Abstract
Gut microbiota alterations manifest as intermittent hypoxia and fragmented sleep, thereby mimicking obstructive sleep apnea–hypopnea syndrome (OSAHS). Here, we sought to perform the first direct survey of gut microbial dysbiosis over a range of apnea–hypopnea indices (AHI) among patients with OSAHS. We obtained fecal samples from 93 patients with OSAHS [5 < AHI ≤ 15 (n=40), 15 < AHI ≤ 30 (n=23), and AHI ≥ 30 (n=30)] and 20 controls (AHI ≤ 5) and determined the microbiome composition via 16S rRNA pyrosequencing and bioinformatics analysis of variable regions 3–4. We measured fasting levels of homocysteine (HCY), interleukin-6 (IL-6), and tumor necrosis factor α (TNF-α). Results revealed gut microbial dysbiosis in several patients with varying severities of OSAHS, reliably separating them from controls with a receiver operating characteristic-area under the curve (ROC-AUC) of 0.789. Functional analysis in the microbiomes of patients revealed alterations; additionally, decreased in short-chain fatty acid (SCFA)-producing bacteria and increased pathogens, accompanied by elevated levels of IL-6. Lactobacillus levels correlated with HCY levels. Stratification analysis revealed that the Ruminococcus enterotype posed the highest risk for patients with OSAHS. Our results show that the presence of an altered microbiome is associated with HCY among OSAHS patients. These changes in the levels of SCFA affect the levels of pathogens that play a pathophysiological role in OSAHS and related metabolic comorbidities.
Keywords: enterotypes, homocysteine, Lactobacillus, microbiota, obstructive sleep apnea-hypopnea syndrome
Introduction
Obstructive sleep apnea–hypopnea syndrome (OSAHS) is characterized by repeated collapse episodes in the upper airway during sleep, resulting in intermittent hypoxia (IH), together with symptoms of sleep fragmentation (SF) and daytime sleepiness. OSAHS is a systemic disorder; it is highly prevalent in Asia and is estimated to occur in 3.7–97.3% of the population [1]. Further, it is one of the most common sleep disorders and is associated with a growing list of OSAHS-induced end-organ morbidities requiring expertise in respiratory medicine, cardiology, neurology, and endocrinology. OSAHS-mediated oxidative stress and endothelial dysfunction are associated with the pathogenesis of atherosclerosis and cardiovascular morbidity [2]. In this context, IH/reoxygenation and SF are the key features of OSAHS-enhanced inflammation and the oxidative stress cascade [3,4]. By activating multiple systemic inflammatory mediators found in immune cells, OSAHS leads to an enhanced, sustained proinflammatory state that contributes to multiorgan morbidities [2], thereby suggesting a linkage with the host’s gut microbiota.
The gut microbiome plays a crucial role in modulating the risk for several chronic diseases, including obesity, metabolic abnormalities, cardiovascular disease, and inflammatory bowel disease (IBD) [5]. Delicate balance in the composition of the gut microbiome is the key to maintain intestinal immunity and whole-body homeostasis [6]. Gram-positive Firmicutes (F) and Gram-negative Bacteroidetes (B) are predominant in the gut microbiome [5]. Imbalance in the Firmicutes/Bacteroidetes (F/B) ratio has been closely related to inflammatory and immune function disorders, obesity, and metabolic disease [7].
Experiments conducted in an animal model of IH mimicking OSAHS and in an animal model of IH + high-fat diet (HFD) have demonstrated alterations in the diversity of gut microbiota. Previous studies have shown an increase in the F/B ratio, which is in turn associated with an increase in the relative abundance of lactate-producing bacteria and a decrease in the relative abundance of short-chain fatty acid (SCFA)-producing bacteria [3,6,8,9]. In addition, chronic SF alters feeding behaviors, thereby promoting obesity and metabolic abnormalities. The host gut microbiome changes, increasing intestinal permeability as well as systemic and adipose tissue inflammation. These changes are accompanied by insulin resistance [4,10]. Although these findings relate to rodent paradigms, we aimed to determine whether these characteristics are also present in humans with OSAHS-related metabolic comorbidities.
Enterotype is another means to investigate the gut microbiota. Major enterotype bacteria may be subdivided into three types: Bacteroides (enterotype 1), Ruminococcus (enterotype 2), and Prevotella (enterotype 3) [11]. Dietary habits affect enterotypes, thereby emphasizing the key role of diet in the composition of the gut microbiome [12]. The balance of the three enterotypes is maintained by various enzymes and is not associated with ethnicity, gender, age, or body mass index (BMI) [11].
Alteration in the gut microbiota has been reported in IH and SF mimicking OSAHS rodent paradigms. However, only few studies have directly investigated the gut microbiota in patients with OSAHS, who often present with metabolic comorbidities. Hence, here, using taxa and enterotype data, we aimed to investigate gut microbial dysbiosis in individuals with varying scores on the apnea–hypopnea index (AHI). This classification differs from IH modeling paradigm, which considers only two groups. In patients with OSAHS, homocysteine (HCY) levels are associated with an increased risk of cardiovascular events and metabolic abnormalities [13]. Therefore, we assessed the correlation between the gut microbiota and HCY levels in relation to OSAHS-related metabolic comorbidities.
Experimental
Subjects
We recruited 113 subjects and examined them by a full night of polysomnography (PSG; SOMNOscreen™ plus PSG+, SOMNOmedics GmbH, Randersacker, Germany), conducted by technologists in a sleep laboratory from 10 p.m. to 8 a.m. at the Department of Pulmonary and Critical Care Medicine. The Institutional Review Board of the Second Affiliated Hospital of Fujian Medical University approved the present study (IRB No. 2017-78). We collected fasting blood and fecal samples the following morning.
OSAHS assessment
All subjects underwent PSG (performed with a computerized polysomnographic system), which included electrocardiography, electroencephalography, electromyography, and electrooculography. After one night of the examination, AHI was calculated as the total number of episodes of apnea (continuous cessation of airflow for at least 10 s) and hypopnea (reduction in airflow for ≥10 s with oxygen desaturation of ≥4%) divided by the total duration of sleep events, according to the diagnostic criteria of the American Academy of Sleep Medicine. AHI ≤ 15 events/h was defined as non-OSAHS (control group), 5 < AHI ≤ 15 as mild OSAHS (Group 1), 15 < AHI ≤ 30 as moderate OSAHS (Group 2), and AHI ≥ 30 as severe OSAHS (Group 3).
Measurement of HCY levels
We determined the level of fasting HCY serum with the Automatic Biochemical Analyzer (TBA-120 FR, Toshiba, Japan) and the HCY assay kit (Yong He Sun Biotech. Ltd., Hunan, China), utilizing the enzymatic cycling method.
Cytokine analysis
Interleukin (IL)-6 (IL-6) and tumor necrosis factor α (TNF-α) were assayed by BD Human Enhanced Sensitivity Cytometric Bead Array Kit (BD Biosciences, New Jersey, U.S.A.) as described previously [14]. The standard coefficient of determination (r2) was greater than 0.995 (detail see in Supplementary Figure S1).
Sampling, DNA extraction, and 16S rRNA gene amplification sequencing
All fresh fecal samples were collected and stored in Microbiome Test Kit (G-BIO Biotech, Inc., Hangzhou, China). Magnetic bead isolation was done to extract genomic DNA using a TIANamp stool DNA kit (TIANGEN Biotech Co., Ltd., Beijing, China), according to the manufacturer’s instructions. The concentration of extracted DNA was determined by a Nanodrop ND-1000 spectrophotometer (Thermo Electron Corporation, U.S.A.), and DNA quality was confirmed using 1.0% agarose gel electrophoresis with 0.5 mg/ml ethidium bromide.
Isolated fecal DNA was used as a template to amplify the V3 and V4 hypervariable regions of the bacterial 16S rRNA gene. The V3 and V4 regions were PCR-amplified (forward primer, 5′-ACTCCTACGGGAGGCAGCAG-3′; reverse primer, 5′-GGACTACHVGGGTWTCTAAT-3′) [15]. The 16S target-specific sequence contained adaptor sequences permitting uniform amplification of a highly complex library ready for downstream next-generation sequencing with Illumina MiSeq (Illumina, U.S.A.). Negative DNA extraction controls (lysis buffer and kit reagents only) were amplified and sequenced as contamination controls. The amplicons were normalized, pooled, and sequenced on the Illumina MiSeq platform, using a V3 reagent kit with 2 × 300 cycles per sample, and with imported and prepared routine data (samsheet) run in the MiSeq sequence program. After sequencing, Q30 scores were ≥70%, the percentage of clusters passing filter (i.e. cluster PF) was ≥80%, and there were at least 30000 clean tags. Finally, image analysis and base calling were conducted with the MiSeq Control Software.
Bioinformatics, predictive function, and statistical analyses
Based on the Quantitative Insights into Microbial Ecology bio-informatic pipeline for performing taxonomy assignment by the operational taxonomic unit (OTU) method, we used 113 sequence data to analyze the fecal microbiota taxa. We predicted the bacterial metabolic function using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States bioinformatics software package and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
We discriminated OSAHS patients from controls with an area under the receiver operating characteristic curve (ROC-AUC) with a patient disease index (PDI) cut-off, wherein ROC-AUC and PDI were calculated using a previous study [16].
Data were expressed as the mean ± standard deviation (S.D.). We analyzed differences in gut microbiota using the Kruskal–Wallis test, as appropriate, and performed principal coordinate analysis (PCoA) on the basis of the Bray–Curtis distance function, using R statistics. We performed other analyses using statistically with SPSS version 19.0 (SPSS Inc., Chicago, IL, U.S.A.) by one-way ANOVA, followed by Scheffe post hoc analyses, we considered a two-sided P<0.05 to be statistically significant. We evaluated the correlation coefficients between gut microbiota and HCY level using Spearman correlation.
Results
Participants’ characteristics
After PSG assessment, we enrolled 93 patients with OSAHS [Group 1 (n=40), Group 2 (n=23), and Group 3 (n=30)] and 20 controls (Table 1). Body weight (P=0.006), BMI (P=0.010), and hip circumference (P=0.025) were significantly higher in the Group 3 than in the control group. Waist circumference [P<0.001 (Control vs. Group 1), P=0.005 (Control vs. Group 2), P<0.001 (Control vs. Group 3)], and waist-to-hip ratio [P<0.001 (Control vs. Group 1), P<0.001 (Control vs. Group 2), P<0.001 (Control vs. Group 3)] were significantly lower among the control group than in the OSAHS groups (Table 1).
Table 1.
Participants’ characteristics
| Control (n=20) | Group 1 (n=40) | Group 2 (n=23) | Group 3 (n=30) | |
|---|---|---|---|---|
| Gender (male/female) | 11/9 | 35/5 | 18/4 | 27/3 |
| Age (years, mean ± S.D.) | 39.0 ± 8.9 | 45.4 ± 11.0 | 48.6 ± 13.8 | 44.4 ± 11.2 |
| Height (cm) | 165.95 ± 8.64 | 166.59 ± 7.64 | 165.65 ± 7.09 | 166.16 ± 7.03 |
| Weight (kg) | 67.26 ± 8.60 | 72.98 ± 11.43 | 73.93 ± 20.50 | 80.77 ± 12.372 |
| BMI (kg.m−2) | 24.31 ± 2.25 | 26.98 ± 4.72 | 26.04 ± 3.69 | 29.04 ± 4.792 |
| Waist circumference (cm) | 81.23 ± 7.58 | 94.18 ± 10.933 | 91.20 ± 7.462 | 98.48 ± 9.413 |
| Hip circumference (cm) | 95.15 ± 6.10 | 99.03 ± 7.71 | 96.20 ± 5.55 | 101.88 ± 9.061 |
| Waist-to-hip ratio | 0.85 ± 0.06 | 0.95 ± 0.053 | 0.95 ± 0.043 | 0.97 ± 4.793 |
| HCY (μmol/l) | 12.50 ± 4.31 | 15.99 ± 6.33 | 17.32 ± 10.57 | 16.41 ± 6.50 |
| History of hypertension [n (%)] | 0 (0) | 16 (40) | 14 (60.9) | 20 (66.7) |
| History of diabetes [n (%)] | 0 (0) | 3 (7.5) | 1 (4.3) | 3 (10) |
| Sleep efficiency (%) | 68.96 ± 15.09 | 68.11 ± 18.44 | 67.84 ± 15.08 | 76.10 ± 17.38 |
| Arousal index (events/h) | 3.28 ± 1.85 | 3.32 ± 1.64 | 3.98 ± 2.00 | 2.44 ± 2.97 |
| AHI (events/h) | 1.91 ± 1.32 | 9.26 ± 3.051 | 19.3 ± 3.843 | 56.69 ± 21.403 |
| Hypopnea index (events/h) | 1.35 ± 1.12 | 6.16 ± 3.241 | 12.09 ± 6.673 | 21.87 ± 15.743 |
| Mean SpO2 (%) | 94.95 ± 2.26 | 94.88 ± 1.40 | 94.39 ± 1.47 | 92.17 ± 3.043 |
| Lowest SpO2 (%) | 91.20 ± 4.27 | 84.95 ± 4.411 | 82.26 ± 7.753 | 71.47 ± 8.803 |
Control: patients with AHI ≤ 5 events/h were considered as non-OSAHS; Group 1: patients with 5 < AHI ≤ 15 were considered as mild OSAHS; Group 2: patients with 15 < AHI ≤ 30 were considered as moderate OSAHS; Group 3: patients with AHI ≥ 30 were considered as severe OSAHS.
1P<0.05.
2P<0.01.
3P<0.001 compared with the Control group by one-way ANOVA with Scheffe test.
Our PSG data indicated that AHI [P=0.014 (Control vs. Group 1), P<0.001 (Control vs. Group 2), P<0.001 (Control vs. Group 3)] and hypopnea index [P=0.036 (Control vs. Group 1), P<0.001 (Control vs. Group 2), P<0.001 (Control vs. Group 3)] were significantly higher in the OSAHS groups than in the control group. The lowest oxygen saturation in the OSAHS groups was lower than that in the control group [P=0.022 (Control vs. Group 1), P<0.001 (Control vs. Group 2), P<0.001 (Control vs. Group 3)]. Mean oxygen saturation was significantly lower in the Group 3 than in the control group (P<0.001; Table 1).
Alterations in taxa among groups
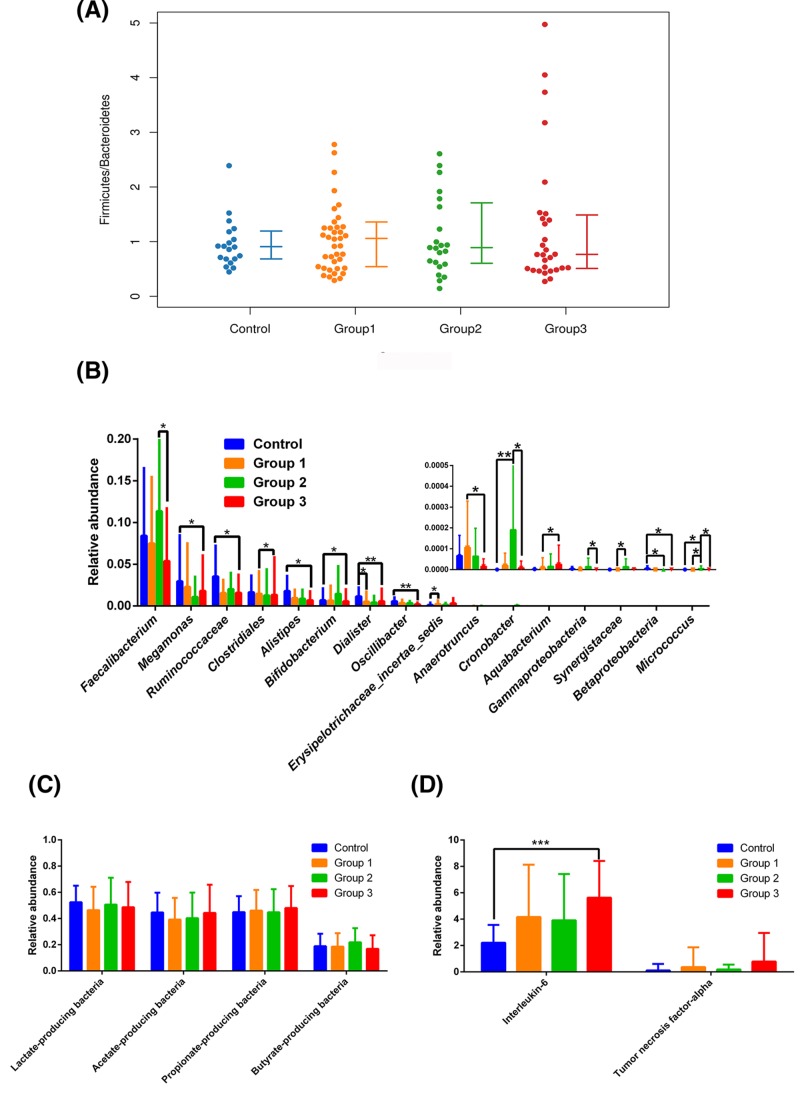
We examined the mean community diversity indices (Supplementary Figure S2) and the structure of the gut microbiome (Supplementary Figure S3). At the phylum level, we found no significant differences in relative abundance, including the F/B ratio (Figure 1A). The relative abundances of the following genera significantly differed among groups: Faecalibacterium (P=0.044), Megamonas (P=0.046), Ruminococcaceae (P=0.048), Clostridiales (P=0.021), Alistipes (P=0.024), Bifidobacterium (P=0.037), Dialister (P=0.007), Oscillibacter (P=0.008), Erysipelotrichaceae (P=0.025), Anaerotruncus (P=0.043), Cronobacter (P=0.006), Aquabacterium (P=0.033), Gammaproteobacteria (P=0.038), Synergistaceae (P=0.032), Betaproteobacteria (P=0.020), and Micrococcus (P=0.007) (Figure 1B; for details, see Supplementary information). We found no significant differences between the control and OSAHS groups in terms of the relative abundances of acetate-, butyrate-, propionate-, or lactate-producing bacteria (Figure 1C).
Figure 1.
Relative abundances of fecal taxa at different levels
The F/B ratio was similar across groups (A). For differences in the fecal microbiota at the genera level (B), statistical analysis was performed by Kruskal–Wallis test. There were no significant differences between the control group and OSAHS groups in the abundance of acetate-, butyrate-, propionate- or lactate-producing bacteria (C). IL-6 was significantly elevated in Group 3 (D), statistical analysis was performed with the Scheffe test. *P<0.05, **P<0.01, ***P<0.001 compared with the control group or OSAHS groups. Control: patients with AHI ≤ 5 events/h were considered as non-OSAHS; Group 1: patients with 5 < AHI ≤ 15 were considered as mild OSAHS; Group 2: patients with 15 < AHI ≤ 30 were considered as moderate OSAHS; Group 3: patients with AHI ≥ 30 were considered as severe OSAHS.
Cytokine analysis
IL-6 of the Group 3 was significantly higher than the control group (P=0.006). However, there was not significantly different in TNF-α among the control group and OSAHS groups (Figure 1D).
Predictive function analysis
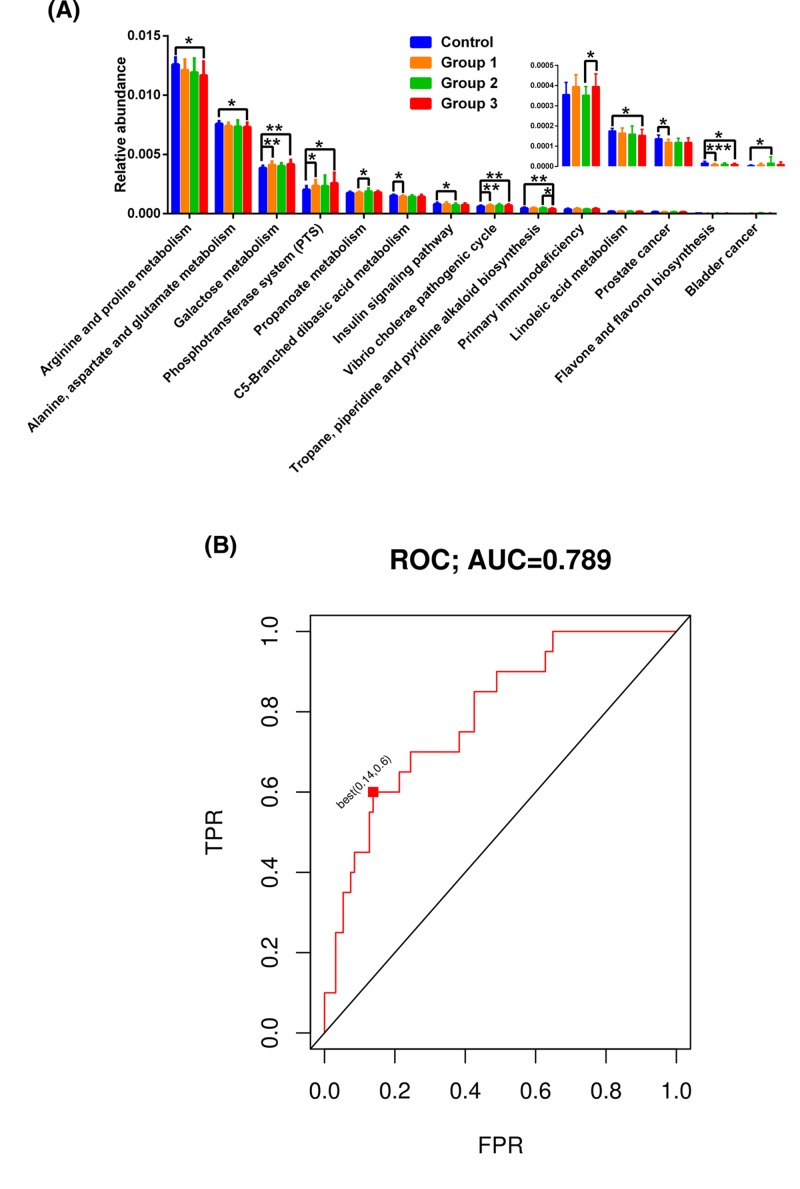
According to KEGG, there were significant differences in the fecal microbiome among these study groups, covering 14 pathways involved in arginine and proline metabolism [P=0.016 (Control vs. Group 3)]; alanine, aspartate, and glutamate metabolism [P=0.049 (Control vs. Group 3)]; galactose metabolism [P=0.002 (Control vs. Group 1), P=0.003 (Control vs. Group 3)]; phosphotransferase system [P=0.037 (Control vs. Group 1), P=0.028 (Control vs. Group 3)]; propanoate metabolism [P=0.027 (Group 1 vs. Group 2)]; C5-branched dibasic acid metabolism [P=0.029 (Control vs. Group 1)]; insulin signaling pathway [P=0.012 (Control vs. Group 2)]; Vibrio cholerae pathogenic cycle [P=0.007 (Control vs. Group 1), P=0.006 (Control vs. Group 3)]; tropane, piperidine, and pyridine alkaloid iosynthesis [P=0.007 (Control vs. Group 3), P=0.031 (Group 2 vs. Group 3)]; primary immunodeficiency [P=0.031 (Group 2 vs. Group 3)]; linoleic acid metabolism [P=0.027 (Control vs. Group 3)]; prostate cancer [P=0.015 (Control vs. Group 1)]; flavone and flavonol biosynthesis [P<0.001 (Control vs. Group 1), P=0.027 (Control vs. Group 3)]; and bladder cancer [P=0.045 (Control vs. Group 2)] (Figure 2A).
Figure 2.
Predictive function analysis and discriminate predictive model for the gut microbiota
Significant KEGG pathways are shown for the fecal microbiome (A), statistical analysis was performed with the Kruskal–Wallis test. *P<0.05, **P<0.01, ***P<0.001, compared with the control group or OSAHS groups. OSAHS patients could be separated from controls using 29 genera, with an ROC-AUC of 0.789 (B). FPR: false positive rate; TPR: true positive rate. Control: patients with AHI ≤ 5 events/h were considered as non-OSAHS; Group 1: patients with 5 < AHI ≤ 15 were considered as mild OSAHS; Group 2: patients with 15 < AHI ≤ 30 were considered as moderate OSAHS; Group 3: patients with AHI ≥ 30 were considered as severe OSAHS.
Association between fecal microbiota and HCY
Neither lactate- nor SCFA-producing bacteria had any correlation with the HCY level, apart from Lactobacillus, which is lactate- and acetate-producing bacterium, with a positive correlation with the HCY level (P=0.041; Table 2).
Table 2.
Correlations between HCY levels and lactate- or SCFA-producing bacteria in non-OSAHS subjects and OSAHS subjects
| P-value | Rho value | |
|---|---|---|
| Atopobium | 0.751 | −0.030 |
| Bacteroides | 0.745 | 0.031 |
| Bifidobacterium | 0.277 | −0.103 |
| Butyrivibrio | 0.738 | 0.032 |
| Clostridium XlVa | 0.804 | −0.024 |
| Clostridium XlVb | 0.918 | 0.010 |
| Clostridium XI | 0.783 | 0.026 |
| Clostridium IV | 0.562 | −0.055 |
| Clostridium XIX | 0.587 | −0.052 |
| Eubacterium | 0.734 | 0.032 |
| Enterococcus | 0.426 | −0.076 |
| Faecalibacterium | 0.597 | −0.050 |
| Fusobacterium | 0.456 | −0.071 |
| Lactobacillus | 0.041 | 0.192 |
| Megasphaera | 0.263 | 0.106 |
| Prevotella | 0.083 | 0.164 |
| Porphyromonas | 0.197 | 0.122 |
| Peptococcus | 0.445 | −0.073 |
| Peptostreptococcus | 0.642 | −0.044 |
| Propionibacterium | 0.418 | 0.077 |
| Roseburia | 0.703 | −0.036 |
| Ruminococcus | 0.900 | −0.012 |
| Streptococcus | 0.185 | −0.126 |
| Veillonella | 0.820 | −0.022 |
Gut microbiota discriminated predictive model
To select predictive features, we discriminated OSAHS patients from controls with ROC-AUC 0.789 using different taxa (Figure 2B). The best predictive point was the false positive rate of 0.14 and the true positive rate of 0.60, with a PDI cut-off of −0.00738 (P=0.00003; data not shown).
Enterotypes analysis
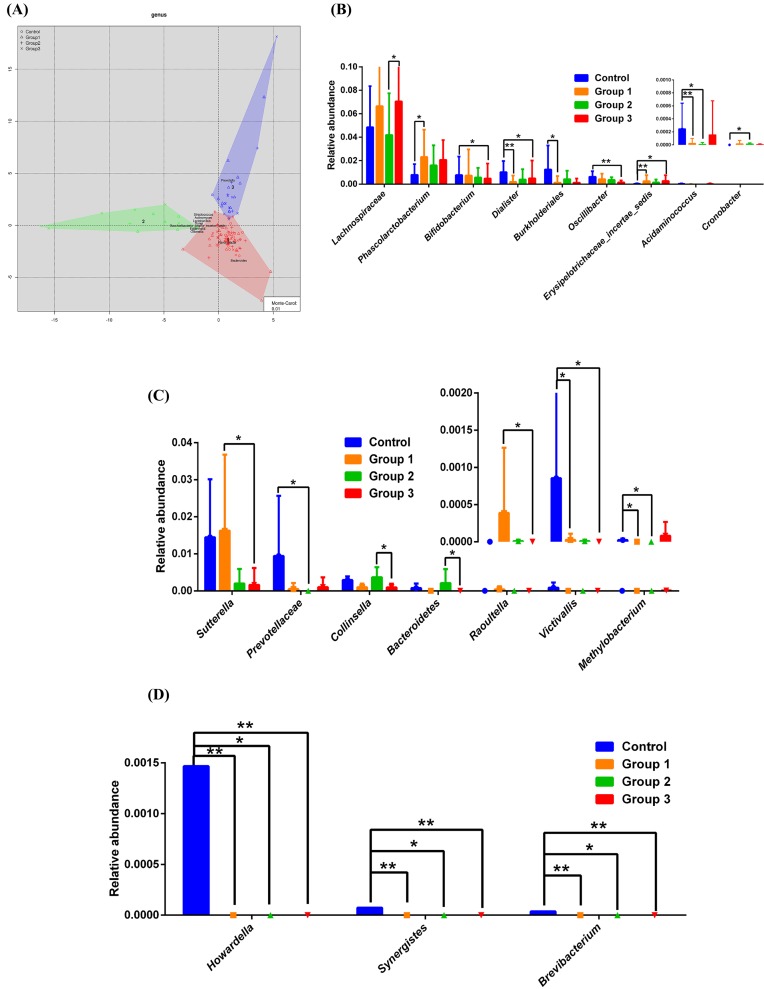
In Figure 3A, we present the distribution of fecal taxa in the control group and OSAHS groups. For enterotype 1, the relative abundances of the genera Lachnospiraceae, Erysipelotrichaceae, Bifidobacterium, Dialister, Burkholderiales, Oscillibacter, Acidaminococcus, Phascolarctobacterium, and Cronobacter were significantly different among four groups (Figure 3B; detail see in Supplementary information). For enterotype 2, the relative abundance of the genera Sutterella, Raoultella, Collinsella, Bacteroidetes, Methylobacterium, Prevotellaceae, and Victivallis were significant different among four groups (Figure 3C; for detail see Supplementary information). For enterotype 3, Howardella, Synergistes, and Brevibacterium were higher in the control group than in the OSAHS groups (Figure 3D; for detail see Supplementary information). After conducting further stratification analysis utilizing the enterotypes, the estimated OSAHS odds ratios were much higher in Ruminococcus enterotype (3.65) and Prevotella enterotype (2.15) for OSAHS risky (Table 3).
Figure 3.
Three enterotypes of fecal taxa in patients with non-OSAHS, compared with OSAHS subjects
Microbiome distribution across three enterotypes (A). The fecal microbiota showed significant differences in the following genera: Bacteroides (enterotype 1) (B), Ruminococcus (enterotype 2) (C), and Prevotella (enterotype 3) (D). Statistical analysis was performed with the Kruskal–Wallis test. *P<.05, **P<0.01 compared with the control group or OSAHS groups. Control: patients with AHI ≤ 5 events/h were considered as non-OSAHS; Group 1: patients with 5 < AHI ≤ 15 were considered as mild OSAHS; Group 2: patients with 15 < AHI ≤ 30 were considered as moderate OSAHS; Group 3: patients with AHI ≥ 30 were considered as severe OSAHS.
Table 3.
Enterotype analysis association with OSAHS risk
| Control N (%) | Group 1 N (%) | Group 2 N (%) | Group 3 N (%) | Odds ratio (95% CI) | |
|---|---|---|---|---|---|
| Enterotype 1 (Bacteroides) | 16 (80.0) | 24 (60.0) | 16 (69.6) | 16 (53.3) | 1.00 |
| Enterotype 2 (Ruminococcus) | 1(5.0) | 5 (12.5) | 3 (13.0) | 6 (20.0) | 3.65 (0.44–30.04) |
| Enterotype 3 (Prevotella) | 3 (15.0) | 11 (27.5) | 4 (17.4) | 8 (26.7) | 2.15 (0.57–8.09) |
Control: patients with AHI ≤ 5 events/h were considered as non-OSAHS; Group 1: patients with 5 < AHI ≤ 15 were considered as mild OSAHS; Group 2: patients with 15 < AHI ≤ 30 were considered as moderate OSAHS; Group 3: patients with AHI ≥ 30 were considered as severe OSAHS.
Discussion
We compared the differences in the gut microbiome between the control and OSAHS groups. The number of candidate taxa associated with gut microbial dysbiosis in OSAHS groups were 2 at the class level, 3 at the order level, 10 at the family level (details in Supplementary Figure S4), and 16 at the genera level. Decrease in the relative abundance of SCFA-producing bacteria and increased levels of proinflammatory cytokines were also observed. These observations were supported by functional analyses of the microbiota. Lactobacillus levels correlated with HCY levels. Our validation cohort showed an ROC-AUC value of 0.789 for these samples, confirming that gut microbiota information can be used to identify patients. The Ruminococcus enterotype was associated with an increased risk for OSAHS. We contend that these findings illustrate the pathophysiological role played by changes in the gut microbiota in OSAHS. These changes might be associated with OSAHS-related metabolic comorbidities.
IH and SF may play pathophysiological roles in changes to the gut microbiota associated with OSAHS. Gut microbial dysbiosis has been demonstrated in several OSAHS-mimicking animal paradigms [3,4,8,9]. However, OSAHS is a systemic and complex disorder. IH, IH + HFD, or SF alone cannot be used to diagnose a patient with OSAHS. Contrary to the results reported previously for animal models [3,10], our results did not show significant differences among Chinese OSAHS patients in terms of Chao richness, Shannon diversity, and Simpson diversity (Supplementary Figure S2). Observations regarding alterations in the gut microbiome in the IH rodent model suggest that although the gut epithelium is remarkably resistant to hypoxia, the absorptive and barrier functions that regulate the intestinal epithelium are sensitive to the level of oxygen inside the gut. By increasing permeability and bacterial translocation and decreasing tight junction integrity, IH/reoxygenation may directly impair cellular function [17,18]. The increasing abundance of anaerobic bacteria noted in the IH model is due to the emergence of anoxic environment in the intestine, which is beneficial for the growth of obligate anaerobic bacteria and endogenous lipopolysaccharide (LPS) production by Gram-negative bacteria [3,8]. Severe circulating endotoxemia was maintained throughout prolonged normoxic recovery after IH exposure in mice [8], suggesting that IH alters the gut microbiome and endotoxin levels.
In addition, SF-induced sleep perturbations led to profound alterations in the gut microbiome, such as increases in the circulating levels of IL-6, neutrophil gelatinase-associated lipocalin, and LPS-binding protein, particularly in patients with obesity [4,19]. These observations suggest the induction of inflammatory processes, perhaps owing to the leakage of microbial metabolites into the circulation, which is similar to our findings in patients with severe OSAHS. SF-induced sleep disturbances also promote the innate immune response, thereby leading to low-grade systemic and adipose tissue inflammation and ultimately resulting in metabolic homeostasis disruption [4]. SF treatment altered the feeding behaviors, eventually promoting obesity and metabolic abnormalities. These findings indicate that SF changes the host’s gut microbiota, increases intestinal permeability, and promotes inflammation in adipose tissue and throughout the body. These changes are accompanied by insulin resistance [4]. Taken together, IH and SF may induce alterations in the intestinal epithelial barrier and increase intestinal permeability, leading to local and systemic inflammatory responses and multi-metabolic abnormalities, such as obesity, hypertension, and diabetes [20,21].
The prevalence of OSAHS in patients with obesity/metabolic syndrome is estimated at 45–60% [22,23]. In our study, patients with OSAHS had a higher waist circumference and waist-to-hip ratio than patients in the control group; in particular, patients with severe OSAHS had higher body weight, BMI, and IL-6 levels. We also observed an increased risk for metabolic abnormalities with increasing age and AHI in males with OSAHS, as reported previously [22]. OSAHS may alter the expression of intestinal epithelial barrier markers, thereby increasing intestinal permeability. Persistently increased permeability may be an important contributor to the development of metabolic complications [21]. Gut permeability is further increased during the development of obesity and metabolic disorders [19]. Obesity involves low-grade inflammation and elevations in the F/B ratio [5]. Although we noted that patients with OSAHS have a higher BMI and greater risk for metabolic abnormalities, we cannot draw a firm conclusion from our data. However, functional analysis of fecal samples collected from patients with OSAHS indicated effects on pathways involved in the down-regulation of amino acid metabolism (arginine and proline metabolism; alanine, aspartate, and glutamate metabolism); the elevation of galactose and propanoate metabolism; the down-regulation of C5-branched dibasic acid metabolism, linoleic acid metabolism, and flavone and flavonol biosynthesis; and the down-regulation of the insulin signaling pathway. These factors are connected to the energy metabolism that promotes obesity, metabolic abnormalities, and inflammation [4,10], indirectly corresponding to the primary immune disruption in our OSAHS groups, as revealed by functional analysis.
The disruption of gut permeability is initially a cascading factor; however, SCFA may regulate gut integrity [24,25]. Our OSAHS groups had lower fecal counts of Faecalibacterium, Megasphaera, Ruminococcaceae, Dialister, and Oscillibacter, which are lactate- or SCFA-producing bacteria [26]. Microbial fermentation of dietary fiber generates SCFA. In some bacterial species, lactate can be converted into butyrate. SCFA-producing bacteria also produce acetate, propionate, and butyrate. A decrease in SCFA production results in intestinal barrier dysfunction [27,28]. SCFA has a direct anti-inflammatory effect on the gut, contributes to mucin synthesis, decreases bacterial translocation, maintains gut integrity, and mitigates inflammation in the intestine [8,24,25]. Moreover, SCFAs participate in immunity, adipogenesis, insulin sensitivity, and oxidative stress [26]. Faecalibacterium, Oscillibacter, and Howardella genera were highly enriched in controls. The levels of Faecalibacterium and Oscillibacter, which contribute to anti-inflammatory activity within the gut [29], were negatively correlated with IBD [25]. We speculate that the relative abundance of anti-inflammation-associated bacteria among the patients with OSAHS at least partly mitigated the degree of inflammation. We also detected an overgrowth of proinflammatory enteric pathogens (the class Gammaproteobacteria and the family Enterobacteriaceae; Supplementary information), such as common Gram-negative bacteria that promote LPS production and thereby increase inflammation [30].
In addition, Lactobacillus and Escherichia are positively correlated with IBD [25]. Altered levels of microbial butyrate and lactate may contribute to OSAHS-induced hypertension [6,9]. Lactate bacteria in the human gastrointestinal tract, including Bifidobacteria, Lactobacilli, Streptococci, and Enterococci, predominantly produce l- and/or d-lactate, which can also be produced by strict anaerobes, such as Eubacterium spp., Selenomonas, Megasphaera, and Veillonella that can convert lactate into acetate and propionate [27]. l-lactate may enter the gut from host tissues. In short-bowel syndrome, d-lactate accumulation may lead to serious outcomes, including neurotoxicity and cardiac arrhythmia. Under normal conditions, lactate is seldom detected in human feces or gut content as a major fermentation product of mixed anaerobic communities [27]. However, plasma lactate concentrations are positively associated with increased blood pressure [31]. Our data reveal that HCY levels are positively correlated with the levels of Lactobacillus, which is a lactate bacterium. In patients with OSAHS, HCY levels are associated with an increased risk for metabolic abnormalities and hypertension [13]. Lactobacillus plantarum has been demonstrated to reduce HCY levels [32] and to reduce the risk for cardiovascular disease [33]. This positive effect may be responsible for the production of SCFA through the bacterial fermentation of fiber [33]. However, the results of our functional analyses of the microbiome did not reflect information about the depletion of vitamins B6 and B12, folates, or methionine in relation to HCY metabolism. Instead, arginine participates in the synthesis of nitric oxide (NO), which declines with increased blood pressure. Arginine levels decreased in our patients with OSAHS, 40–67% of whom suffer from hypertension. Arginine, methionine, HCY, and vitamins influence NO synthesis, eventually affecting the endothelial function that controls vascular homeostasis [34]. Furthermore, diet seemed to play a key role in the synthesis of HCY and NO as well as the composition of the gut microbiota. We suggest that Lactobacillus plays important roles in OSAHS-related metabolic comorbidities; however, this hypothesis requires additional evidence.
On the other side, enterotype analysis has been proposed as a useful method to understand human gut microbial communities, including Bacteroides, Ruminococcus, and Prevotella enterotypes, irrespective of ethnicity, gender, age, or BMI [11]. The IH rodent model may be classified as enterotypes Bacteroides and Prevotella [3]. Using this model, researchers found an increase in the abundance of Prevotella and Desulfovibrio; however, this trend was not observed in our human data described above. Two types of mucus-degrading bacteria deplete the mucosa on the epithelial layer of the gut when exposed to IH, resulting in altered intestinal permeability [3]. However, we conducted stratification analysis using various enterotypes to reveal that the Ruminococcus enterotype posed the highest risk for OSAHS. Although the Prevotella enterotype belongs to the phyla Bacteroidetes, the accompanying endotoxins render the host prone to low-grade inflammation, triggering the progression of OSAHS. The Prevotella enterotype is associated with diets high in carbohydrates (fiber) [12]. Therefore, we speculated that could promote microbial fermentation and produce SCFA to protect the host.
In contrast, the Bacteroides enterotype is associated with Western-style diets, including the consumption of high amounts of protein and fat [12]. According to our findings, this enterotype comprises the putative anti-inflammation-associated bacteria Lachnospiraceae, Phascolarctobacterium, Dialister, Oscillibacter, Acidaminococcus, and Bifidobacterium [26,35,36] to a greater degree than the pathobiont Burkholderiales. Their gut microbiota protects the intestinal mucosa, maintains intestinal permeability, and attenuates the inflammatory response, ultimately reducing the risk for OSAHS. Patients with autism spectrum disorder with the Ruminococcus enterotype have high levels of the enriched bacteria Sutterella spp. and Ruminococcus spp. [37]. Patients with OSAHS also have increased putative pathobionts (e.g. Raoultella spp., Methylobacterium), which can trigger infection [38,39] and decrease the relative abundance of Collinsella in smokers with IBD [40], as observed in our patients. However, the non-OSAHS patients in this enterotype may maintain constant health, leading to the speculation that Prevotellaceae, Bacteroidetes, and Victivallis spp. are putative SCFA-producing bacteria [41,42] and occur to a lesser degree in patients with OSAHS than in normal controls.
There are certain limitations to our study. The first limitation is the small sample size, particularly for the Ruminococcus enterotype; the present data must therefore be conservatively interpreted. Second, BMI should not affect the enterotype theory [11]. A future prospective study should include controls and OSAHS groups with comparable BMI. Third, although the gut bacterial profiles are influenced by dietary habits [5,12], we did not collect or analyze data regarding the participants’ dietary habits; this will need to be corrected in the future because long-term dietary differences could cause differences in a patient’s microbiome and confer different health risks. Fourth, we did not comprehensively analyze lactate and endotoxins in blood; thus, even greater caution is required when interpreting alterations in the relative abundance of microbiota taxa with regard to their effects on the pathogenesis of OSAHS.
Conclusion
IH and SF may cause gut microbial dysbiosis. Changes in the gut microbiome are associated with reduced SCFA production and increased pathogen levels, and those changes induce alterations in the levels of intestinal epithelial barrier markers and increase intestinal permeability, leading to local and systemic inflammatory responses and metabolic comorbidities. Our findings show that changes in the gut microbiota have a pathophysiological role in OSAHS and suggest that changes in the gut microbiome may be associated with the pathophysiology of metabolic comorbidities in patients with OSAHS.
Clinical perspectives
Gut microbial dysbiosis is found in varying degrees in patients with OSAHS. We performed enterotypes stratification analysis to identify a panel of gut microbiome biomarkers that could be used as a non-invasive test with which to accurately diagnose OSAHS.
Decreases in the relative abundance of SCFA-producing bacteria and elevated pathogens, accompanied by elevated levels of proinflammatory cytokines, are associated with the pathophysiology of OSAHS-related metabolic comorbidities.
Lactobacillus levels were correlated with HCY levels. Elucidating the interactions between microbiomes and body homeostasis may shed some new insight into the pathophysiology of metabolic comorbidities in OSAHS.
Acknowledgments
We thank all the participants and their families who took part in the present study. We thank the assistance provided by Huan Wu (G-BIO Biotech, Inc., Hangzhou, China) for performing the bioinformatics analysis.
Abbreviations
- AHI
apnea–hypopnea index
- BMI
body mass index
- F/B
Firmicutes/Bacteroidetes
- HCY
homocysteine
- HFD
high-fat diet
- IBD
inflammatory bowel disease
- IH
intermittent hypoxia
- IL-6
interleukin-6
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
lipopolysaccharide
- NO
nitric oxide
- OSAHS
obstructive sleep apnea–hypopnea syndrome
- PSG
polysomnography
- ROC-AUC
area under the receiver operating characteristic curve
- SCFA
short-chain fatty acid
- SF
sleep fragmentation
- TNF-α
tumor necrosis factor α
Competing interests
The authors declare that they have no financial or personal relationships with others who might inappropriately influence the results or interpretation in this manuscript.
Funding
We thank the Fujian Provincial Health and Family Planning Commission, China [grant number 2018-CX-36]; Fujian Province Science and Technology Project, China [grant numbers 2018J01290 and 2015J01445]; and Quanzhou Science and Technology Project, China [grant numbers 2018Z107, 2018N003S, 2017Z016, 2013Z55]; and the Young people training project from Fujian Province Health System [grant number 2014JQNJC20]. This article was also subsidized by Strait Exchanges Postdoctoral Funding Schemes of Fujian Province, China, and academic funding of the Second Affiliated Hospital of Fujian Medical University [grant number BSH001].
Author contribution
Conception and design: C.-Y.K., H.-P.Z., and Y.-M.Z. Acquisition of data: C.-Y.K., Q.-Q.L., H.-Z.S., J.-M.F., J.-H.Y., A.-K.H., and Y.-Q.L. Analysis and interpretation of data: C.-Y.K., D.C., H.-Z.S., H.-P.Z., and Y.-M.Z. Drafting/revising of the article: C.-Y.K., D.C., H.-P.Z., and Y.-M.Z.
References
- 1.Mirrakhimov A.E., Sooronbaev T. and Mirrakhimov E.M. (2013) Prevalence of obstructive sleep apnea in Asian adults: a systematic review of the literature. BMC Pulm. Med. 13, 10 10.1186/1471-2466-13-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavie L. (2015) Oxidative stress in obstructive sleep apnea and intermittent hypoxia-revisited-the bad ugly and good: implications to the heart and brain. Sleep Med. Rev. 20, 27–45 10.1016/j.smrv.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 3.Moreno-Indias I., Torres M., Montserrat J.M., Sanchez-Alcoholado L., Cardona F., Tinahones F.J.. et al. (2015) Intermittent hypoxia alters gut microbiota diversity in a mouse model of sleep apnoea. Eur. Respir. J. 45, 1055–1065 10.1183/09031936.00184314 [DOI] [PubMed] [Google Scholar]
- 4.Poroyko V.A., Carreras A., Khalyfa A., Khalyfa A.A., Leone V., Peris E.. et al. (2016) Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci. Rep. 6, 35405 10.1038/srep35405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R.K., Chang H.W., Yan D., Lee K.M., Ucmak D., Wong K.. et al. (2017) Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 15, 73 10.1186/s12967-017-1175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robles-Vera I., Toral M., Romero M., Jiménez R., Sánchez M., Pérez-Vizcaíno F.. et al. (2017) Antihypertensive effects of probiotics. Curr. Hypertens. Rep. 19, 26 10.1007/s11906-017-0723-4 [DOI] [PubMed] [Google Scholar]
- 7.Boulangé C.L., Neves A.L., Chilloux J., Nicholson J.K. and Dumas M.E. (2016) Impact of the GM on inflammation, obesity, and metabolic disease. Genome Med. 8, 42 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Indias I., Torres M., Sanchez-Alcoholado L., Cardona F., Almendros I., Gozal D.. et al. (2016) Normoxic recovery mimicking treatment of sleep apnea does not reverse intermittent hypoxia-induced bacterial dysbiosis and low-grade endotoxemia in mice. Sleep 39, 1891–1897 10.5665/sleep.6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durgan D.J., Ganesh B.P., Cope J.L., Ajami N.J., Phillips S.C., Petrosino J.F.. et al. (2016) Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension 67, 469–474 10.1161/HYPERTENSIONAHA.115.06672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farré N., Farré R. and Gozal D. (2018) Sleep apnea morbidity: a consequence of microbial-immune cross-talk? Chest 154, 754–759 10.1016/j.chest.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 11.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R.. et al. (2011) Enterotypes of the human gut microbiome. Nature 473, 174–180 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlon M.A. and Bird A.R. (2015) The impact of diet and lifestyle on gut microbiota and human health. Nutrients 7, 17–44 10.3390/nu7010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monneret D., Tamisier R., Ducros V., Garrel C., Levy P., Baguet J.P.. et al. (2012) The impact of obstructive sleep apnea on homocysteine and carotid remodeling in metabolic syndrome. Respir. Physiol. Neurobiol. 180, 298–304 10.1016/j.resp.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 14.Kao Y.C., Ko C.Y., Wang S.C. and Liu Y.P. (2016) Protective effects of quetiapine on metabolic and inflammatory abnormalities in schizophrenic patients during exacerbated stage. Chin. J. Physiol. 59, 69–77 [DOI] [PubMed] [Google Scholar]
- 15.Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R.M.. et al. (2014) An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6 10.1186/2049-2618-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L.. et al. (2014) Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 10.1038/nature13568 [DOI] [PubMed] [Google Scholar]
- 17.Xu D.Z., Lu Q., Kubicka R. and Deitch E.A. (1999) The effect of hypoxia/reoxygenation on the cellular function of intestinal epithelial cells. J. Trauma 46, 280–285 10.1097/00005373-199902000-00014 [DOI] [PubMed] [Google Scholar]
- 18.Taylor C.T. and Colgan S.P. (2007) Hypoxia and gastrointestinal disease. J. Mol. Med. 85, 1295–1300 10.1007/s00109-007-0277-z [DOI] [PubMed] [Google Scholar]
- 19.Kheirandish-Gozal L., Peris E., Wang Y., Tamae Kakazu M., Khalyfa A., Carreras A.. et al. (2014) Lipopolysaccharide-binding protein plasma levels in children: effects of obstructive sleep apnea and obesity. J. Clin. Endocrinol. Metab. 99, 656–663 10.1210/jc.2013-3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barceló A., Esquinas C., Robles J., Piérola J., De la Peña M., Aguilar I.. et al. (2016) Gut epithelial barrier markers in patients with obstructive sleep apnea. Sleep Med. 26, 12–15 10.1016/j.sleep.2016.01.019 [DOI] [PubMed] [Google Scholar]
- 21.Grootjans J., Thuijls G., Verdam F., Derikx J.P., Lenaerts K. and Buurman W.A. (2010) Non-invasive assessment of barrier integrity and function of the human gut. World J. Gastrointest. Surg. 2, 61–69 10.4240/wjgs.v2.i3.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parish J.M., Adam T. and Facchiano L. (2007) Relationship of metabolic syndrome and obstructive sleep apnea. J. Clin. Sleep Med. 3, 467–472 [PMC free article] [PubMed] [Google Scholar]
- 23.Romero-Corral A., Caples S.M., Lopez-Jimenez F. and Somers V.K. (2010) Interactions between obesity and obstructive sleep apnea: implications for treatment. Chest 137, 711–719 10.1378/chest.09-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz Y. and Moya-Pérez A. (2014) Microbiota, inflammation and obesity. Adv. Exp. Med. Biol. 817, 291–317 10.1007/978-1-4939-0897-4_14 [DOI] [PubMed] [Google Scholar]
- 25.Moustafa A., Li W., Anderson E.L., Wong E.H.M., Dulai P.S., Sandborn W.J.. et al. (2018) Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin. Transl. Gastroenterol. 9, e132 10.1038/ctg.2017.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macfarlane G.T. and Macfarlane S. (2012) Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 95, 50–60 10.5740/jaoacint.SGE_Macfarlane [DOI] [PubMed] [Google Scholar]
- 27.Duncan S.H., Louis P. and Flint H.J. (2004) Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70, 5810–2817 10.1128/AEM.70.10.5810-5817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durgan D.J. (2017) Obstructive sleep apnea-induced hypertension: role of the gut microbiota. Curr. Hypertens. Rep. 19, 35 10.1007/s11906-017-0732-3 [DOI] [PubMed] [Google Scholar]
- 29.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., Gratadoux J.J.. et al. (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maes M., Kubera M. and Leunis J.C. (2008) The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuroendocrinol. Lett. 29, 117–124 [PubMed] [Google Scholar]
- 31.Juraschek S.P., Bower J.K., Selvin E., Subash Shantha G.P., Hoogeveen R.C., Ballantyne C.M.. et al. (2015) Plasma lactate and incident hypertension in the atherosclerosis risk in communities study. Am. J. Hypertens. 28, 216–224 10.1093/ajh/hpu117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barreto F.M., Colado Simão A.N., Morimoto H.K., Batisti Lozovoy M.A., Dichi I. and Helena da Silva Miglioranza L. (2014) Beneficial effects of Lactobacillus plantarum on glycemia and homocysteine levels in postmenopausal women with metabolic syndrome. Nutrition 30, 939–942 10.1016/j.nut.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 33.Naruszewicz M., Johansson M.L., Zapolska-Downar D. and Bukowska H. (2002) Effect of Lactobacillus plantarum 299v on cardiovascular disease risk factors in smokers. Am. J. Clin. Nutr. 76, 1249–1255 10.1093/ajcn/76.6.1249 [DOI] [PubMed] [Google Scholar]
- 34.Menzel D., Haller H., Wilhelm M. and Robenek H. (2018) L-Arginine and B vitamins improve endothelial function in subjects with mild to moderate blood pressure elevation. Eur. J. Nutr. 57, 557–568 10.1007/s00394-016-1342-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumas-Bilak E., Carlier J.P., Jean-Pierre H., Mory F., Teyssier C., Gay B.. et al. (2007) Acidaminococcus intestini sp. nov., isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 57, 2314–2319 10.1099/ijs.0.64883-0 [DOI] [PubMed] [Google Scholar]
- 36.Wu F., Guo X., Zhang J., Zhang M., Ou Z. and Peng Y. (2017) Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp. Ther. Med. 14, 3122–3126 10.3892/etm.2017.4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Christophersen C.T., Sorichm M.J., Gerber J.P., Angley M.T. and Conlon M.A. (2013) Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 4, 42 10.1186/2040-2392-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai C.C., Cheng A., Liu W.L., Tan C.K., Huang Y.T., Chung K.P.. et al. (2011) Infections caused by unusual Methylobacterium species. J. Clin. Microbiol. 49, 3329–3331 10.1128/JCM.01241-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yumoto T., Naito H., Ihoriya H., Tsukahara K., Ota T., Watanabe T.. et al. (2018) Raoultella planticola bacteremia-induced fatal septic shock following burn injury. Ann. Clin. Microbiol. Antimicrob. 17, 19 10.1186/s12941-018-0270-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opstelten J.L., Plassais J., van Mil S.W., Achouri E., Pichaud M., Siersema P.D.. et al. (2016) Gut microbial diversity is reduced in smokers with Crohn’s disease? Inflamm. Bowel. Dis. 22, 2070–2077 10.1097/MIB.0000000000000875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Temuujin U., Chi W.J., Park J.S., Chang Y.K., Song J.Y. and Hong S.K. (2012) Identification and characterization of a novel β-galactosidase from Victivallis vadensis ATCC BAA-548, an anaerobic fecal bacterium. J. Microbiol. 50, 1034–1040 10.1007/s12275-012-2478-6 [DOI] [PubMed] [Google Scholar]
- 42.Poeker S.A., Geirnaert A., Berchtold L., Greppi A., Krych L., Steinert R.E.. et al. (2018) Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model (PolyFermS). Sci. Rep. 8, 4318 10.1038/s41598-018-22438-y [DOI] [PMC free article] [PubMed] [Google Scholar]