Many plant pathogens secrete toxins that disable their host (1). Some of these have general phytotoxic properties and are active toward a broad range of plant species. Others are host-selective, affecting only certain plant varieties or genotypes. These host-selective toxins (HSTs) can act as agents of virulence of pathogens to toxin-sensitive hosts, so determining host range or specificity (1, 2). Nearly all HSTs that have been described are low molecular weight secondary metabolites produced by fungi. The genes required for the synthesis of these molecules are often tightly clustered in the fungal genome and are coordinately regulated (3). In a recent issue of PNAS, Pedley and Walton (4) report that the regulation of synthesis of a host-selective cyclic peptide toxin by the maize pathogen Cochliobolus carbonum is controlled by a novel pathway-specific transcription factor that may be unique to plant pathogenic fungi.
C. carbonum causes northern leaf spot and ear mold disease of maize. Race 1 (Tox2+) isolates of this fungus produce a HST known as HC-toxin, a cyclic tetrapeptide with the structure cyclo(d-Pro-l-Ala-d-Ala-l-Aeo), where Aeo is 2-amino-9,10-epoxi-8-oxodecanoic acid (1). Genetic analysis has shown that pathogenicity of C. carbonum race 1 is determined by a single locus known as TOX2, which also confers the ability to produce HC-toxin (Fig. 1) (5, 6). The precise effects of HC-toxin on toxin-sensitive plants are not fully understood, but there is evidence to indicate that one possible mode of action is by inhibition of histone deacetylases (7). Histone deacetylases reversibly deacetylate the core histones of chromatin and so may influence the expression of genes by means of their effects on chromatin structure. An attractive model is that inhibition of histone deacetylases by HC-toxin may interfere with expression of defense genes, so allowing fungal invasion to progress (1, 7).
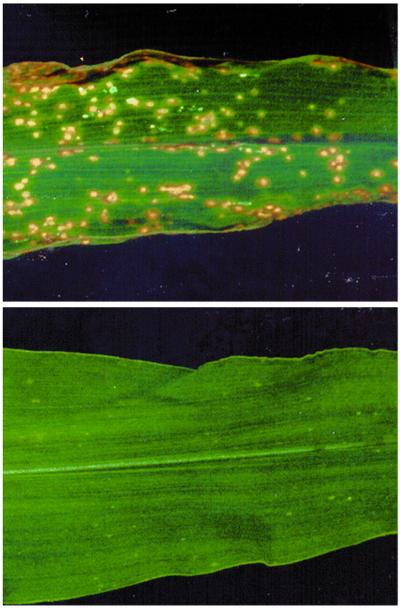
Figure 1.
Disease symptoms caused by a Tox2+ (Upper) and a Tox2− (Lower) isolate of C. carbonum.
Extensive molecular genetic analysis of the TOX2 locus by Walton and coworkers has led to the characterization of seven genes that are implicated in toxin synthesis. These include genes encoding a nonribosomal peptide synthetase (HTS1) (8), a putative HC-toxin efflux pump (TOXA) (9), a fatty acid synthase β subunit believed to be required for synthesis of the Aeo side chain (TOXC) (10), a predicted branched-chain amino acid transaminase (TOXF) (11), and an alanine racemase (TOXG) (12). The TOX genes are clustered and are present in multiple copies in toxin-producing isolates of C. carbonum, but are absent from strains that do not produce HC-toxin (13, 14). A seventh gene, TOXE, is required for HC-toxin biosynthesis and for expression of at least three of the other TOX2 genes, including the gene encoding the candidate HC-toxin efflux pump (15).
In a recent issue Pedley and Walton (4) demonstrate that TOXE is a novel transcription factor that coordinates expression of other genes at the TOX2 locus. TOXE expressed in Escherichia coli is able to bind to the promoters of other TOX2 genes in in vitro assays. Sequence comparisons of these promoters identified a conserved 10-bp motif that was present in one or two copies in each. The requirement of this motif (the “tox-box”) for binding of TOXE was demonstrated by mutational analysis. The absence of an obvious tox-box motif in the TOXE promoter suggests that TOXE does not regulate itself. These in vitro results were confirmed by the demonstration that TOXE can drive gene expression in yeast in a tox-box-dependent fashion.
TOXE is an unusual protein that defines a new class of transcription factors. It has an N-terminal basic region that matches the consensus sequence of the basic leucine zipper (bZIP) family of transcriptional regulators. Pedley and Walton have shown experimentally that this sequence is required for DNA binding (4). However, TOXE lacks a leucine zipper and so is not a true bZIP protein. TOXE appears to be unique in that in addition to the consensus bZIP basic DNA-binding domain it also contains four ankyrin repeats, motifs that are often associated with protein–protein interactions. C-terminal deletion mutants of TOXE lacking the ankyrin repeats are unable to bind DNA, indicating that both the basic DNA-binding domain and the ankyrin repeats are required for DNA binding. Deletion analysis experiments also identified a discrete domain in the mid- to C-terminal region of the protein that may encompass one of the ankyrin repeats and that is required for transcriptional activation. Because the only obvious effects of mutation of TOXE are loss of ability to synthesize HC-toxin and associated loss of pathogenicity, TOXE appears to be a specific transcriptional regulator for HC-toxin biosynthetic genes. It is not clear why fungal genes for secondary metabolite biosynthesis tend to be clustered along with their transcriptional regulators. One possibility is that these genes are “selfish clusters” that may depend in part on horizontal gene transfer for their dispersal and survival (3). It remains to be seen whether the presence of tox-box motifs in promoters of other C. carbonum genes will enable the identification of additional TOX genes.
Genes for secondary metabolite biosynthesis in other fungi are also subject to regulation by pathway-specific transcription factors, but the regulators that have been described for these pathways so far are all distinct from TOXE (1, 2, 4, 15). The only other related gene that has been identified to date is BAP1 from the fungal pathogen of tomato, Cladosporium fulvum (16). BAP1 was cloned by using a degenerate oligonucleotide designed to match the conserved basic DNA-binding domain of the bZIP-type YAP1 transcription factors found in yeasts. BAP1, like TOXE, lacks a leucine zipper but contains C-terminal ankyrin-like repeats. The term “bANK proteins” has been proposed for the class of proteins that so far consists of TOXE and BAP1 (16). Cladosporium fulvum does not appear to synthesize toxins during infection of tomato plants, and the function of BAP1 has not yet been tested by mutation. In yeast, yAP1 confers resistance to oxidative stress and cytotoxic compounds and, since at least one of the genes regulated by TOXE may encode a toxin efflux pump, a common theme could be the regulation of genes involved in self-protection mechanisms.
Although C. carbonum race 1 is highly aggressive to susceptible maize lines, most lines are resistant. Resistance is conferred by the dominant gene Hm1. In 1991 Walton and coworkers developed a method for generating radiolabeled HC-toxin with high specific activity by feeding d-[3H]alanine to fungal cultures (17). When this substrate was incubated with shoot extracts from maize seedlings segregating for Hm1 it was found that only lines that were homozygous or heterozygous for Hm1 were able to convert HC-toxin to a single product that lacked biological activity (8-hydroxy-HC-toxin), suggesting that detoxification was the basis of disease resistance (18). The enzyme responsible for this detoxification was named HC-toxin reductase. This finding paved the way for the isolation of the Hm1 gene by transposon tagging by using a screen for insertional inactivation of Hm1 based on HC-toxin reductase activity (19).
Hm1 was the first disease-resistance gene to be cloned from plants. However, it is regarded as somewhat unorthodox by many of those working in the disease-resistance field. The vast majority of other disease-resistance genes that have been cloned and characterized over the last 10 years are involved in specific interactions of a rather different kind, namely the classical gene-for-gene interactions that involve recognition of pathogens expressing specific avirulence determinants (20). Recognition leads to the activation of plant defense responses and hence disease resistance. Many avirulence determinants are proteins or peptides, although in some cases it is clear that the products of some avirulence genes are involved in the synthesis of low molecular weight secondary metabolites that can be recognized by plants. Examples include the avrD gene of Pseudomonas syringae, which is required for the synthesis of low molecular weight molecules known as syringolides (21, 22), and the ACE1 gene of the rice blast fungus Magnaporthe grisea, which is predicted to encode a polyketide synthase.†
Evidence is emerging to suggest that the genomes of plant pathogenic fungi are rich in genes that are likely to be involved in the synthesis of secondary metabolites such as nonribosomal peptide synthases and polyketide synthases, whereas saprophytes appear to be deficient in such genes (23). Further genome mining is likely to unveil new clusters of coregulated genes that are required for the synthesis of novel secondary metabolites, some of which may be determinants of virulence and/or host range. It seems likely, then, that so far we have seen only the tip of the iceberg. Secondary metabolites may play a far more significant role in determining the outcome of plant–pathogen interactions than has previously been anticipated.
Acknowledgments
The Sainsbury Laboratory is supported by the Gatsby Charitable Foundation.
Footnotes
See companion article on page 14174 in issue 24 of volume 98.
Böhnert, H. U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J.-L. & Lebrun, M.-H., Abstracts of the 10th International Congress on Molecular Plant-Microbe Interactions, July 10–14, 2001, Madison, WI, abstr. 362.
References
- 1.Walton J D. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Idnurm A, Howlett B J. Mol Plant Pathol. 2001;2:241–255. doi: 10.1046/j.1464-6722.2001.00070.x. [DOI] [PubMed] [Google Scholar]
- 3.Walton J D. Fung Genet Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- 4.Pedley K F, Walton J D. Proc Natl Acad Sci USA. 2001;98:14174–14179. doi: 10.1073/pnas.231491298. . (First Published November 6, 2001; 10.1073/pnas.231491298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson R R, Ullstrap A J. Phytopathology. 1961;51:1–2. [Google Scholar]
- 6.Scheffer R P, Nelson R R, Ullstrap A J. Phytopathology. 1967;57:1288–1289. [Google Scholar]
- 7.Brosch G, Ransom R, Lechner T, Walton J D, Loidl P. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panaccione D G, Scott-Craig J S, Pocard J-A, Walton J D. Proc Natl Acad Sci USA. 1992;89:6590–6594. doi: 10.1073/pnas.89.14.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitkin J W, Panaccione D G, Walton J D. Microbiology. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J-H, Walton J D. Mol Plant-Microbe Interact. 1997;10:207–214. doi: 10.1094/MPMI.1997.10.2.207. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y-Q, Ahn J-H, Walton J D. Microbiology. 1999;145:3539–3546. doi: 10.1099/00221287-145-12-3539. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Y-Q, Walton J D. J Biol Chem. 2000;275:4906–4911. doi: 10.1074/jbc.275.7.4906. [DOI] [PubMed] [Google Scholar]
- 13.Ahn J-H, Walton J D. Plant Cell. 1996;8:887–897. doi: 10.1105/tpc.8.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahn, J.-H., Cheng, Y.-Q. & Walton, J. D., Fung. Genet. Biol., in press.
- 15.Ahn J-H, Walton J D. Mol Gen Genet. 1998;260:462–469. doi: 10.1007/pl00008632. [DOI] [PubMed] [Google Scholar]
- 16.Bussink H-J, Clark A, Oliver R. Eur J Plant Pathol. 2001;107:655–659. [Google Scholar]
- 17.Meeley R B, Walton J D. Plant Physiol. 1991;97:1080–1086. doi: 10.1104/pp.97.3.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meeley R B, Johal G S, Briggs S P, Walton J D. Plant Cell. 1992;4:71–77. doi: 10.1105/tpc.4.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johal G S, Briggs S P. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 20.Hulbert S H, Webb C A, Shavannor S M, Sun Q. Ann Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi D Y, Tamaki S J, Keen N T. Mol Plant-Microbe Interact. 1990;3:94–102. doi: 10.1094/mpmi-3-094. [DOI] [PubMed] [Google Scholar]
- 22.Smith M J, Mazzola E P, Sims J J, Midland S L, Keen N T, Burton V, Stayton M M. Tetrahedron Lett. 1993;34:223–226. [Google Scholar]
- 24.Yoder O C, Turgeon B G. Curr Opin Plant Biol. 2001;4:315–321. doi: 10.1016/s1369-5266(00)00179-5. [DOI] [PubMed] [Google Scholar]