Abstract
Countering the obesity pandemic will require better understanding of disease mechanisms and development of new diagnostic methods. Small molecule metabolites excreted in urine can be important biomarkers of disease progression and treatment. However, with multiple pathways involved, it has been challenging to identify key pathway(s) that closely follow disease features such as body fat. We employed a high-fat diet (HFD) mouse model of obesity with the goal of determining changes in urinary metabolite profile related to body fat using proton nuclear magnetic resonance (1H NMR). Several urinary metabolites with significantly lower levels in HFD compared to control mice have been identified. Specifically, major changes were found in metabolites from tricarboxylic acid (TCA) cycle, amino acid, nicotinamide, and choline metabolism including 2-hydroxydlutarate, cis-aconitate, trans-aconitate, alanine, creatine, trigonelline, dimethylamine, and trimethylamine. However, levels of only two metabolites, namely dimethylamine and trimethylamine, showed significant reverse correlation with total body fat. These metabolites derive from choline processing by gut microbiota and may be prospective biomarkers indicative of accumulation of body fat in obesity.
Keywords: Biochemistry, Bioinformatics, Microbiology, Physiology
1. Introduction
Obesity has become a worldwide pandemic [1]. Among multitude of factors contributing to obesity diet plays one of the key roles, however there is lack of reliable biomarkers to ascertain contribution from dietary factors [2]. High-fat diet (HFD) mouse models have been a valuable tool to investigate pathogenic mechanisms and to evaluate prospective therapies. In particular, a number of studies using these models have focused on changes in metabolome and specific metabolic pathways affected in obesity [3, 4, 5]. These efforts include a more recent focus on the changes in metabolites derived from gut microbiota [3, 4, 6, 7, 8]. The studies revealed a broad spectrum of metabolic alterations associated with HFD-induced obesity. In particular, metabolites derived from dietary choline processing by bacteria in the gut appear to be affected in obesity. However, the findings have been controversial exhibiting either opposing trends or absence of changes associated with high-fat diet [3, 4, 6, 7, 8]. Moreover, the relation of these metabolic changes to disease phenotype such as total body fat is not fully understood due, in part, to the short duration of the HFD regiments (<15 weeks) and, as a result, relatively small increases in body weight [3, 4, 6, 7, 8].
To address this knowledge gap, we set out to identify those biomarkers within complex metabolic perturbations in mouse model of HFD-induced obesity that specifically correlate with body fat. The experimental design included a prolonged duration of the HFD regimen (30 weeks), focus on urinary metabolome, a three-step statistical analysis for determining differences in metabolite levels between HFD and control groups and correlation of these levels with total body fat in individual mice. This approach allowed us to identify two products of dietary choline processing by gut microbiota, namely dimethylamine and trimethylamine, which demonstrated statistically significant results in all the analyses. We propose that these metabolites are prospective biomarkers that can follow the levels of body fat in HFD-induced obesity.
2. Materials and methods
2.1. Animal studies
All institutional and national guidelines for the care and use of laboratory animals were followed. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Mississippi Medical Center. Studies were performed on male C57BL/6J mice placed on a high-fat diet consisting of 60% of the calories coming from fats (Teklad custom diet #06414, Envigo Teklad Diets, Madison, WI). Mice were fed the high-fat diet for 30 weeks beginning at 4 weeks of age (HFD group, n = 6). Control mice (n = 6) were fed a standard laboratory diet consisting 17% of calories derived from fat (Teklad 22/5 Rodent Diet, Envigo Teklad Diets, Madison, WI). At the end of experiment, urine samples were collected using individual metabolic cages. Urine samples were stored at −80 °C until analysis.
2.2. Determination of total body fat and other physiological parameters
Mice were euthanized after 30 weeks from the start of the experiment and their body lengths and weights were recorded. Animal hearts, kidneys and livers were removed and weighted using analytical balances. Similarly, epydidymal, visceral and total fat pads were removed and weighted in both control and HFD mice.
2.3. NMR experiments
NMR spectra were acquired using a 14.0 T Bruker magnet equipped with a Bruker AV-III console operating at 600.13 MHz. All spectra were acquired in 3-mm NMR tubes using a Bruker 5-mm TCI cryogenically cooled NMR probe operating at 298 K. Samples were prepared as 200 μL solutions that included 100 μL of urine, 41 μL of combination of 70 mM sodium phosphate buffer, TSP, and NaN3, and 59 μL of 90%–10% H2O/D2O which served as the 2H lock solvent. TSP (3-(trimethylsilyl)propionic-2,2,3,3-d4 acid) in the buffer solution served as the zero ppm chemical shift reference.
For 1D 1H NMR, experiments were performed using a one-dimensional nuclear Overhauser (1D-NOE) pulse sequence with presaturation solvent suppression to suppress the signal associated with water. The 1D-NOE experiment filters NMR signals associated with broad line widths such as those arising from proteins that might be present in urine samples and adversely affect spectral quality. Experimental conditions included: 32K data points, 13 ppm sweep width, a recycle delay of 3 seconds, a mixing time of 150 ms and 32 scans.
2.4. NMR data analysis
Principal component analysis (PCA) was performed using the AMIX program (Bruker Biospin Corp., Billerica MA). This method requires NMR data to be distributed in chemical shift bins (0.01 ppm width) while eliminating the area associated with the water solvent suppression (4.6–5.1 ppm) and glucose (3.2–3.58, 3.62–4.108 and 5.22–5.276 ppm). Buckets with less than 5% variance were omitted prior to PCA. PCA reduces the dimensionality of the data and summarizes the similarities and differences between multiple NMR spectra using scores plots. Buckets that corresponded to statistically significant P scores were compared to ppm values of the peaks that were identified from 1D & 2D NMR assignments. Candidate metabolites were quantified by measuring the sum of the areas under all the characteristic resonance peaks corresponding to a given metabolite.
2.5. Identification of metabolites
Metabolites were identified by comparison of acquired spectra with standard spectra using the Chenomx Profiler program (Fig. S1). Metabolite identities were further confirmed using two dimensional 1H-1H COSY and 1H-13C HQSC (see Fig. S2 and Table S2 for spectral assignments of selected metabolites and Table S3 for their PubChem IDs and identification levels). For 2D 1H-1H COSY, experimental conditions included 2048 × 512 data matrix, 13 ppm sweep width, recycle delay of 1.5 seconds and 8 scans per increment. The data were processed using squared sinebell window function, symmetrized, and displayed in magnitude mode. Multiplicity-edited HSQC experiments were acquired using a 1024 × 256 data matrix, a J(C-H) value of 145 Hz, which resulted in a multiplicity selection delay of 34 ms, a recycle delay of 1.5 seconds and 32 scans per increment along with GARP decoupling on 13C during the acquisition time (150 ms). The data were processed using a π/2 shifted squared sine window function and displayed with CH/CH3 signals phased positive and CH2 signals phased negative.
2.6. Statistical analyses
The primary statistical analysis of the NMR data was performed as previously described [9, 10]. The P scores were calculated and automatically divided into four groups by AMIX software based on significance level. These groups were color coded within PCA loadings plots according to P score values, thus generating a heat map representation of the data. The variability within each experimental group was expressed as means ± SEM. The secondary statistical analysis was performed using one-way ANOVA followed by post-hoc Tukey test. Statistical significance was determined after taking into account Bonferroni correction for multiple comparisons. Finally, Pearson correlation analysis of metabolite levels with total body fat was performed using SigmaStat 4.0.
3. Results and discussion
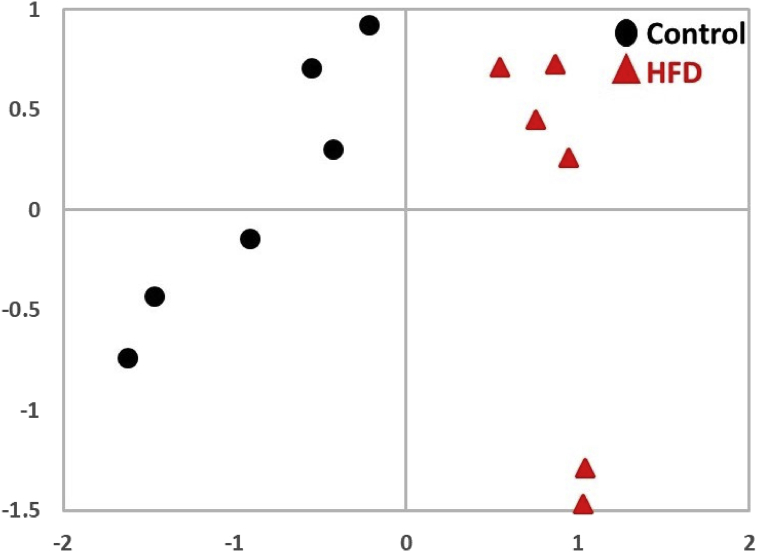
Compared to controls, HFD mice exhibited significant increase in body weight, body fat, and weight of major organs after 30 weeks, consistent with obesity (Table 1). Small molecule metabolites were analyzed in mouse urine using 1H NMR spectroscopy. Principal component analysis (PCA) of the NMR spectra revealed a clear separation of the data points into NFD (control) and HFD clusters (Fig. 1). Because there was an HFD-induced increase in the intensity of NMR peaks corresponding to glucose, these specific proton NMR signals were removed from the spectra before the analysis.
Table 1.
Body weights, organ weights and fat deposits in control and HFD mice. The data are presented as mean ± SEM. Asterisks indicate significant differences in HFD vs. control; *P < 0.05 and **P < 0.01; n = 6.
| Parameter | Control mice | HFD mice |
|---|---|---|
| Body weight, g | 28.5 ± 1.1 | 51.6 ± 1.0** |
| Body length, cm | 9.8 ± 0.2 | 10.3 ± 0.2** |
| Heart weight, mg | 151.2 ± 6.7 | 176.5 ± 6.4** |
| Kidney weight, mg | 323.2 ± 15.1 | 355.2 ± 21.0* |
| Liver weight, g | 1.2 ± 0.1 | 2.2 ± 0.2** |
| Epydidymal fat, g | 0.52 ± 0.10 | 2.16 ± 0.39** |
| Visceral fat, g | 0.70 ± 0.08 | 2.52 ± 0.38** |
| Total fat, g | 1.21 ± 0.17 | 4.68 ± 0.72** |
Fig. 1.
Principle component analysis (PCA) of the 1H NMR data from control and HFD mice. PCA scores are plotted for control (black circles) and HFD (red triangles) groups.
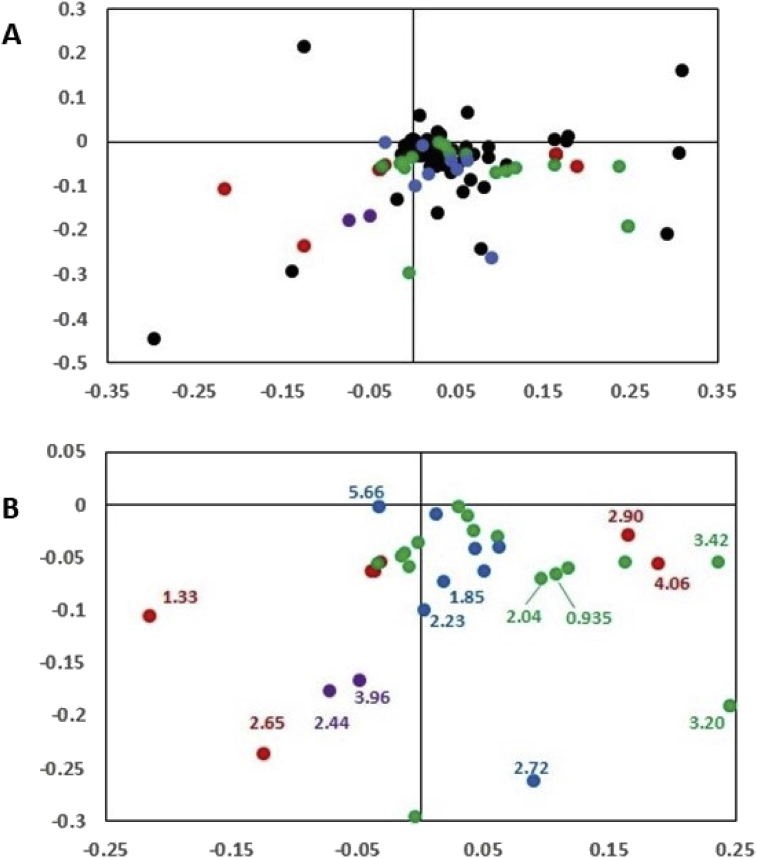
The spectral features that contributed to the observed data clustering were identified by dividing the NMR spectra into 0.01 ppm intervals (buckets) and analyzing signal intensities in each bucket using AMIX software. The P score analysis identified multiple buckets with significant differences between control and HFD samples (Fig. 2). Importantly, this unbiased analysis of the spectral differences was performed before the identities of specific metabolites were determined. This allowed for identification of only those urinary metabolites, whose levels significantly changed in HFD compared to control urine samples. The analysis identified 23 candidate metabolites shown in Table S1. Additional statistical analysis of integrated peak areas including Bonferroni correction for multiple comparisons demonstrated that out of these 23 candidate metabolites, only eleven have shown significant changes, i.e. all were significantly decreased in HFD vs. control urine (Table 2 and Fig. 3, metabolites indicated by numbers 1 through 11). Manual analyses of 1H NMR spectra using Chenomx Profiler program did not find any additional metabolites with significant changes in urinary levels, thus confirming the results. Interestingly, the levels of only two metabolites, i.e. dimethylamine (DMA) and trimethylamine (TMA), significantly correlated with total body fat (Table 3, top two entries). Thus, urinary levels of these two metabolites, were significantly decreased in HFD vs. control (Table 2) and showed the strongest reverse correlation with total body fat in HFD mice but not in control mice (Table 3). Urinary levels of a third metabolite, trans-aconitate, have also shown a strong reverse correlation with total body fat but have not reached statistical significance (Table 3).
Fig. 2.
Loadings plot for 1H NMR data from control and HFD mice. (A) Heat maps are color-coded according to bucket P score values as described under Materials and Methods: P < 8.3 × 10−5 (red), 8.3 × 10−5 < P < 1.6 × 10−4 (purple), 1.6 × 10−4 < P < 2.5 × 10–4 (green), 2.5 × 10–4 < P < 3.2 × 10−3 (blue), and P > 3.2 × 10−3 (black). The numbers indicate ppm values of the selected NMR peaks with P < 3 × 10−3 for which metabolite assignments have been made. (B) Zoomed-in areas from the loadings plot in panel A.
Table 2.
Relative levels of urinary metabolites with significant differences between control and HFD mice. Data presented as relative metabolite concentrations in urine after 30 weeks of HFD (mean ± SEM). Relative metabolite levels represent integrated peak areas normalized to urinary creatinine and using TSP as a standard. P values based on comparison of control vs. HFD groups (n = 6); Bonferroni-adjusted P value cut-off was 2.2 × 10−3 to adjust for multiple comparisons of initial 23 candidate metabolites identified after primary statistical analysis (see Fig. S1).
| Metabolite | 1H NMR peak position (ppm) | Relative level in control urine | Relative level in HFD urine | P value (HFD vs. control) |
|---|---|---|---|---|
| Dimethylamine | 2.7 | 0.705 ± 0.018 | 0.345 ± 0.016 | 8.10E-04 |
| Trimethylamine | 2.9 | 5.28 ± 0.239 | 1.47 ± 0.051 | 1.30E-04 |
| trans-Aconitate | 6.6 | 0.116 ± 0.002 | 0.042 ± 0.004 | 1.20E-04 |
| Alanine | 1.5, 3.8 | 1.105 ± 0.033 | 0.512 ± 0.015 | 1.02E-05 |
| Creatine | 3.0, 3.9 | 2.60 ± 0.066 | 0.98 ± 0.047 | 3.38E-05 |
| Trigonelline | 4.4, 8.1, 8.8, 9.1 | 0.547 ± 0.025 | 0.213 ± 0.013 | 6.60E-04 |
| cis-Aconitate | 3.1 | 0.407 ± 0.017 | 0.171 ± 0.012 | 7.10E-04 |
| 2-Hydroxyglutarate | 1.8, 2.0, 2.2, 4.0 | 7.82 ± 0.184 | 5.10 ± 0.149 | 9.10E-04 |
| Succinate | 2.4 | 1.02 ± 0.039 | 0.23 ± 0.026 | 4.19E-05 |
| Acetate | 1.9 | 3.13 ± 0.219 | 0.79 ± 0.056 | 7.10E-04 |
| Benzoate | 7.5, 7.6, 7.9 | 0.449 ± 0.019 | 0.076 ± 0.007 | 9.49E-06 |
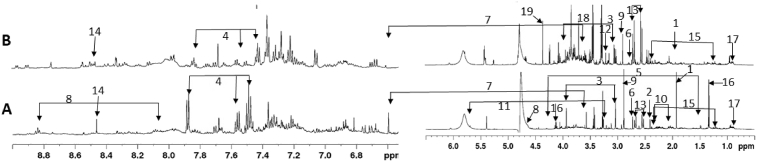
Fig. 3.
Representative 600 MHz 1H NMR spectra of urine samples collected from control (A) and HFD (B) mice. Shown are identified metabolites with significantly different levels in HFD vs. control groups: 1: acetate, 2: succinate, 3: creatine, 4: benzoate, 5: alanine, 6: dimethylamine, 7: trans-aconitate, 8: trigonelline, 9: trimethylamine, 10: 2-hydroxygluterate, and 11: cis-aconitate. Other selected metabolites found in the urine that passed primary but not secondary statistical tests: 12: choline, 13: citrate, 14: formate, 15: 3-hydroxyisovalerate, 16: lactate, 17: 2-oxovalerate, 18: propanoate, 19: sarcosine.
Table 3.
Pearson correlation analysis of total body fat vs. levels of urinary metabolites in HFD and control mice. Pearson correlation coefficient was determined for eleven urinary metabolites with significantly different levels in HFD vs. control mice. Asterisk indicates significant correlation; *P < 0.05 (n = 6).
| Metabolite | HFD group |
Control group |
Pathway | ||
|---|---|---|---|---|---|
| Pearson correlation coefficient | P value | Pearson correlation coefficient | P value | ||
| Dimethylamine | −0.878 | 0.0214* | 0.325 | 0.529 | Choline degradation by gut microbiota |
| Trimethylamine | −0.886 | 0.0188* | 0.276 | 0.597 | |
| Trigonelline | −0.242 | 0.644 | 0.0992 | 0.852 | Nicotinamide metabolism |
| Alanine | −0.702 | 0.120 | −0.311 | 0.549 | Amino acid metabolism |
| Creatine | 0.675 | 0.142 | −0.469 | 0.348 | |
| trans-Aconitate | −0.775 | 0.0702 | 0.375 | 0.464 | TCA cycle |
| cis-Aconitate | 0.0616 | 0.908 | 0.236 | 0.652 | |
| 2-Hydroxyglutarate | 0.503 | 0.310 | −0.296 | 0.570 | |
| Succinate | 0.299 | 0.564 | −0.306 | 0.556 | |
| Acetate | −0.238 | 0.650 | −0.671 | 0.145 | |
| Benzoate | 0.0264 | 0.960 | −0.292 | 0.575 | Aromatic amino acid degradation by gut microbiota |
Potential change in renal function (nephropathy) reported in HFD-induced obese animals [11, 12], is unlikely to affect the results since other small molecule urinary metabolites showed no significant correlation with body fat (Table 3). In addition, the potential effect of variations in urine volume was excluded by normalizing metabolite levels to the corresponding urinary creatinine concentration in each individual animal. The HFD mice also exhibited increases in fasting blood glucose and as a result increases in glucose excreted in urine. To prevent the interference of glucose peaks with data analysis, glucose spectral signals were specifically removed in both control and HFD samples. Thus, the observed decrease in urinary levels of DMA and TMA indeed reflects metabolic changes related to HFD that follow body fat deposits.
There is a growing body of evidence that the metabolites produced by gut microbiota are important modulators of host physiology [13]. Moreover, factors such as diet have significant impact on gut microbiome, thus introducing additional complexity into host-microbiome relationship [14, 15]. Of special interest is metabolism by gut microbiota of dietary choline that has been shown to promote fat accumulation in model obesity [16, 17]. This microbial pathway specifically produces precursors DMA and TMA that are converted by the host liver to trimethylamine oxide (TMAO), a compound implicated in pathogenesis of obesity [18]. Recently, there have been several reports connecting HFD-induced obesity with TMAO, DMA and TMA in animal models. The results are controversial with studies showing an increase [4, 7], a decrease [3, 6], or no difference [8] in the levels of these metabolites in HFD models vs. controls. The reason for these discrepancies is unclear and may have to do with relatively short duration of HFD regimens in these studies. In the present long HFD duration study, we have found that two specific metabolites, DMA and TMA, which are products of choline processing by gut microbiota [4, 14, 15] show significantly lower urinary levels in HFD-induced obesity and a strong negative correlations with total body fat in HFD mice. These two metabolites may be prospective biomarkers indicative of body fat accumulation in obesity.
Declarations
Author contribution statement
Donald F Stec: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Calisa Henry: Performed the experiments; Analyzed and interpreted the data.
David E Stec: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data.
Paul Voziyan: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by grants R01DK65138 (P.V.) and R25DK096999 from the National Institutes of Health; grant P20GM-104357-02 from the National Institute of General Medical Sciences (D.E.S.); and grant S10RR019022 from the National Institutes of Health for funding the purchase of the 600 MHz NMR spectrometer used in this study. Ms. Calisa Henry was supported by the AspirnautTM program from the Center for Matrix Biology at Vanderbilt University Medical Center.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Meldrum D.R., Morris M.A., Gambone J.C. Obesity pandemic: causes, consequences, and solutions-but do we have the will? Fertil. Steril. 2017;107:833–839. doi: 10.1016/j.fertnstert.2017.02.104. [DOI] [PubMed] [Google Scholar]
- 2.Hilger-Kolb J., Bosle C., Motoc I., Hoffmann K. Associations between dietary factors and obesity-related biomarkers in healthy children and adolescents - a systematic review. Nutr. J. 2017;16:85. doi: 10.1186/s12937-017-0300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z.Y., Ding L.L., Li J.M., Xu B.L., Yang L., Bi K.S., Wang Z.T. (1)H-NMR and MS based metabolomics study of the intervention effect of curcumin on hyperlipidemia mice induced by high-fat diet. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M., Lu B., Li Y., Wang Y., Zheng H., Zhong D., Liao Z., Wang M., Ma F., Liao Q., Xie Z. Metabolomics insights into the modulatory effects of long-term compound polysaccharide intake in high-fat diet-induced obese rats. Nutr. Metab. 2018;15:8. doi: 10.1186/s12986-018-0246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao J., Huo T.I., Cheng H.Y., Tsai J.C., Liao J.W., Lee M.S., Qin X.M., Hsieh M.T., Pao L.H., Peng W.H. Gallic acid ameliorated impaired glucose and lipid homeostasis in high fat diet-induced NAFLD mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montoliu I., Cominetti O., Boulange C.L., Berger B., Siddharth J., Nicholson J., Martin F.P. Modeling longitudinal metabonomics and microbiota interactions in C57BL/6 mice fed a high fat diet. Anal. Chem. 2016;88:7617–7626. doi: 10.1021/acs.analchem.6b01343. [DOI] [PubMed] [Google Scholar]
- 7.Barouei J., Bendiks Z., Martinic A., Mishchuk D., Heeney D., Hsieh Y.H., Kieffer D., Zaragoza J., Martin R., Slupsky C., Marco M.L. Microbiota, metabolome, and immune alterations in obese mice fed a high-fat diet containing type 2 resistant starch. Mol. Nutr. Food Res. 2017;61 doi: 10.1002/mnfr.201700184. [DOI] [PubMed] [Google Scholar]
- 8.Men L., Pi Z., Zhou Y., Wei M., Liu Y., Song F., Liu Z. Urine metabolomics of high-fat diet induced obesity using UHPLC-Q-TOF-MS. J. Pharm. Biomed. Anal. 2017;132:258–266. doi: 10.1016/j.jpba.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Goodpaster A.M., Romick-Rosendale L.E., Kennedy M.A. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Anal. Biochem. 2010;401:134–143. doi: 10.1016/j.ab.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Stec D.F., Wang S., Stothers C., Avance J., Denson D., Harris R., Voziyan P. Alterations of urinary metabolite profile in model diabetic nephropathy. Biochem. Biophys. Res. Commun. 2015;456:610–614. doi: 10.1016/j.bbrc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury S.S., Lecomte V., Erlich J.H., Maloney C.A., Morris M.J. Paternal high fat diet in rats leads to renal accumulation of lipid and tubular changes in adult offspring. Nutrients. 2016;8 doi: 10.3390/nu8090521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei P., Lane P.H., Lane J.T., Padanilam B.J., Sansom S.C. Glomerular structural and functional changes in a high-fat diet mouse model of early-stage Type 2 diabetes. Diabetologia. 2004;47:1541–1549. doi: 10.1007/s00125-004-1489-1. [DOI] [PubMed] [Google Scholar]
- 13.Lin R., Liu W., Piao M., Zhu H. A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids. 2017;49:2083–2090. doi: 10.1007/s00726-017-2493-3. [DOI] [PubMed] [Google Scholar]
- 14.Albenberg L.G., Wu G.D. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146:1564–1572. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin H., An Y., Hao F., Wang Y., Tang H. Correlations of fecal metabonomic and microbiomic changes induced by high-fat diet in the pre-obesity state. Sci. Rep. 2016;6:21618. doi: 10.1038/srep21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G., Zhang L., Li T., Lopaschuk G., Vance D.E., Jacobs R.L. Choline deficiency attenuates body weight gain and improves glucose tolerance in ob/ob mice. J. Obes. 2012;2012:319172. doi: 10.1155/2012/319172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs R.L., Zhao Y., Koonen D.P., Sletten T., Su B., Lingrell S., Cao G., Peake D.A., Kuo M.S., Proctor S.D., Kennedy B.P., Dyck J.R., Vance D.E. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schugar R.C., Willard B., Wang Z., Brown J.M. Postprandial gut microbiota-driven choline metabolism links dietary cues to adipose tissue dysfunction. Adipocyte. 2018;7:49–56. doi: 10.1080/21623945.2017.1398295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.