Abstract
Artemisinin, an important compound produced by Artemisia annua, is the active ingredient in the treatment of malaria. Jasmonic acid, one of the phytohormones, is an important elicitor of artemisinin biosynthesis by enhancing transcription levels of transcription factors. SPL transcription factors are plant-specific transcription factors of plant growth, development, and secondary metabolism regulation. However, to date, the SPL transcription factors that regulate artemisinin biosynthesis is currently unclear. Here, we show that an SPL transcription factor can positively regulate artemisinin biosynthesis by binding to the promoter of artemisinin biosynthetic pathway genes. We screened AaSPL2 by gene expression profiles analysis in 14 SPL transcription factors. We demonstrated that AaSPL2 can activate the promoter of DBR2 by dual-LUC assy. Moreover, in the AaSPL2 overexpression plants, the artemisinin content was increased by 33–86%, and in the AaSPL2 -RNAi transgenic plants, artemisinin content was decreased by 33–65%. These data suggest that AaSPL2 and DBR2 interact with a “GTAC” cis-element in the DBR2 promoter, mediating the transcriptional activation of DBR2 in response to JA and resulting in the improvement on artemisinin content.
Keywords: SPL transcription factor, jasmonic acid, transcript regulation, artemisinin, Artemisia annua
Introduction
Malaria is a global infectious disease that 3.3 billion people are easy to infect it. Artemisinin combination therapy (ACT) has been the frontline treatment for malaria in recent years (World Health Organization, 2014). Artemisinin is produced in the glandular trichomes of Artemisia annua L (Duke and Paul, 1993). The artemisinin biosynthetic pathway is well studied (Figure 1). In recent years, the semisynthesis of artemisinin following microbial production of artemisinic acid has already succeeded (Paddon et al., 2013). Because the price of artemisinin is fluctuating, the microbial deriving artemisinin may supplement the market but not replace agricultural production (Peplow, 2016). Our aim is to improve the content of artemisinin and reduce production costs.
FIGURE 1.
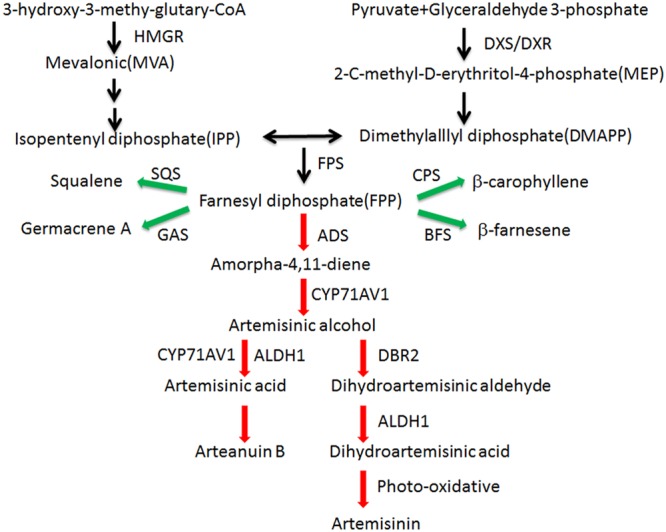
Artemisinin biosynthetic pathways in A. annua. Artemisinin derived from MVA and MEP pathways. Enzymes marking in red are the artemisinin biosynthetic pathway genes and enzymes marking in green are the artemisinin biosynthetic branch-pathway genes. ADS: amorpha-4,11-diene synthase (AF138959); ALDH1: aldehyde dehydrogenase 1 (FJ809784); BFS: β-farnesene synthase (AY835398); CPS:β-caryophyllene synthase (AF472361); CYP71AV1: amorphadiene-12-hydroxylase (DQ453967); DBR2: artemisinic aldehyde Δ11(13) reductase (EU704257); DXR: 1-deoxy-Dxylulose-5-phosphate reductoisomerase (AF182287); DXS: 1-deoxy-D-xylulose-5-phosphate synthase (AF182286); FPS: farnesyl diphosphate synthase (U36376); GAS: germacrene A synthase (DQ447636); HMGR:3-hydroxy-3-methyl-glutaryl coenzyme A reductase (AF142473); SQS: squalene synthase (AY445505).
In general, the average content of A. annua lies between 0.5–1.2%, while the purification yields are typically in the range of 50–80% of the crude extract (Kindermans et al., 2007). Because of the short supply and high costs of artemisinin, some strategies are employed to increase artemisinin production, such as blocking branch pathway genes (Zhang et al., 2009; Chen et al., 2011; Lv et al., 2016b), regulating transcription factors levels (Yu et al., 2015; Lv et al., 2016a) and modulating trichome numbers (Shi et al., 2017b; Yan et al., 2018). Semisynthesis of artemisinin by fermentation from engineered yeast is a feasible method of production of a precursor molecule, DHAA, which is easy to convert to artemisinin (Westfall et al., 2012).
Plant phytohormones increasing the secondary metabolites can be defined: the terpenoids, alkaloids and phenylpropanoids. In general, jasmonic acid (JA) is an important conserved elicitor of plant secondary metabolism. It triggers extensive transcriptional reprogramming and leads to an increase in the expression level of the gene encoding the key enzyme for secondary metabolism biosynthetic pathway (De Geyter et al., 2012). JA can induce the mRNA levels of ORCA3 in Catharanthus roseus, which is an important transcription factor for the regulation of terpenoid indole alkaloid biosynthesis (Zhang et al., 2011). ERF189 can be induced by JA in tobacco and it regulates the nicotine biosynthesis by directly binding to the GCC-box in the promoter region of putrescine N-methyltransferase (PMT) (Shoji et al., 2010). JA is involved in the biosynthesis of artemisinin as well. Previously, JA increased the expression levels of transcription factor AaWRKY1, which enhanced artemisinin content by directly binding to the promoter of ADS (Ma et al., 2009). We have found that AaNAC1 was induced by JA and played an important role in the modulation of artemisinin biosynthesis (Lv et al., 2016a). Many transcription factors can be induced by JA and many are involved in artemisinin biosyntheses, such as AaERF1/AaERF2, AaORA, and AaMYC2. The AP2/ERF transcription factors AaERF1/AaERF2 and AaORA positively regulate the biosynthesis of artemisinin (Yu et al., 2012; Lu et al., 2013). The bHLH transcription factor AaMYC2 is a positive regulator for artemisinin biosynthesis by binding to the G-box-like motifs within the promoters of the artemisinin biosynthetic pathway gene CYP71AV1 and DBR2 (Shen et al., 2016).
SQUAMOSA Promoter-Binding Protein-Like (SPL) genes are functionally diverse, including modulation of grain size, grain shape, branching, vegetative phase change, shoot regeneration and flowering time (Wang et al., 2009; Jiao et al., 2010; Zhang et al., 2015; Si et al., 2016). Recently, some studies indicate that SPL transcription factors can regulate secondary metabolism biosynthesis. AtSPL9 can regulate anthocyanin accumulation by inhibiting expression levels of anthocyanin biosynthetic pathway genes through destabilization of the Myb-bHLH-TTG1 complex (Gou et al., 2011). At the same time, AtSPL9 can regulate the content of (E)-β-caryophyllene in Arabidopsis thaliana by binding to the promoter of TPS21 (Yu et al., 2015). PpSPL1 negatively regulates the anthocyanin level in the peach flesh by inhibiting the transactivation activity of NAC transcription factors BL and PpNAC1 through destabilization of the Myb-bHLH-TTG1 complex (Zhou et al., 2015).
Here, we also provide evidence that SPL transcription factors can regulate the artemisinin content by binding to the promoter of artemisinin biosynthesis gene DBR2. We provide evidence that overexpression of AaSPL2 enhances artemisinin content, indicating that AaSPL2 is a positive transcription factor of artemisinin biosynthesis in A. annua.
Materials and Methods
Plant Material
Seeds of “Huhao 1” (Shen et al., 2016), which is a high artemisinin yielding A. annua strain, were first surface-sterilized in 75% ethanol for 2 min, followed by 10% (v/v) NaOCl (sodium hypochlorite) solution for 10 min, and rinsed three times with sterile water. Subsequently, the seeds were planted on MS medium (Murashige and Skoog, 1962) with 3.0% sucrose (m/v) and 0.8% agar (m/v) and cultured with a photoperiod of 16 h light/8 h dark and light at 8,000 Lux at 28°C. Three weeks later, the leaves of young seedlings of A. annua (about 5–7 cm in height) were cut and used as explants for transformation with the Agrobacterium tumefaciens-mediated leaf disk transformation method (Zhang et al., 2009).
Seeds of tobacco (Nicotiana benthamiana) were sown directly on soil (vermiculite: perlite: peat moss = 7: 0.5: 2), 1 week later, the tobacco seedlings were transferred into the new pots and cultured with the same conditions as A. annua. Five weeks later, the leaves of tobacco can be used for A. tumefaciens infiltration assay.
Construction and Transformation of A. annua
The ORF of AaSPL2 was introduced into the BamHI and SacI sites of pCAMBIA2300 (CAMBIA, Canberra, Australia) under the driven of CaMV35S promoter to generate CaMV35S:: AaSPL2 (overexpression vector). For RNAi interference assay, a 300 bp AaSPL2 fragment was fused into PENTR vector via the BP reaction (Invitrogen, United States) and then introduce into the pHELLSGATE12 vector via the LR reaction (Invitrogen, United States) to generate the final RNAi- AaSPL2 vector.
The overexpression constructs OX-AaSPL2 and RNAi constructs AaSPL2-RNAi were introduced into A. tumefaciens strain EHA105 by a conventional freezing-and-melting method (Zhang et al., 2009). The method of transformation of A. annua was according to the previous description (Lv et al., 2016b).
Plant Hormone Treatments
MJ was purchased from Sigma-Aldrich (St Louis, MO, United States). For plant hormone treatment, 4-week-old A. annua seedlings were treated with 100 μM MJ (Maes et al., 2011; Yu et al., 2012; Shen et al., 2016). Then the treated plants were sampled at time point 0, 1, 3, 6, 9, 12, and 24 h for RNA extracting and Q-PCR analysis.
Isolation of Genomic DNA and PCR Analysis
The cetyltrimethylammonium bromide (CTAB) method was employed to obtain the genomic DNA of the transgenic plants (Allen et al., 2006). The forward primers of AaTGA6 were used to detect the transgenic plants. A 20 bp sequence of the vector was designed as reverse primers. The 20 ml PCR with 10 pmol forward primer, 10 pmol reverse primer, 50 ng of plant genomic DNA, 7 ml of sterilized water and 10 ml of rTaq DNA polymerase mix (TaKaRa, Japan) was performed. The amplification procedure consisted of a 5-min incubation at 95°C, followed by 30 cycles of 95°C for 30 s, and 60 s at 55°C for primer annealing and 72°C for 600 s elongation.
RNA Isolation and Real-Time PCR Analysis
Young leaves of A. annua plants were used for RNA isolation by plant RNA Kit (Tiangen) according to the manufacturer’s instructions. The isolated total RNA was reverse-transcribed into cDNA using the TaKaRa cDNA synthesis kit. qRT-PCR assays using SYBR Green PCR Master Mix kit (Tiangen) as a detector were performed in Roch LightCycler®96 system according to the manufacturer’s instructions. The comparative cycle threshold (CT) method was estimated relative transcript levels of samples and means of triplicate independent PCRs. The β-actin gene of A. annua was used to estimate relative mRNA levels. The qRT-PCR primers are listed in Supplementary Table S1. The process for amplification was 5 min at 95°C, followed by 40 cycles of 95°C for 10 s, and 40 s at 60°C for primer annealing and elongation. Then a dissociation stage for 20 s at 95°C, 40 s at 60°C, and 20 s at 95°C.
The 2-ΔΔCT method was used for estimating the relative gene expression level according to the following formula (Livak and Schmittgen, 2001):
Yeast One-Hybrid Assay
To analyze the specific binding of AaSPL2 to GTAC-box of the DBR2 promoters, the full length of AaSPL2 was fused into the EcoRI–XhoI sites of the activation domain of the vector pB42AD (Clontech). Equal volume of the GTAC-box forward primer (10 μM) and reverse primer (10 μM) (Table 1) were mixed and incubated at these conditions: 95°C for 1 min, 85°C for 1 min, 75°C for 1 min, 65°C for 1 min, 55°C for 1 min, 45°C for 1 min, 35°C for 1 min, 25°C for 1 min. The reaction primer products were inserted into P178 vector (gifted by Dr. Yang Hongquan of Shanghai Jiao Tong University, Shanghai, China) with a single restriction site XhoI to form the reporter vector. Vectors containing the transcription factor gene and 3 × GTAC-box were introduced into the yeast strain EGY48 by LiAc mediated transformation method (Gietz and Schiestl, 2007). Yeast cells were selected on the synthetic dropout (SD) base medium with deficient in Trp and Ura for 3 days at 30°C. Then yeast cells were resuspended with sterile water and dropped on an 5-bromo-4-chloro-3-indolyl-beta-Dgalactopyrano-side (X-gal) medium plate (Zhang et al., 2015). The sequences of the 3 × GTAC-box fragment are listed in Supplementary Table S1.
Table 1.
The character of SPLs in A. annua.
| Gene name | Accession number | Gene length | PI | MW | miR156 target site |
|---|---|---|---|---|---|
| AaSPL1 | MH090670 | 2484 | 4.9 | 206.73 | |
| AaSPL2 | MH090671 | 954 | 5.09 | 79.08 | |
| AaSPL3 | MH090672 | 2379 | 4.92 | 195.68 | |
| AaSPL4 | MH090673 | 879 | 5.1 | 72.71 | ✓ |
| AaSPL5 | MH090674 | 423 | 5.3 | 33.52 | ✓ |
| AaSPL6 | MH090675 | 2865 | 4.89 | 234.37 | |
| AaSPL7 | MH090676 | 1131 | 5.09 | 93.74 | ✓ |
| AaSPL8 | MH090677 | 1167 | 5.04 | 97.21 | ✓ |
| AaSPL9 | MH090678 | 906 | 5.1 | 74.37 | ✓ |
| AaSPL10 | MH090679 | 1284 | 5.02 | 107.01 | ✓ |
| AaSPL11 | MH090680 | 447 | 5.07 | 35.32 | |
| AaSPL12 | MH090681 | 516 | 5.27 | 40.57 | ✓ |
| AaSPL13 | MH090682 | 1197 | 5.03 | 99.08 | ✓ |
| AaSPL14 | MH090683 | 555 | 5.23 | 44.87 | ✓ |
Dual-LUC Assay
For the dual-LUC assay, the promoter of DBR2 was fused to firefly luciferase gene on the plasmid pGreenII 0800-LUC. The sequenced plasmid was introduced into A. tumefaciens strain EHA105. Then incubated Agrobacterium cells were harvested by centrifugation and resuspended in MS medium (containing 10 mM MES and 150 mM acetosyringone) to an OD600 nm of 0.6. After 3 h incubation at room temperature, bacteria (reporter strain DBR2pro::LUC mixed with effector strain 35Spro::AaSPL2 1:1) were injected into tobacco leaves. Forty-eight hours’ later, the injected tobacco leaves were collected for dual-LUC assay. The dual-Luciferase Reporter Assay system of Promega was used for dual-Luc assays according to the manufacturer’s instructions.
Quantification of Artemisinin Using HPLC-ELSD (HPLC-Evaporative Light Scattering Detection)
The upper parts leaves of A. annua were collected at pre-flowering stage and dried at 50°C for 24 h. then all the leaves were ground into powder. A sample of 0.1 g powder was suspended in ethanol (2 mL) for ultrasonication. Sampling was carried out in triplicate. Thirty min later, Samples were centrifuged for 15 min, 4°C, at 12 000 g. The supernatant was filtered through a 0.22 mm pore size filter and then analyzed by the Waters Alliance 2695 HPLC system. Artemisinin standard (98% pure) was purchased from Sigma. The approach for detecting artemisinin using HPLC-ELSD has already been described (Zhang et al., 2009).
Results
Characterization of AaSPLs in A. annua
The SQUAMOSA promoter-binding protein (SBP) genes contain a conserved domain, which encodes 76 amino acid residues in length for the interaction with DNA (Cardon et al., 1999). To identify the SPLs in A. annua, the putative conserved SBP domain was used for searching the transcriptome database (Graham et al., 2010; Hao et al., 2017) using the HMMER3.0 search program1 (Finn et al., 2011). Through molecular cloning, we identified a total of 14 SPLs (AaSPL1∼AaSPL14) in A. annua with the GenBank accession number (Table 1). The open reading frame (ORF) of the SPL is varied in length from 447bp to 2865bp. All the SPLs encode protein products 149 to 955 amino acids with the calculated isoelectric point2 of 4.89 to 5.3. The details of the gene character of SPLs are shown in the Table 1.
MiR156 plays important role in modulation of transcription levels of targeting SPLs for translational repression or cleavage (Wang et al., 2009; Wu et al., 2009). So the miR156 in A. annua has been studied recently (Pani et al., 2011). To confirm the targets of miR156, we screened all of the SPLs in A. annua. Nine of SPLs have target site of miR156 and they are predicted to be the potential targets of miR156 (Table 1).
Expression Patterns of SPLs in A. annua
JA plays a crucial role in inducing the artemisinin accumulation (Jing et al., 2009; Maes et al., 2011; Yu et al., 2012). We hypothesized that JA may be used for screening the SPL which takes part in modulating artemisinin biosynthesis in A. annua. First, we checked mRNA levels of AaSPL1∼ AaSPL14 at different time points (0, 1, 3, 6, 9, 12, and 24 h) after treatment with JA (100 μM). To this end, the expression intensity and hierarchical clustering (HCL) of SPL genes were formatted as a heatmap by Mev4.9.0 software.3 Exogenous application of JA to A. annua seedlings resulted in strong activation of both AaSPL2 and AaSPL13 (Figure 2A), indicating that both of them may play a role in the regulation of artemisinin.
FIGURE 2.
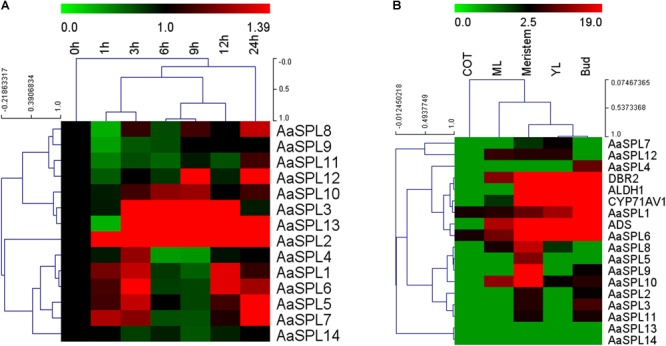
Heat map representation of expression profiles of SPL transcription factors. (A) The heat map represents the SPL transcription factors expression by Q-PCR under the treatment of JA at time point of 0,1,3,6,9,12, and 24 h. (B) Expression data for SPL genes, ADS, CYP71AV1, DBR2, and ALDH1 in cotyledon, young leaves, mature leaves, buds and meristems. Expression data were detected by Q-PCR and the results were visualized using Mev 4.9 software. β-actin was used as a reference. Red indicates high expression; green, low expression.
In the process of artemisinin biosynthesis, the artemisinin biosynthetic pathway genes ADS, CYP71AV1, DBR2, and ALDH1 mRNA levels were accumulated in the young leave tissues (YL) and buds, confirming previous studies that artemisinin was produced in the glandular trichomes (Maes et al., 2011). To confirm the expression files of SPLs of A. annua, we tested SPL expression levels in the RNA-sequence of different tissues (Graham et al., 2010). As shown in Figure 2B, heatmap results indicated that the mRNA levels of AaSPL2 are higher in bud compared with AaSPL13, therefore, we focus our attention on AaSPL2 according to the results of the expression profiles of SPLs.
Characterization of AaSPL2
SPL transcription factors are known to regulate the biosynthesis of secondary metabolites, such as sesquiterpene, flavonoid (Gou et al., 2011; Yu et al., 2015). We hypothesized that SPL may modulate the biosynthesis of sesquiterpene lactone artemisinin by directly or indirectly regulation of artemisinin biosynthetic pathway genes. To investigate the gene function and control mechanism, a 954-bp segment of the AaSPL2 ORF region was isolated from cDNA. The full-length cDNA of AaSPL2 encodes a polypeptide of 318 amino acid residues with a calculated mass of 79 kDa and a pI of 5.09 (Table 1). We also detected the gene expression level of AaSPL2 in leaf0∼leaf7 (L0∼L7) (Figure 3A). The Q-PCR results indicated that AaSPL2 expressed at a high level in the L0, L4, L5, and L6 (Figure 3B).
FIGURE 3.
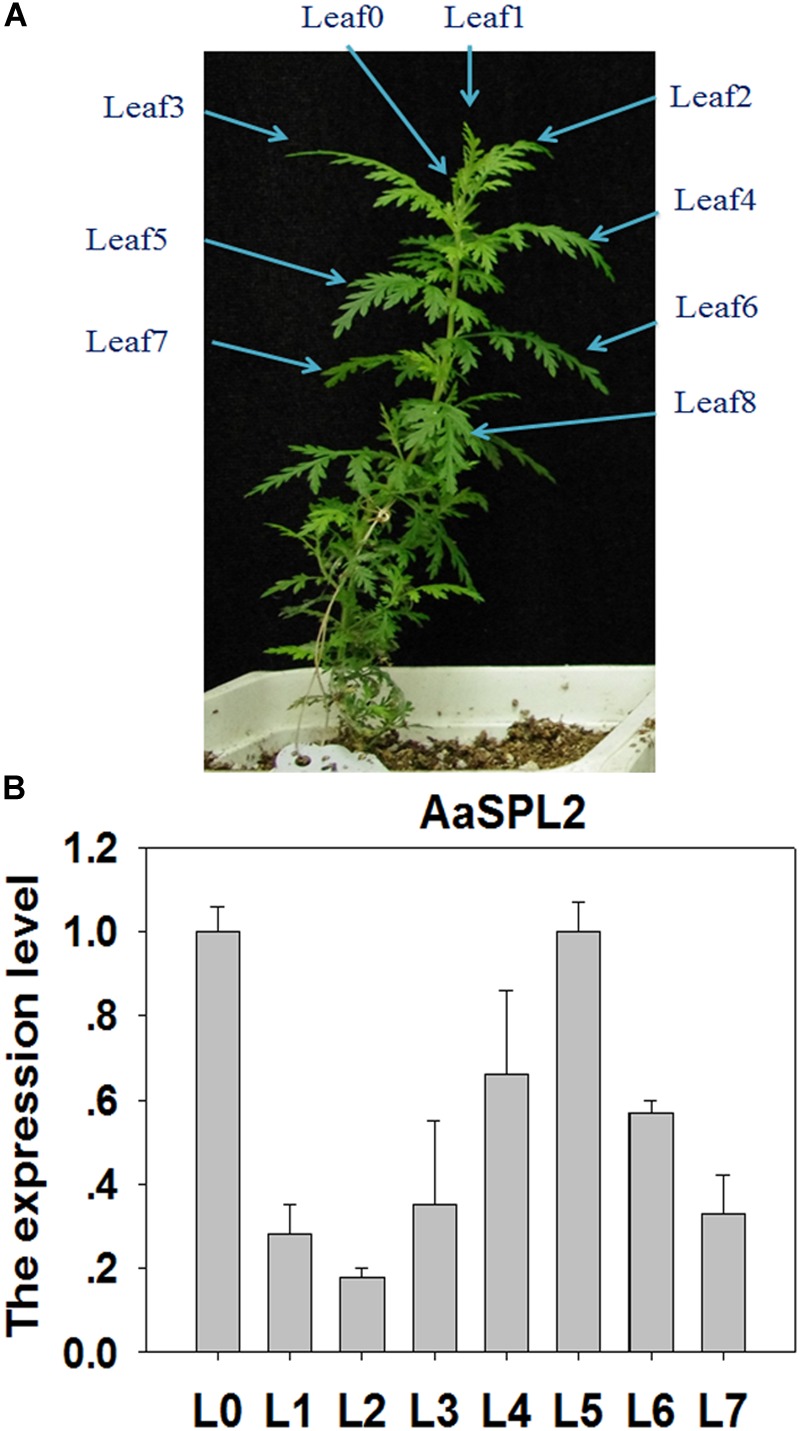
Expression profiles of AaSPL2 in the different leaves. (A) L0 to L7 were labeled on the 6-week-old A. annua seedling. (B) Expression of AaSPL2 in the different leaves. L0 to L7 means leaves 0 to leaves 7. The expression level is normalized by Leaf0 value. Bars represent means ± SE (n = 3).
Phylogenetic Analysis of AaSPL2 Proteins
The analysis on full-length nucleotide sequence showed that AaSPL2 contains a conserved SBP domain in the 289–471 regions. A phylogenetic tree by using nucleic acid sequences was built to investigate the phylogenetic relationships among the SPL proteins in A. annua, Arabidopsis and rice other species. The results indicated that AaSPL2 belongs to the subfamily of AtSPL2/10/11 transcription factors (Figure 4). AtSPL2/10/11 has been reported to modulate vegetative phase change and trichome development (Shikata et al., 2009), suggesting that AaSPL2 has a potential role in the regulation of phase transition or trichome initiation in A. annua.
FIGURE 4.
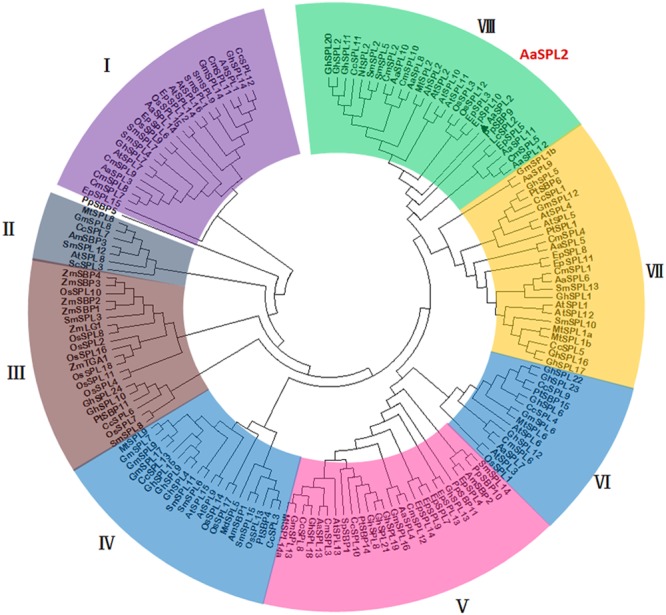
Phylogenetic tree of SPL transcription factors from selected plant species. The tree was constructed using MEGA 7 software with Neighbor–Joining method: bootstrap analysis of 1,000 replicates. The numbers at the nodes indicate the bootstrap values. The bar at the bottom indicates the relative divergence of the sequences examined. AaSPL2 was marked with a black triangle.
Subcellular Localization of AaSPL2
To investigate the subcellular localization of the AaSPL2 protein, a YFP was fused to the C-terminus of AaSPL2 and the AaSPL2-YFP construct was expressed under the control of the double 35S Cauliflower mosaic virus (CaMV) promoter. Agroinfiltration method was performed to infiltrate N. tabacum leaves (Zhang et al., 2015). Controls showed GFP expressed throughout the cytoplasm (Figure 5), whereas the AaSPL2-YFP fusion was targeted to the nucleus (Figure 5).
FIGURE 5.
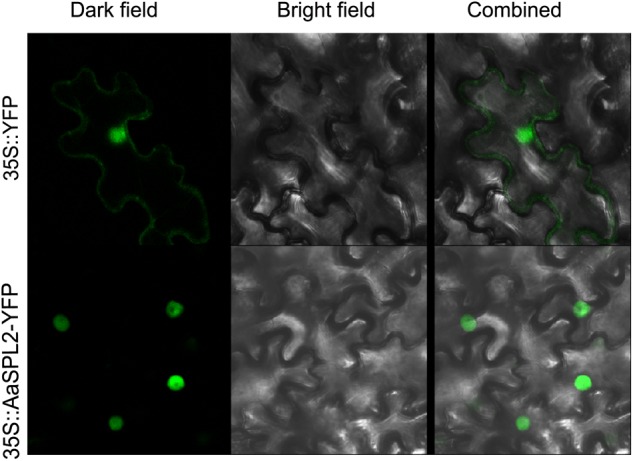
Subcellular localizations of AaSPL2 in N. benthamiana leaves. Constructs 35S::AaSPL2–YFP were introduced into Agrobacterium and transferred into N. benthamian by infiltration. Constructs 35S::YFP were used as control.
AaSPL2 Activates the Transcription of DBR2 in vivo
The SQUAMOSA promoter binding protein transcription factors can activate the transcription of MADS transcription factors by binding to “GTAC” cis-element in the promoter (Franco-Zorrilla et al., 2014). There are 9 “GTAC” cis-elements in the promoter of DBR2 (Jiang et al., 2014). Thus, we hypothesized that AaSPL2 may potentially induce the transcription of DBR2. First, a dual luciferase assay (dual-LUC) was employed to detect if the expression of DBR2 can be regulated by the product of the transcription factor AaSPL2 (Figure 6A). The results of dual-LUC indicated that AaSPL2 activates the transcription of DBR2 (Figure 6B).
FIGURE 6.
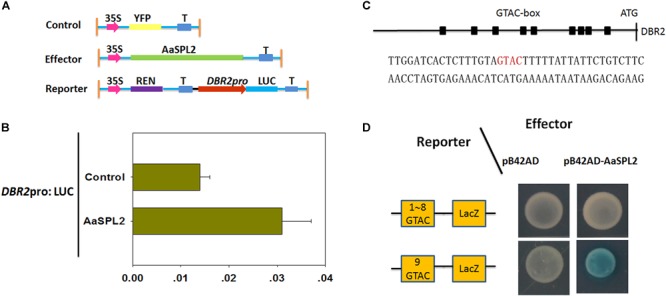
AaSPL2 promotes transactivation of DBR2. (A) A Schematic representation of control, effectors and reporters used in the dual-LUC assays is shown. The LUC reporter constructs harboring DBR2 promoters were used as reporter (DBR2pro: LUC). (B) Effector constructs harboring the AaSPL2 full length were driven by the CaMV35S promoter. The LUC activities were normalized by Renilla (REN) luciferase activities. The YFP driven by 35S promoter was used as negative control. (C) A Schematic representation of the “GTAC” elements in the DBR2 promoters. Black box indicates the “GTAC” elements in the DBR2 promoters. The DNA sequences used in the Y1H assays is shown. (D) Yeast one-hybrid assay indicates AaSPL2 binding to the promoter of DBR2. A Schematic in the left representation of the reporter constructs was used in the yeast one-hybrid assay. Right panel represents the yeast cells. Yeast strains were diluted with sterile water and 6 μL of each dilution was plated on synthetic dropout media without Trp and Ura (SD-T-U) plate plus X-gal.
To further substantiate this finding, we tested whether AaSPL2 is able to bind to the “GTAC” box in the DBR2 promoters. Therefore, the ninth “GTAC” elements were cloned into the reporter vector p178 (Figure 6C). The results of the Y1H indicated that AaSPL2 can bind to the “GTAC” of the DBR2 promoters directly (Figure 6D).
Overexpression of AaSPL2 in A. annua Increases Artemisinin Biosynthesis
The function of SPL transcription factors is diverse. To investigate the functional role of AaSPL2, we overexpressed AaSPL2 (abbreviated as AaSPL2-OX) in A. annua. The transgenic plants were detected by PCR. Overexpression of AaSPL2 in A. annua resulted in enhanced AaSPL2 expression upon wild-type (Figure 7). The expression levels of artemisinin biosynthetic pathway genes ADS, CYP71AV1, DBR2, and ALDH1 were also detected. Similar results were obtained with the expression levels of ADS, CYP71AV1, DBR2, and ALDH1 (Figure 7). Therefore, to investigate whether AaSPL2 is a positive regulator for artemisinin biosynthesis, the artemisinin content was detected. In the AaSPL2 overexpression plants, the artemisinin and DHAA content was increased to 33–86% and 26–159%, respectively (Figure 8A and Supplementary Figure S1A). In the AaSPL2-RNAi transgenic plants, the mRNA levels of AaSPL2 and artemisinin biosynthetic pathway genes all decreased (Figure 7). The artemisinin and DHAA content was decreased by 33–65% and 3–16%, respectively (Figure 8B and Supplementary Figure S1B). Thus, AaSPL2 is a positive regulator for artemisinin biosynthesis. The transgenic plants displaying normal phenotype overexpressed in Arabidopsis (Supplementary Figure S2) and in A. annua (Supplementary Figure S3).
FIGURE 7.
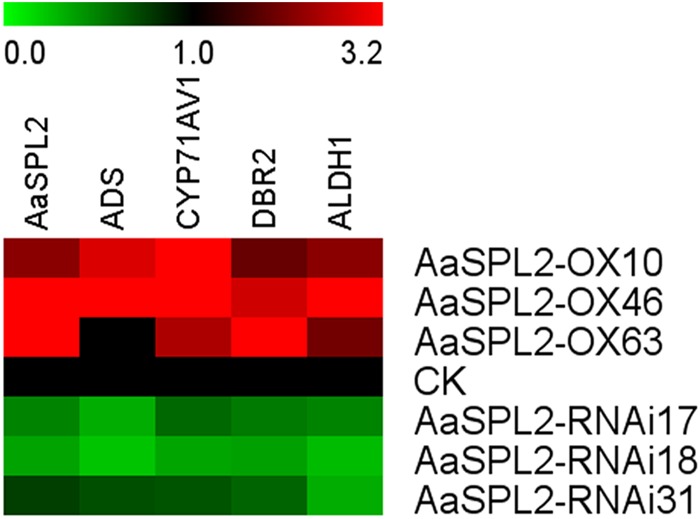
Heat map showing the expression levels of AaSPL2 and artemisinin biosynthesis pathway genes. Expression data were detected by Q-PCR and the results were visualized using Mev 4.9 software. β-actin was used as a reference. Red indicates high expression; green, low expression.
FIGURE 8.
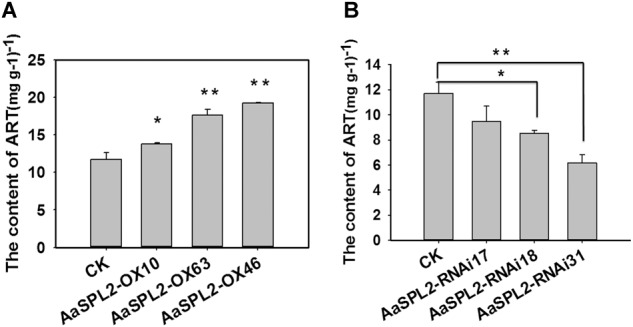
Artemisinin content in the transgenic plants was determined by HPLC. (A) Artemisinin content was detected in the AaSPL2 overexpression plants. (B) Artemisinin content was detected in the AaSPL2 RNAi plants. Error bars indicate SD (n = 3). ∗p < 0.05, ∗∗p < 0.01.
Discussion
MJ Induced Artemisinin Biosynthesis
Since JA and its cyclic precursors and derivatives play important roles in inducing the genes encoding enzymes involving in the biosynthesis of secondary metabolites in the plant (Van der Fits and Memelink, 2000; De Boer et al., 2011). So JA becomes the hotspot as the secondary metabolites inducer. Previously, it was shown that overexpression of artemisinin biosynthetic pathway genes (ADS, CYP71AV1, DBR2, ALDH1, CPR) can redirect carbon flux to the artemisinin pathway and increase artemisinin contents for several folds (Chen et al., 2013; Shi et al., 2017a). JA enhances artemisinin biosynthetic pathway genes mRNA levels and plays the same roles as overexpression artemisinin biosynthetic pathway genes (Maes et al., 2011). JA improved artemisinin content by increasing mRNA levels of transcription factors as well, which regulate gene expression level through specific binding to cis-acting elements in the target genes promoter. Many transcription factors are involved in JA signaling cascades and regulate the biosynthesis of artemisinin. Such as AP2/ERF transcription factors AaERF1 and AaERF2, they bind to the promoter of ADS and CYP71AV1 directly (Yu et al., 2012). Another AP2/ERF transcription factor AaORA (Lu et al., 2013), which increases artemisinin content by improving artemisinin biosynthetic pathway genes mRNA levels. AaMYC2 and AaNAC1 activate the expression level of artemisinin biosynthesis pathway genes CYP71AV1, DBR2, and ADS, respectively (Lv et al., 2016a; Shen et al., 2016). AaSPL2 mRNA level was enhanced under the treatment of JA and the content of artemisinin was increased in the AaSPL2 overexpression plants also (Figure 2). AtSPL9 attended the JA pathway through interacting with known JA signaling factor (such as JAZ3) to regulate biosynthesis of secondary metabolism glucosinolate and β-caryophyllene (Yu et al., 2015; Mao et al., 2017). On the other hand, JA improved artemisinin content by increasing of trichome number (Maes et al., 2011). Artemisinin was specifically synthesized in the glandular trichome and was related to the trichome number. The increase of trichome number may decrease feedback Inhibition of artemisinin content (Arsenault et al., 2010). In our study, trichome number changes not obviously (Supplementary Figure S3F), suggesting that AaSPL2 may not take part in trichome regulation.
SPL Transcription Factors Regulation of Terpenoid Products
Through genome-wide identification, a total of 14 SPLs have been found in Salvia miltiorrhiza (Zhang et al., 2014). There are 19 SPL genes in the rice and 16 SPLs in Arabidopsis (Birkenbihl et al., 2005; Manning et al., 2006). A systematical study reported that 59 SPLs were identified in the Genome-wide of G. hirsutum. SPLs play important roles in fiber initiation, branching, root growth, grain size and yield, suggesting that SPL transcription factors play key roles in G. hirsutum growth and development. The reasons for so many SPLs in the G. hirsutum are the tandem duplication events or by assembly error (Cai et al., 2018). In this study, a total of 14 SPLs were cloned. AaSPL2 mainly expressed in the buds (Figure 2B) and induced by MJ (Figure 2A), play important role in artemisinin biosynthesis (Figure 8). However, AaSPL3 has the same expression patterns with AaSPL2 (Figure 2A,B), therefore, it is a potential gene for improving artemisinin content.
SPL transcription factors can regulate gene expression through binding to cis-regulatory specific core “GTAC” consensus sequences in the promoters of their target genes. Previously, the core “GTAC” consensus sequence of DFR has been identified as the binding motif of SPL9 (Gou et al., 2011). Plant terpenoids have important physiological and ecological functions, such as serving as signals in biocommunications and interactions, and phytoalexins in defense against pests and pathogens (Sharma et al., 2017). Terpene synthase 21 (TPS21) is responsible for the vast majority of floral sesquiterpenes and it can catalyze the conversion of FPP into β-caryophyllene. Transcription factor SPL9 directly binds to TPS21 promoters and activates its expression to regulate the biosynthesis of β-caryophyllene (Yu et al., 2015). Artemisinin is a sesquiterpene, hence, we postulated that SPL in A. annua may function similarly as an SPL in Arabidopsis. The phylogenetic tree indicated that AaSPL2 may homologous to the AtSPL3 in Arabidopsis.
The expression patterns indicated that AaSPL2 mainly expressed in the buds (Figure 2), which is partly same as artemisinin biosynthetic genes expression patterns, so AaSPL2 in A. annua may play the role in the regulation of sesquiterpene artemisinin. AaSPL2-OX and AaSPL2-RNAi results show that SPL2 can modulate artemisinin (Figure 8) and DHAA accumulation (Supplementary Figure S1). The fact that the overexpression of AaSPL2 partially increases the transcription levels of artemisinin biosynthetic gene ADS, CYP71AV1, DBR2, and ALDH1 supported the hypothesis that enhancing of AaSPL2 expression level may be a plausible mechanism for increase artemisinin content.
Author Contributions
ZL, KT, WC, and LZ conceived and designed the entire research plans. BP, YL, and ZL performed the most of the experiments. BP and ZL provided the technical assistance. ZL wrote the manuscript. ZL, YW, and LZ helped with the organization and editing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- ADS
amorpha-4,11-diene synthase
- ALDH1
aldehyde dehydrogenase 1
- CYP71AV1
amorphadiene 12-hydroxylase
- DBR2
artemisinic aldehyde Δ11(13) reductase
- DHAA
dihydroartemisinic acid
- YFP
yellow fluorescent protein
Funding. This work was funded by National Science and Technology Major Project of the Ministry of Science and Technology of China (2017ZX09101002-003-002), China Postdoctoral Science Foundation (Grant No. 46108), and China Transgenic Research Program (Grant No. 2016ZX08002001-006).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00409/full#supplementary-material
DHAA content in the transgenic plants was determined by HPLC. (A) DHAA content was detected in the AaSPL2 overexpression plants. (B) DHAA content was detected in the AaSPL2 RNAi plants. Error bars indicate SD (n = 3). ∗∗p < 0.01.
The phenotype of AaSPL2 overexpressed in Arabidopsis.
The phenotype of AaSPL2 overexpressed in A. annua. (A) A schematic representation of overexpression vector and RNAi vector used in the A. annua transformation is shown. (B) the phenotype of overexpression and RNAi transgenic plants. (C) Glandular trichome of control. (D) Glandular trichome of overexpression plants. (E) Glandular trichome of RNAi plants. (F) Trichome number of A. annua. Scale bar represents 0.5 mm.
Primers used in this study.
References
- Allen G. C., Flores-Vergara M. A., Krasynanski S., Kumar S., Thompson W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1 2320–2325. 10.1038/nprot.2006.384 [DOI] [PubMed] [Google Scholar]
- Arsenault P. R., Vail D., Wobbe K. K., Erickson K., Weathers P. J. (2010). Reproductive development modulates gene expression and metabolite levels with possible feedback inhibition of artemisinin in Artemisia annua. Plant Physiol. 154 958–968. 10.1104/pp.110.162552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R. P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain, overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352 585–596. 10.1016/j.jmb.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Cai C., Guo W., Zhang B. (2018). Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci. Rep. 8:762. 10.1038/s41598-017-18673-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237 91–104. 10.1016/S0378-1119(99)00308-X [DOI] [PubMed] [Google Scholar]
- Chen J. L., Fang H. M., Ji Y. P., Pu G. B., Guo Y. W., Huang L. L., et al. (2011). Artemisinin biosynthesis enhancement in transgenic Artemisia annua plants by downregulation of the (-caryophyllene synthase dene. Planta Med. 77 1759–1765. 10.1055/s-0030-1271038 [DOI] [PubMed] [Google Scholar]
- Chen Y., Shen Q., Wang Y., Wang T., Wu S., Zhang L., et al. (2013). The stacked over-expression of FPS, CYP71AV1 and CPR genes leads to the increase of artemisinin level in Artemisia annua L. Plant Biotechnol. Rep. 7 287–295. 10.1007/s11816-012-0262-z [DOI] [Google Scholar]
- De Boer K., Tilleman S., Pauwels L., Vanden B. R., De Sutter V., Vanderhaeghen R., et al. (2011). APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 66 1053–1065. 10.1111/j.1365-313X.2011.04566.x [DOI] [PubMed] [Google Scholar]
- De Geyter N., Gholami A., Goormachtig S., Goossens A. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 17 349–359. 10.1016/j.tplants.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Duke S. O., Paul R. N. (1993). Development and fine structure of the glandular trichomes of Artemisia annua L. Int. J. Plant Sci. 154 107–118. 10.2307/2995610 [DOI] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server, interactive sequence similarity searching. Nucleic Acids Res. 39 W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., López-Vidriero I., Carrasco J. L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111 2367–2372. 10.1073/pnas.1316278111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H. (2007). Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2 38–41. 10.1038/nprot.2007.15 [DOI] [PubMed] [Google Scholar]
- Gou J. Y., Felippes F. F., Liu C. J., Weigel D., Wang J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23 1512–1522. 10.1105/tpc.111.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I. A., Besser K., Blumer S., Branigan C. A., Czechowski T., Elias L., et al. (2010). The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327 328–331. 10.1126/science.1182612 [DOI] [PubMed] [Google Scholar]
- Hao X., Zhong Y., Fu X., Lv Z., Shen Q., Yan T., et al. (2017). Transcriptome analysis of genes associated with the artemisinin biosynthesis by jasmonic acid treatment under the light in Artemisia annua. Front. Plant Sci. 8:971. 10.3389/fpls.2017.00971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Lu X., Qiu B., Zhang F., Shen Q., Lv Z., et al. (2014). Molecular cloning and characterization of a trichome-specific promoter of artemisinic aldehyde Δ11 (13). reductase (DBR2). in Artemisia annua. Plant Mol. Biol. Rep. 32 82–91. 10.1007/s11105-013-0603-2 [DOI] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42 541–544. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Jing F., Zhang L., Li M., Tang Y., Wang Y., Wang Y., et al. (2009). Abscisic acid (ABA). Treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia 64 319–323. 10.2478/s11756-009-0040-8 [DOI] [Google Scholar]
- Kindermans J., Pilloy J., Olliaro P., Gomes M. (2007). Ensuring sustained ACT production and reliable artemisinin supply. Malar. J. 6:125. 10.1186/1475-2875-6-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu X., Zhang L., Zhang F., Jiang W., Shen Q., Zhang L., et al. (2013). AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198 1191–1202. 10.1111/nph.12207 [DOI] [PubMed] [Google Scholar]
- Lv Z., Wang S., Zhang F., Chen L., Hao X., Pan Q., et al. (2016a). Overexpression of a novel NAC domain-containing transcription factor (AaNAC1). Enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 57 1961–1971. 10.1093/pcp/pcw118 [DOI] [PubMed] [Google Scholar]
- Lv Z., Zhang F., Pan Q., Fu X., Jiang W., Shen Q., et al. (2016b). Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield Artemisinin. Plant Cell Physiol. 57 588–602. 10.1093/pcp/pcw014 [DOI] [PubMed] [Google Scholar]
- Ma D., Pu G., Lei C., Ma L., Wang H., Guo Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50 2146–2161. 10.1109/TPWRD.2004.829125 [DOI] [PubMed] [Google Scholar]
- Maes L., Van Nieuwerburgh F. C. W., Zhang Y., Reed D. W., Pollier J., Vande Casteele S. R. F., et al. (2011). Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189 176–189. 10.2307/40960883 [DOI] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A. J., King G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38 948–952. 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- Mao Y. B., Liu Y. Q., Chen D. Y., Chen F. Y., Fang X., Hong G. J., et al. (2017). Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 8:13925. 10.1038/ncomms13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Paddon C. J., Westfall P. J., Pitera D. J., Benjamin K., Fisher K., Mcphee D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496 528–532. 10.1038/nature12051 [DOI] [PubMed] [Google Scholar]
- Pani A., Mahapatra R. K., Behera N., Naik P. K. (2011). Computational identification of sweet wormwood (Artemisia annua). microRNA and their mRNA targets. Genomics Proteomics Bioinformatics 9 200–210. 10.1016/S1672-0229(11)60023-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplow M. (2016). Synthetic biology’s first malaria drug meets market resistance. Nature 530 389–390. 10.1038/530390a [DOI] [PubMed] [Google Scholar]
- Sharma E., Anand G., Kapoor R. (2017). Terpenoids in plant and arbuscular mycorrhiza-reinforced defence against herbivorous insects. Ann. Bot. 119 791–801. 10.1093/aob/mcw263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Lu X., Yan T., Fu X., Lv Z., Zhang F., et al. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210 1269–1281. 10.1111/nph.13874 [DOI] [PubMed] [Google Scholar]
- Shi P., Fu X., Liu M., Shen Q., Jiang W., Li L., et al. (2017a). Promotion of artemisinin content in Artemisia annua by overexpression of multiple artemisinin biosynthetic pathway genes. Plant Cell Tissue Organ Cult. 129 251–259. 10.1007/s11240-017-1173-z [DOI] [Google Scholar]
- Shi P., Fu X., Shen Q., Liu M., Pan Q., Tang Y., et al. (2017b). The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. 1 261–276. 10.1111/nph.14789 [DOI] [PubMed] [Google Scholar]
- Shikata M., Koyama T., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50 2133–2145. 10.1093/pcp/pcp148 [DOI] [PubMed] [Google Scholar]
- Shoji T., Kajikawa M., Hashimoto T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22 3390–3409. 10.1105/tpc.110.078543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L., Chen J., Huang X., Gong H., Luo J., Hou Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48 447–456. 10.1038/ng.3518 [DOI] [PubMed] [Google Scholar]
- Van der Fits L., Memelink J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289 295–297. 10.1126/science.289.5477.295 [DOI] [PubMed] [Google Scholar]
- Wang J. W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138 738–749. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- Westfall P. J., Pitera D. J., Lenihan J. R., Eng D., Woolard F. X., Regentin R., et al. (2012). Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. U.S.A. 109 E111–E118. 10.1073/pnas.1110740109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2014). World Malaria Report 2014. Geneva: World Health Organization; 10.30875/9f925144-en [DOI] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Li L., Xie L., Chen M., Shen Q., Pan Q., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 2 567–578. 10.1111/nph.15005 [DOI] [PubMed] [Google Scholar]
- Yu Z., Wang L., Zhao B., Shan C., Zhang Y., Chen D., et al. (2015). Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and patchouli (Pogostemon cablin). by the miR156-targeted SPL transcription factors. Mol. Plant 8 98–110. 10.1093/mp/ssu127 [DOI] [PubMed] [Google Scholar]
- Yu Z. X., Li J. X., Yang C. Q., Hu W. L., Wang L. J., Chen X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5 356–365. 10.1093/mp/ssr087 [DOI] [PubMed] [Google Scholar]
- Zhang H., Hedhili S., Montiel G., Zhang Y., Chatel G., Pré M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67 61–71. 10.1111/j.1365-313X.2011.04575.x [DOI] [PubMed] [Google Scholar]
- Zhang L., Jing F., Li F., Li M., Wang Y., Wang G., et al. (2009). Development of transgenic Artemisia annua (Chinese wormwood). Plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol. Appl. Biochem. 52 199–207. 10.1042/BA20080068 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wu B., Zhao D., Li C., Shao F., Lu S. (2014). Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J. Integr. Plant Biol. 56 38–50. 10.1111/jipb.12111 [DOI] [PubMed] [Google Scholar]
- Zhang T., Lian H., Tang H., Dolezal K., Zhou C., Yu S., et al. (2015). An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 27 349–360. 10.1105/tpc.114.135186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Wang H., Gu C., Dare A. P., Espley R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82 105–121. 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DHAA content in the transgenic plants was determined by HPLC. (A) DHAA content was detected in the AaSPL2 overexpression plants. (B) DHAA content was detected in the AaSPL2 RNAi plants. Error bars indicate SD (n = 3). ∗∗p < 0.01.
The phenotype of AaSPL2 overexpressed in Arabidopsis.
The phenotype of AaSPL2 overexpressed in A. annua. (A) A schematic representation of overexpression vector and RNAi vector used in the A. annua transformation is shown. (B) the phenotype of overexpression and RNAi transgenic plants. (C) Glandular trichome of control. (D) Glandular trichome of overexpression plants. (E) Glandular trichome of RNAi plants. (F) Trichome number of A. annua. Scale bar represents 0.5 mm.
Primers used in this study.


