to an increase in the basal azithromycin resistance levels, combined with lesser efflux pump activity, at least in regard to PAβN-inhibitible efflux pumps. Another possibility is the presence of different patterns of overexpressed efflux pumps. In this line, selecting azithromycin resistant mutants in the presence of PAβN a similar scenario was observed (MIC of 32–16 mg/L with no further PAβN effect). In all these mutants the presence of an overexpressed OmpW was observed24. In fact, OmpW has been associated with EmrE, an efflux pump belonging to the small multidrug resistance (SMR) family9,25. Furthermore, the overexpression of EmrE has been related to E. coli grown in the presence of erythromycin26.
In agreement with the presence of up to 7 gene copies and the subsequent need for multiple mutated alleles to visualize an effect on macrolide resistance9, in the present study no mutations in the 23S rRNA gene were observed in the 66 isolates analysed. Regarding L4 and L22, the alterations detected seem to have a minor role in the development of azithromycin resistance, and most might be gene polymorphisms without antibiotic resistance relevance. Regarding the alterations at L4 and L22 observed, to our knowledge only the alterations at amino acid codon K82, D94 and K98 of L22 have previously been described in in vitro obtained E. coli macrolide-resistant mutants but always concomitantly with other L22 amino acid alterations27. The L22 alteration L46Q was present in 3 cases, all having a MIC ≥ 32 mg/L. Although in one case the addition of PAβN resulted in a MIC of 2 mg/L, and another was concomitantly present with the mph(A) gene, a possible slight effect of this alteration on macrolide susceptibility cannot be ruled out.
Regarding MRGs, in our series the relevant role of Mph(A) is undoubtable. This finding is in accordance with what has been previously described in E. coli and other Enterobacteriaceae9,28–31. Those isolates with the mph(A) gene presented the highest percentages of azithromycin resistance both in the presence and the absence of PAβN. Nonetheless, relevant differences were observed in the MIC levels among isolates carrying the mph(A) gene. Thus, while 2 mph(A)-carrying isolates had a MICI of 8 mg/L which decreased to MICPAβN of 0.25 and 1 mg/L, another 11 isolates in which no other MRG was detected had a MICI > 256 mg/L which in no case decreased below the breakpoint considered in the presence of PAβN. This heterogeneity may be observed on analysing together different studies performed either in E. coli or other closely related Enterobacteriaceae9,28–30. Different explanations may be proposed, including differences related to expression levels which may be due to the number of copies of the gene related to its genetic environment (e.g.: plasmids with different sizes and copy numbers), with alterations at the promotor sequence or with the presence of other undetected MRGs.
The remaining MRGs, seemed to have a marginal role in azithromycin resistance. In fact, the cumulative MIC curve of these isolates was close to that of wt microorganisms. Nonetheless, those isolates presenting the mph(A) together with another MRG ranked among those most resistant and less affected by the addition of PAβN, suggesting a slight contribution of other MRGs to final MIC levels when mph(A) gene is present. This finding was also showed when cumulative MICs were established.
Of these MRGs, among Enterobacteriaceae, the Msr(A) has only been described in E. coli and Enterobacter spp.20,32. In the present study, the msr(A) gene was detected in isolates having MICI of 8 mg/L, supporting the loss of activity of this gene when cloned in E. coli33. The other ATP binding transporter studied, Msr(D), it was detected independently of the presence of Mef(A). Moreover, in no case the mef(A) and the msr(D) genes were detected together. To our knowledge this is the first description of the msr(D) gene alone, since it has always been described concomitantly with mef(A)9. Nevertheless, the presence of polymorphisms in the mef(A) primers annealing region cannot be ruled out. While the effect of Msr(D) on the final MIC levels was within the range of those previously described, this dissociation might result in impaired Mef(A)34. Contrary to what was observed in the present study, Mef(A) has been described to be frequent in Enterobacteriaceae31. This difference may be related to the geographical origin of the samples.
This is the first description of Erm(A) in Enterobacteriaceae9,35. While no data on erm(A) functionality in Enterobacteriaceae has been found, previous studies have described an impairment in the expression levels of erm(C)36, which, if combined with a limited gene copy number, might result in a marginal influence on azithromycin MIC levels such as those detected in present study. Regarding Erm(B), the concomitant presence with mph(A) detected here in 3 isolates, has also been previously described30.
Also Ere(A) had a minimal role in the resistance to azithromycin in the present isolates. This finding is in accordance with the proposed lack of activity of Ere(A) in azithromycin37.
There is controversy about the ability of Mph(B) to hydrolyse azithromycin. Thus, while Chesneau and col38. have described its inability to confer azithromycin resistance, other authors have established a similar activity on hydrolysing erythromycin and azithromycin39. The only isolate of our study that possessed the mph(B) gene exhibited an azithromycin MIC of 16 mg/L in the absence of PAβN.
Despite this marginal role of most MRGs in the final azithromycin MIC, the detection of 6 out of 10 MRGs among commensal E. coli is noteworthy because of their role as a gene-reservoir40,41. Conjugation studies showed that only the mph(A) or erm(B) genes were transferred alone or together. Additionally, in one case in which no MRG was previously detected, transconjugants were obtained showing the presence of an undetermined MRG. In fact other MRGs have been described in E. coli9,35. However, it should be noted that the conjugation assay was designed to detect the transference of high levels of azithromycin resistance (>32 mg/L), and thus, if the resistance levels associated with transferable MRGs was lower, the transference of these elements would probably remain undetected.
Although the presence of non-sought mechanisms of azithromycin resistance, similar to observed in the isolate 3491, may not be discharged, and their presence may influences final MIC as observed when mph(A) was present concomitantly with other MRG. The fact that the cumulative MIC curves of those isolates presenting target mutations or MRG other than mph(A) were only slightly higher than those belonging to wt isolates (on special when role of efflux pumps was discounted with the use of PAβN) confirms the spurious or merely complementary role of these mechanisms as primary azithromycin-resistance cause in E. coli and highlight the relevant role of mph(A).
Thus, the present data showed that the mph(A) gene, is by far, the most effective mechanism of azithromycin resistance present, leading to MIC values higher than 32 mg/L in 93% of the cases, while 88.9% of isolates without mechanisms of resistance remained with MIC levels <32 mg/L. Therefore the use of 32 mg/L seems adequate to suspect the presence of mph(A) and in general of non-wt E. coli isolates. Nonetheless, the presence of sporadic E. coli isolates possessing Mph(A) with MIC values of 8–16 mg/L was also showed. Therefore studies are needed to determine the possible need for more conservative breakpoint.
In summary, the present data demonstrate the presence of azithromycin resistance among intestinal, either pathogenic or not, E. coli from the area of Lima, highlighting the need for susceptibility data to adequately use this antimicrobial agent. Moreover, the relevant and hidden role of efflux pumps in the intrinsic levels of azithromycin resistance is highlighted, showing the potential clinical utility of efflux pumps inhibitors. The present data indicate that the majority of isolates harbouring mph(A) will have MICs ≥ 32 mg/L. These data, combined with other epidemiological data will be useful to establish an E. coli ECOFF value. Clinical data will be needed to establish breakpoints for azithromycin in E. coli.
Materials and Methods
Bacterial strains
Three hundred forty-three diarrhoeagenic (259 isolates, including 78 EAEC, 41 ETEC, 20 DAEC and 120 EPEC) or commensal (84 isolates) E. coli isolates from faeces samples collected in previous studies from children under 5 years of age in periurban areas of Lima (Peru) were recovered from frozen stocks to be included in the study. The uidA gene of all grown isolates was amplified as previously described by Walk and colleagues as a quality control42.
In all cases the previous studies in which were collected the E. coli isolates were approved by the Ethical Committee of the Universidad Peruana Cayetano Heredia, faeces were sampled after informed consent was obtained from parents and/or children legal guardians and all experiments were performed in accordance with relevant guidelines and regulations.
Antimicrobial susceptibility testing
The MIC of azithromycin was determined by the agar dilution method in accordance with the CLSI guidelines17 in the absence (MICI) and presence (MICPAβN) of 20 mg/L of PAβN20,41. The effect of 20 mg/L of PAβN on the viability of microorganisms was also assessed. The PAβN effect on the MIC levels was considered when MICI/MICPAβN > 2. The isolates with a MIC > 256 mg/L that remained unaltered or decreased to 256 mg/L when PAβN was added were not considered in the statistical analysis.
Ribosomal target gene amplification and DNA sequencing
In a random selected subset of 263 (rplD and rplV genes) and 66 samples (23S rRNA) the presence of point mutations was established by PCR (Table 6), as previously described23. The amplified products were recovered with Wizard SV Gel and the PCR Clean Up System (Promega, Madison, Wi) following the manufacturer’s instructions and thereafter sequenced (Macrogen, Seoul, Korea).
Table 6.
Oligonucleotids used in the study.
| Target | Primers | Size (bp) | Ann. (°C) | Ref. | ||
|---|---|---|---|---|---|---|
| Gene | Prot | Forward (5′ → 3′) | Reverse (5′ → 3′) | |||
| Macrolide Resistance Genes | ||||||
| ere(A) | EreA | GCCGGTGCTCATGAACTTGAG | CGACTCTATTCGATCAGAGGC | 420 | 60 | 20 |
| erm(A) | ErmA | TCTAAAAAGCATGTAAAAGAAA | CGATACTTTTTGTAGTCCTTC | 533 | 52 | 20 |
| erm(B) | ErmB | GAAAAAGTACTCAACCAAATA | AGTAACGGTACTTAAATT | 639 | 45 | 20 |
| erm(C) | ErmC | TCAAAACATAATATAGATAAA | GCTAATATTGTTTAAATCGTCAAT | 642 | 45 | 20 |
| mef(A) | MefA | AGTATCATTAATCACTAGTGC | TTCTTCTGGTACTAAAAGTGG | 345 | 54 | 20 |
| mef(B) | MefB | ATGAACAGAATAAAAAATTG | AAATTATCATCAACCCGGTC | 1255 | 45 | 20 |
| mph(A) | MphA | GTGAGGAGGAGCTTCGCGAG | TGCCGCAGGACTCGGAGGTC | 403 | 60 | 20 |
| mph(B) | MphB | ATTAAACAAGTAATCGAGATAGC | TTTGCCATCTGCTCATATTCC | 868 | 50 | 20 |
| msr(A) | MsrA | GCACTTATTGGGGGTAATGG | GTCTATAAGTGCTCTATCGTG | 384 | 58 | 20 |
| msr(D) | MsrD | CCCCAGTTGGACGAAGTAA | TTGTTTTTCCGATTCCATTAC | 781 | 50 | 20 |
| Macrolide Chromosomal Targets | ||||||
| rplD | L4 | GGCAAGAAAATGGCAGGTCAGATGG | TTCCATCGCAGTAGACGCTTTTTCA | 845 | 56 | 23 |
| rplV | L22 | CGGTGGAAAGCGGAGACAAGAAGCC | ACCAGTTTTGCGTCCAGTTCAGGCT | 925 | 56 | 23 |
| rrlH a | — | TAAGGTAGCGAAATTCCTTGTCG | TGATGCGTCCACTCCGGTC | 756 | 61 | 23 |
| Other | ||||||
| rep b | — | GCGCCGICATGCGGCATT | — | MB | 40 | 23 |
| uidA | CATTACGGCAAAGTGTGGGTCAAT | CCATCAGCACGTTATCGAATCCTT | 658 | 55 | 42 | |
DNA: Amplified gene or DNA fragment; Prot: Encoded protein; Size: Amplified product size; Ann: Annealing temperature; MB: Multiband (having different and no related sizes).
aEncode the 23S rRNA; bPrimer designed to amplify the space between Repetitive Extragenic Palindromic (REP) sequences.
Transferable azithromycin resistance mechanism detection
The presence of 10 established MRGs (erm(A), erm(B), erm(C), ere(A), mph(A), mph(B), msr(A), msr(D), mef(A) and mef(B) genes) was sought in all isolates by PCR (Table 6). In all cases negative and positive controls (microorganisms carrying the MRGs included in the study) were used to validate the results. Additionally random selected positive PCRs were sequenced.
Conjugation assays
A total of 66 isolates with a MIC ≥ 32 mg/L were selected to determine the transferability of the MRGs. The conjugation was carried out in Luria-Bertani broth (Conda, Madrid, Spain) with azide-resistant E. coli J53 as a recipient strain. Transconjugants were selected in plates containing 150 mg/L of sodium azide and 32 mg/L of azithromycin. In order to avoid considering possible contaminations the relationship of transconjugants and the respective recipient strain was established by REP-PCR23. The amplification of the MRGs present in the donor and derived transconjugant strains was performed by PCR as mentioned previously.
Statistical analysis
The Fisher exact test was used for statistical analysis. P values ≤ 0.05 were considered significant. A microorganism was considered “wt” when no sought mechanism of resistance other than PAβN inhibitable efflux pumps was identified.
Acknowledgements
JR was supported by program I3, of the ISCIII (grant number: CES11/012). We want to thank Dr. Virve I. Enne for kindly providing the P286 10–99C3, P126 11–99C1 and P473 10–99C3 strains with the mef(B) gene; Dr. Ferran Navarro for kindly providing the P0008 (erm(B)), P0037 (erm(A)) and P00 (mef(A)) strains; Dr. Yolanda Saénz for kindly providing the 2381 (erm(B)), 1920 (erm(C)), 1576 (erm(C) and msr(A)) and 2929 (msr(A)) strains. We also thank Dr. J. Calvo Montes (Hospital Marqués de Valdecilla, Santander, Spain) and Mar Olga Perez-Moreno (Hospital Verge de la Cinta, Tortosa, Spain) for kindly providing E. coli J53-AzR. The authors thank Donna Pringle for editorial assistance. “ISGlobal is a member of the CERCA Programme, Generalitat de Catalunya”.
Author Contributions
C.G., T.J.O. and J.R. designed the experiments; C.G., L.R.-R. and J.M. developed the laboratory studies; C.G. and J.R. analysed the data; C.G. and J.R. wrote the manuscript draft. All authors read the manuscript critically, provide suggestions and approved the final version.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lanata CF, et al. Global causes of diarrheal disease mortality in children, 5 years of age: a systematic review. PLoS One. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379:2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Oral rehydration salts. Production of the new ORS. WHO Document Production Services, Geneva, Switzerland (2006).
- 4.Gonzales L, et al. Prevalence, seasonality and severity of disease caused by pathogenic Escherichia coli in children with diarrhoea in Bolivia. J Med Microbiol. 2013;62:1697–1706. doi: 10.1099/jmm.0.060798-0. [DOI] [PubMed] [Google Scholar]
- 5.Mandomando IM, et al. Etiology of diarrhea in children younger than 5 years of age admitted in a rural hospital of southern Mozambique. Am J Trop Med Hyg. 2007;76:522–527. doi: 10.4269/ajtmh.2007.76.522. [DOI] [PubMed] [Google Scholar]
- 6.Benmessaoud R, et al. Antimicrobial resistance levels among diarrhoeagenic micro-organisms recovered from children under-5 with acute moderate-to-severe diarrhoea in Rabat, Morocco. J Glob Antimicrob Resist. 2016;7:34–36. doi: 10.1016/j.jgar.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Ochoa TJ, et al. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. Am J Trop Med Hyg. 2009;8:296–301. doi: 10.4269/ajtmh.2009.81.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrar WE. Antibiotic resistance in developing countries. J Infect Dis. 1985;152:1103–1106. doi: 10.1093/infdis/152.6.1103. [DOI] [PubMed] [Google Scholar]
- 9.Gomes C, et al. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit Rev Microbiol. 2017;43:1–30. doi: 10.3109/1040841X.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 10.Lubbert C. Antimicrobial therapy of acute diarrhoea: a clinical review. Expert Rev Anti Infect Ther. 2016;14:193–206. doi: 10.1586/14787210.2016.1128824. [DOI] [PubMed] [Google Scholar]
- 11.Cohen R, Raymond J, Gendrel D. Antimicrobial treatment of diarrhea/acute gastroenteritis in children. Arch Pediatr. 2017;24(12S):S26–29. doi: 10.1016/S0929-693X(17)30515-8. [DOI] [PubMed] [Google Scholar]
- 12.Klontz KC, Singh N. Treatment of drug-resistant Shigella infections. Expert Rev Anti Infect Ther. 2015;13:69–80. doi: 10.1586/14787210.2015.983902. [DOI] [PubMed] [Google Scholar]
- 13.Erdman SM, Buckner EE, Hindler JF. Options for treating resistant Shigella species infections in children. J Pediatr Pharmacol Ther. 2008;13:29–43. doi: 10.5863/1551-6776-13.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yates J. Traveler’s diarrhea. Am Fam Physician. 2005;71:2095–2100. [PubMed] [Google Scholar]
- 15.Gascón J. Epidemiology, etiology and pathophysiology of traveler’s diarrhea. Digestion. 2006;73(Suppl 1):102–108. doi: 10.1159/000089785. [DOI] [PubMed] [Google Scholar]
- 16.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty six informational supplement. CLSI document M100-S25. CLSI, Wayne (2016).
- 18.Sjölund-Karlsson M, et al. Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob Agents Chemother. 2011;55:3985–3989. doi: 10.1128/AAC.00590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du D, et al. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 20.Palma N, et al. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteremia in hospitalized children. J Global Antimicrob Resist. 2017;11:28–33. doi: 10.1016/j.jgar.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Gonzales C, Bada C, Rojas R, Bernaola G, Chavez C. Guía de práctica clínica sobre el diagnóstico y tratamiento de la diarrea aguda infecciosa en pediatría Perú − 2011. Rev Gastroenterol Peru. 2011;31:258–277. [PubMed] [Google Scholar]
- 22.Guarino A, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition/European Society for Pediatric Infectious Diseases evidence-based guidelines for the management of acute gastroenteritis in children in Europe: update 2014. J Pediatr Gastroenterol Nutr. 2014;59:132–152. doi: 10.1097/MPG.0000000000000375. [DOI] [PubMed] [Google Scholar]
- 23.Gomes C, et al. In vitro development and analysis of Escherichia coli and Shigella boydii azithromycin-resistant mutants. Microb Drug Resist. 2013;19:88–93. doi: 10.1089/mdr.2012.0036. [DOI] [PubMed] [Google Scholar]
- 24.Gomes C, et al. Which mechanisms of azithromycin resistance are selected when efflux pumps are inhibited? Int J Antimicrob Agents. 2013;42:307–311. doi: 10.1016/j.ijantimicag.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Beketskaia MS, Bay DC, Turner RJ. Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux. J Bacteriol. 2014;196:1908–1914. doi: 10.1128/JB.01483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 27.Diner EJ, Hayes CS. Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics. J Mol Biol. 2009;386:300–315. doi: 10.1016/j.jmb.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lluque A, et al. Virulence factors and mechanisms of antimicrobial resistance in Shigella strains from periurban areas of Lima (Peru) Int J Med Microbiol. 2015;305:480–490. doi: 10.1016/j.ijmm.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma. Q, et al. A waterborne outbreak of Shigella sonnei with resistance to azithromycin and third-generation cephalosporins in China in 2015. Antimicrob Agents Chemother. 2017;61:e00308–17. doi: 10.1128/AAC.00308-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen MCP, et al. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis. 2009;15:1648–1650. doi: 10.3201/eid1510.090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ojo KK, et al. The mef(A) gene predominates among seven macrolide resistance genes identified in gram-negative strains representing 13 genera, isolated from healthy Portuguese children. Antimicrob Agents Chemother. 2004;48:3451–3456. doi: 10.1128/AAC.48.9.3451-3456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojo KK, et al. Staphylococcus efflux msr(A) gene characterized in Streptococcus, Enterococcus, Corynebacterium, and Pseudomonas isolates. Antimicrob Agents Chemother. 2006;50:1089–1091. doi: 10.1128/AAC.50.3.1089-1091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka M, Jánosi L, Endou K, Nakajima Y. Cloning and sequences of inducible and constitutive macrolide resistance genes in Staphylococcus aureus that correspond to an ABC transporter. FEMS Microbiol Lett. 1999;181:91–100. doi: 10.1111/j.1574-6968.1999.tb08830.x. [DOI] [PubMed] [Google Scholar]
- 34.Nunez-Samudio V, Chesneau O. Functional interplay between the ATP binding cassette Msr(D) protein and the membrane facilitator superfamily Mef(E) transporter for macrolide resistance in Escherichia coli. Res Microbiol. 2013;164:226–235. doi: 10.1016/j.resmic.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Roberts MC. Tetracycline and MLS nomenclature; https://faculty.washington.edu/marilynr/. [accessed on June 21 2018].
- 36.Thakker-Varia S, Ranzini AC, Dubin DT. Ribosomal RNA methylation in Staphylococcus aureus and Escherichia coli: effect of the “MLS” (erythromycin resistance) methylase. Plasmid. 1985;14:152–161. doi: 10.1016/0147-619X(85)90075-7. [DOI] [PubMed] [Google Scholar]
- 37.Morar M, Pengelly K, Koteva K, Wright GD. Mechanism and diversity of the erythromycin esterase family of enzymes. Biochemistry. 2012;51:1740–1751. doi: 10.1021/bi201790u. [DOI] [PubMed] [Google Scholar]
- 38.Chesneau O, Tsvetkova K, Courvalin P. Resistance phenotypes conferred by macrolide phosphotransferases. FEMS Microbiol Lett. 2007;269:317–322. doi: 10.1111/j.1574-6968.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi K, et al. Identification of functional amino acids in the macrolide 2’-phosphotransferase II. Antimicrob Agents Chemother. 1999;43:2063–2065. doi: 10.1128/AAC.43.8.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey JK, Pinyon JL, Anantham S, Hall RM. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol 2010. 2010;59:1331–1339. doi: 10.1099/jmm.0.022475-0. [DOI] [PubMed] [Google Scholar]
- 41.Pons MJ, et al. Analysis of quinolone-resistance in commensal and diarrheagenic Escherichia coli isolates from infants in Lima, Peru. Trans R Soc Trop Med Hyg. 2014;108:22–28. doi: 10.1093/trstmh/trt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walk ST, et al. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 2009;75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]