Abstract
Background
In China, lung cancer is also the most commonly diagnosed cancer with a lower 5‐year survival rate, leading to high social burdens. Recently, many studies highlighted the importance of inflammation in the initiation and progression of cancer. The goal of this study was to investigate the association between interleukin‐4 (IL‐4, OMIM#147780) single nucleotide polymorphisms (SNPs) and lung cancer susceptibility.
Methods
A case‐control study was conducted in a Chinese population including 199 male patients with lung cancer and 266 healthy men. Six SNPs selected from the HapMap database were genotyped using Agena MassARRAY. Genetic models and haplotype analyses were utilized to evaluate the association between SNPs and lung cancer risk.
Results
In our findings, rs2243250 was associated with a decreased lung cancer risk under the log‐additive model (odds ratio, OR = 0.71, 95% confidence interval, CI = 0.51–0.97, p = 0.030), and the G/G genotype of rs2227284 conferred a negative effect; the risk of lung cancer under the codominant (OR = 0.19, 95% CI = 0.04–0.87, p = 0.040) and recessive models (OR = 0.20, 95% CI = 0.04–0.88, p = 0.012) after adjusted by age.
Conclusions
These data indicated potential associations between IL‐4 polymorphisms and lung cancer susceptibility. That may help to improve the understanding of the relationship between inflammation and lung cancer in the future.
Keywords: interleukin‐4, lung cancer, rs2227284, rs2243250
1. INTRODUCTION
Currently, lung cancer is the most common cancer around the world and the leading cause of cancer‐related deaths (Stewart & Wild, 2014). In China, lung cancer is also the most commonly diagnosed cancer with a lower 5‐year survival rate, and the estimated age‐standardized mortality rate in 2015 for lung cancer was 610.2 per 100,000 population including 432.4/100,000 men and 177.8/100,000 women (Chen et al., 2016), which was much higher than the world average. According to the research, an estimated 733.3 per 100,000 new lung cancer patients occurred in China in 2015, including 509.5/100,000 males and 224.0/100,000 females (Chen et al., 2016). Comparing with the previous data, the incidence of lung cancer in China was on the rise, leading to high social and economic burdens (Hong et al., 2015). Therefore, it is necessary to enhance the efficiency of prevention and early diagnosis of lung cancer.
Many definite risk factors have been found to be linked to lung cancer risk, such as tobacco use, environmental pollution, food, genetics, and chronic obstructive pulmonary disease (Hong et al., 2015). Also, researchers revealed that genetic factors have an important role in the etiology of lung cancer, such as TP53 (OMIM#191170), RB1 (OMIM# 614041), and so forth (Granville & Dennis, 2005; Yin et al., 2016). Recently, much attention of genetic susceptibility studies of cancer has been focused on single nucleotide polymorphisms (SNPs) in candidate genes. In 2010, Timofeeva et al. reported that polymorphisms in genes involved in Phase I and Phase II of xenobiotic metabolism are related to the risk of early‐onset lung cancer (Timofeeva et al., 2010). In 2015, Leng et al. conducted a genome‐wide association study, and they reported that a functional 15q12 variant was identified as a risk factor for lung cancer (Leng et al., 2015).
Many clinical and epidemiologic studies have suggested strong association between inflammation and lung cancer (Barreiro et al., 2013; Sohal, Mahmood, & Walters, 2014). Tumor cells could produce several cytokines and chemokines, some of which have been implicated to mediate different steps in the pathway leading to carcinogenesis (Gomes, Teixeira, Coelho, Araújo, & Rui, 2014). Interleukin‐4 (IL‐4) is an anti‐inflammatory cytokine related to the growth of some tumors, such as colon, breast, and lung (Gomes et al., 2012; Toi, Bicknell, & Harris, 1992). In lung cancer, IL‑4 (OMIM#147780) polymorphisms have been associated with a reduced risk of non‐small cell lung cancer (NSCLC) among the Portuguese and Chinese populations (Gomes et al., 2012; Gu, Shen, & Zhang, 2014). However, the overall information about association between IL‐4 polymorphisms and lung cancer risk is poor.
In this study, we performed a case‐control study to evaluate the association between SNPs distributed in the IL‐4 gene and lung cancer susceptibility in a Chinese population. Six SNPs were genotyped and analyzed in an attempt to identify new susceptibility locus that may provide guidance for early diagnosis for lung cancer.
2. MATERIALS AND METHODS
2.1. Ethical compliance
The study protocol was abided by the Declaration of Helsinki and its later amendments or comparable ethical standards. Additionally, this study was approved by the Clinical Research Ethics of Hainan General Hospital for Approval of Research Involving Human Subjects and written informed consent was obtained from all individuals included in the study.
2.2. Study subjects and sample collection
A total of 199 male patients with lung cancer and 266 healthy men were included in this study. The demographic characteristics of the lung cancer patients recruited from the First Affiliated Hospital of Xi'an Jiao tong University were shown in Table 1. The inclusions of the case groups were: (a) males; (b) diagnosed with confirmed lung cancer patients; (c) did not have history of any other cancers; (d) never received chemotherapy or radiotherapy treatment before. Healthy controls with no evidence of lung or other cancers were selected from healthy men who did medical examination during the same period.
Table 1.
Characteristics of the male individuals in cases and controls
| Group | N | Mean | Standard deviation (SD) | Mean ± SD | p‐value | |
|---|---|---|---|---|---|---|
| Age | Case | 199 | 59.31 | 9.642 | 59.31 ± 9.642 | p < 0.001* |
| Control | 266 | 48.06 | 12.555 | 48.06 ± 12.555 |
p‐values were calculated by Welch's t tests.
Five milliliters of venous blood samples were collected from each subject into tubes containing EDTA, then centrifuged and stored at −80°C.
2.3. SNP selection and genotyping
Six SNPs (rs2243250, rs2227284, rs2243267, rs2243270, rs2243283, rs2243289) with a minor allele frequency (MAF) > 0.05 in IL‐4 gene (its GenBank reference is NC_000005.10) were selected from theHapMap database. Genomic DNA was extracted from blood sample by GoldMag‐Mini Whole Blood Genomic DNA Purification Kit (GoldMag Ltd. Xi'an. China) according to manufacturer's protocol, and the concentration of DNA was measured by Nanodrop 2000. MassARRAY Assay Design 3.0 Software (Agena, San Diego, CA, USA) was utilized to design Multiplex SNP MassEXTEND assays (Yang et al., 2005). SNPs genotyping were performed based on Agena MassARRAY RS1000 (Agena, Inc.) according to the manufacturer's protocol, and the Agena Typer Software, version 4.0 (Agena, Inc.) was used to manage and analyze the data as earlier described (Gabriel, Ziaugra, & Tabbaa, 2009; Thomas et al., 2007).
2.4. Statistical analysis
Data were analyzed using SPSS version 18.0 statistical software (SPSS Inc., Chicago, IL, United States) and Excel 20.0 (Microsoft Corp., Redmond, WA, United States) (Zhou et al., 2018). Distribution differences in age and gender between cases and controls were compared by using Welch's t test and Chi‐square test, respectively (Adamec, 1964). Hardy–Weinberg Equilibrium (HWE) was assessed for the frequency of each SNP using a goodness‐of‐fit χ2 test in the control subjects. Logistic regression analysis was used to determine the association between SNPs and the risk of lung cancer by calculating ORs and 95% CIs (Bland & Altman, 2000; Zheng et al., 2017). Web‐based software SNPStats (https://www.snpstats.net/start.htm?q=snpstats/start.htm) was used to analyze the association under five genetic models (codominant, dominant, over‐dominant, recessive, and log‐additive) with an adjustment of age (Valls & Iniesta, 2006). Haploview software package (version 4.2) and the SHEsis software platform were used to conduct linkage disequilibrium analysis and haplotype‐based associations (Barrett, Fry, Maller, & Daly, 2005; Shi & He, 2005). All p values in this study were two‐tailed, and p < 0.05 was considered statistically significant.
SNP–SNP interactions were investigated by using the MDR (Multifactor dimensionality reduction) software package (version 3.0.2). The best model with the maximization of cross‐validation consistency was selected (Ritchie et al., 2001). In addition, traditional statistical methods were performed to examine the results from MDR analyses, and p < 0.05 was considered statistically significant (Lin et al., 2013).
3. RESULTS
3.1. Demographic information of participants
A total of 199 patients with lung cancer and 266 healthy individuals were enrolled in this study, and all the participants are males. Demographic information of participants is listed in Table 1. The mean age of the participants was 48.06 ± 12.555 years in the control group and 59.31 ± 9.642 years in the case group. In addition, there were significant differences in age distribution between the case and control groups (p < 0.05).
3.2. Association between SNPs in IL‐4 and lung cancer risk
Six SNPs (rs2243250, rs2227284, rs2243267, rs2243270, rs2243283, rs2243289) selected from the HapMap were genotyped, Table 2 summarized the frequency information of tested SNPs among the individuals in the case and control groups. In the control groups, all six SNPs were conformed to HWE (p > 0.05). Then, we conducted two‐sided Pearson chi‐square tests to identify differences in allele frequency distributions between cases and controls. As shown in Table 3, rs2243250 (HGVS: NM_000589.3:g.132673462C>T) was related to a decreased risk of lung cancer (OR = 0.692, 95% CI = 0.500–0.958, p = 0.026), and similar results (OR = 0.722, 95% CI = 0.521–1.001, p = 0.050) were observed in rs2243267 (HGVS: NM_000589.3: g.132678194G>C). No associations were observed between the other four SNPs and lung cancer. After Bonferroni correction, none of the SNPs showed statistically significant associations.
Table 2.
Basic information of the SNPs in IL‐4
| SNP ID | Position | Role | Allele | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A/B) | AA | AB | BB | AA | AB | BB | A (MAF) | B | A (MAF) | B | |||
| rs2243250 | 132,009,154 | Promoter | C/T | 7 | 57 | 135 | 19 | 89 | 158 | 0.178 | 0.822 | 0.239 | 0.761 |
| rs2227284 | 132,012,725 | Intron | G/T | 2 | 49 | 148 | 13 | 66 | 186 | 0.133 | 0.867 | 0.174 | 0.826 |
| rs2243267 | 132,013,886 | Intron | G/C | 7 | 57 | 135 | 17 | 89 | 160 | 0.178 | 0.822 | 0.231 | 0.769 |
| rs2243270 | 132,014,109 | Intron | A/G | 7 | 57 | 134 | 17 | 89 | 160 | 0.179 | 0.821 | 0.231 | 0.769 |
| rs2243283 | 132,016,593 | Intron | G/C | 7 | 55 | 137 | 14 | 76 | 175 | 0.173 | 0.827 | 0.196 | 0.804 |
| rs2243289 | 132,018,132 | Intron (boundary) | A/G | 7 | 58 | 134 | 16 | 90 | 160 | 0.181 | 0.819 | 0.229 | 0.771 |
SNP: single‐nucleotide polymorphism; MAF: minor allele frequency; A/B: minor/major alleles frequencies in the controls and cases.
The GenBank reference of IL‐4: NC_000005.10.
Table 3.
IL‐4 polymorphisms and lung cancer risk
| SNP ID | HWE p‐value | ORs | 95% CI | Pearson Chi‐square p‐value | p‐value after Bonferroni correction |
|---|---|---|---|---|---|
| rs2243250 | 0.236 | 0.69 | 0.500–0.958 | 0.026 | 0.156 |
| rs2227284 | 0.050 | 0.73 | 0.507–1.055 | 0.093 | 0.558 |
| rs2243267 | 0.387 | 0.72 | 0.521–1.001 | 0.050 | 0.300 |
| rs2243270 | 0.387 | 0.73 | 0.524–1.007 | 0.054 | 0.324 |
| rs2243283 | 0.170 | 0.86 | 0.614–1.203 | 0.376 | 1.000 |
| rs2243289 | 0.489 | 0.74 | 0.536–1.028 | 0.072 | 0.432 |
SNP: single‐nucleotide polymorphism; HWE: Hardy–Weinberg equilibrium; ORs: odds ratios; 95% CI: 95% confidence interval.
The GenBank reference of IL‐4: NC_000005.10.
p‐values were calculated by Pearson χ2 test.
Additionally, we assessed the association under five different genetic models (dominant, recessive, log‐additive, codominant, and over‐dominant) by logistic regression analysis. As shown in Table 4, rs2243250 was associated with a decreased lung cancer risk under the log‐additive model (OR = 0.71, 95% CI = 0.51–0.97, p = 0.030); the G/G genotype of rs2227284 (HGVS: NM_000589.3:g.132677033T>G) conferred a protective effect on the lung cancer risk in the codominant (OR = 0.19, 95% CI = 0.04–0.87, p = 0.040); and recessive model (OR = 0.20, 95% CI = 0.04–0.88, p = 0.012). With adjustment by age, G/G genotype of rs2227284 still significantly decreased lung cancer risk by 0.20‐fold (OR = 0.20, 95% CI = 0.04–0.94, p = 0.020).
Table 4.
Logistic regression analysis of the association between polymorphisms in IL‐4 and lung cancer
| SNP ID | Model | Genotype | Control | Case | OR (95% CI) | p‐value |
|---|---|---|---|---|---|---|
| rs2243250 | Codominant | T/T | 158 (59.4%) | 135 (67.8%) | 1.00 | 0.084 |
| C/T | 89 (33.5%) | 57 (28.6%) | 0.75 (0.50–1.12) | |||
| C/C | 19 (7.1%) | 7 (3.5%) | 0.43 (0.18–1.06) | |||
| Dominant | T/T | 158 (59.4%) | 135 (67.8%) | 1.00 | 0.061 | |
| C/T‐C/C | 108 (40.6%) | 64 (32.2%) | 0.69 (0.47–1.02) | |||
| Recessive | T/T‐C/T | 247 (92.9%) | 192 (96.5%) | 1.00 | 0.084 | |
| C/C | 19 (7.1%) | 7 (3.5%) | 0.47 (0.20–1.15) | |||
| Over‐dominant | T/T‐C/C | 177 (66.5%) | 142 (71.4%) | 1.00 | 0.270 | |
| C/T | 89 (33.5%) | 57 (28.6%) | 0.80 (0.54–1.19) | |||
| Log‐additive | — | — | — | 0.71 (0.51–0.97) | 0.030 | |
| rs2227284 | Codominant | T/T | 186 (70.2%) | 148 (74.4%) | 1.00 | 0.040 |
| G/T | 66 (24.9%) | 49 (24.6%) | 0.93 (0.61–1.43) | |||
| G/G | 13 (4.9%) | 2 (1%) | 0.19 (0.04–0.87) | |||
| Dominant | T/T | 186 (70.2%) | 148 (74.4%) | 1.00 | 0.320 | |
| G/T‐G/G | 79 (29.8%) | 51 (25.6%) | 0.81 (0.54–1.23) | |||
| Recessive | T/T‐G/T | 252 (95.1%) | 197 (99%) | 1.00 | 0.012 | |
| G/G | 13 (4.9%) | 2 (1%) | 0.20 (0.04–0.88) | |||
| Overdominant | T/T‐G/G | 199 (75.1%) | 150 (75.4%) | 1.00 | 0.940 | |
| G/T | 66 (24.9%) | 49 (24.6%) | 0.98 (0.64–1.51) | |||
| Log‐additive | — | — | — | 0.74 (0.52–1.06) | 0.100 |
SNP: single‐nucleotide polymorphism; HWE: Hardy–Weinberg equilibrium; ORs: odds ratios; 95% CI: 95% confidence interval.
The GenBank reference of IL‐4: NC_000005.10.
p‐values were calculated by logistic regression analysis without adjustment.
3.3. Association between haplotypes in IL‐4 and lung cancer risk
We further performed linkage disequilibrium analysis for six SNPs in the logistic regression model. The haplotype structure of the IL‐4 gene was analyzed, and single LD block consisting of all six SNPs were detected (Figure 1). Four common haplotypes were listed in Table 5. Interestingly, we found that the Crs2243250Grs2227284Grs2243267Ars2243270Crs2243283Ars2243289 haplotype was associated with a significantly decreased lung cancer risk (OR = 0.63, 95% CI = 0.42–0.96, p = 0.034). After adjustment of age, the association was still significant (OR = 0.67, 95% CI = 0.46–0.97, p = 0.035).
Figure 1.
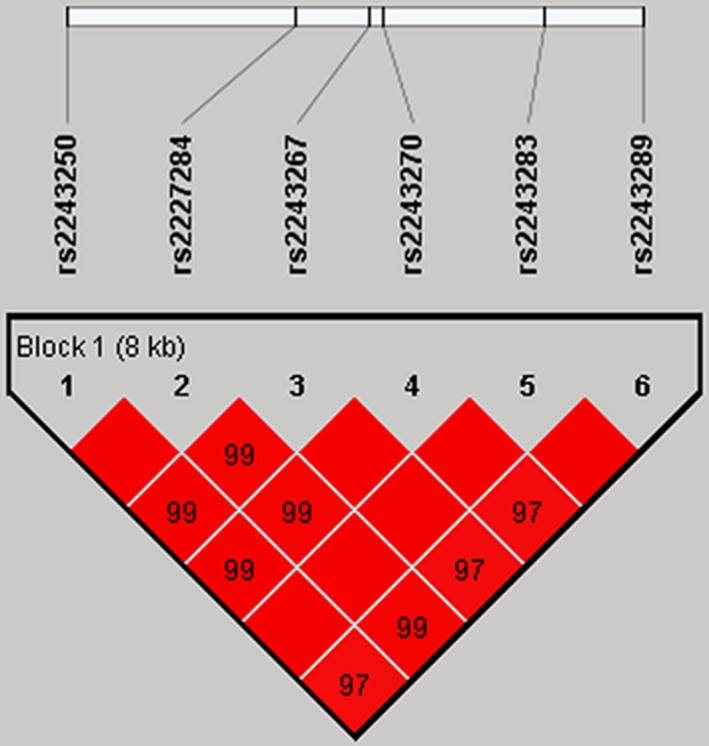
Linkage disequilibrium analysis for six SNPs in the logistic regression model. Standard color schemes indicate different levels of LD. Bright red: LOD > 2, D’ = 1
Table 5.
IL‐4 haplotype frequencies and the association with the risk of lung cancer
| rs2243250 | rs2227284 | rs2243267 | rs2243270 | rs2243283 | rs2243289 | Freq | Crude analysis | adjusted by age | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p a‐value | OR (95% CI) | p b‐value | ||||||||
| 1 | T | T | C | G | C | G | 0.598 | 1.00 | — | 1.00 | — |
| 2 | T | T | C | G | G | G | 0.185 | 0.76 (0.54–1.08) | 0.120 | 0.78 (0.53–1.14) | 0.200 |
| 3 | C | G | G | A | C | A | 0.150 | 0.67 (0.46–0.97) | 0.035 | 0.63 (0.42–0.96) | 0.034 |
| 4 | C | T | G | A | C | A | 0.050 | 0.73 (0.40–1.33) | 0.300 | 0.87 (0.44–1.73) | 0.690 |
| Rare | * | * | * | * | * | * | 0.013 | 0.38 (0.10–1.45) | 0.160 | 0.38 (0.08–1.68) | 0.200 |
OR: odd ratio; 95% CI: 95% confidence interval. The GenBank reference of IL‐4: NC_000005.10.
p a‐values were calculated by Wald test without adjustment.
p b‐values were calculated by Wald test adjusted for age.
indicates the rare haplotypes with lower frequencies.
3.4. SNP–SNP interaction and lung cancer risk
MDR software package was used to detect the potential interactions between the analyzed SNPs in relation to lung cancer risk. Table 6 summarized the cross‐validation consistency and the prediction error of best models. Obviously, the three‐locus model (rs2243250, rs2243284, rs2243283) had a maximum testing accuracy of 56.4% and a maximum cross‐validation consistency (10/10) that was significant at p < 0.05 level, after determined empirically by permutation testing.
Table 6.
Predication of lung cancer risk factors in MDR analysis
| Best model | Training accuracy (%) | Testing accuracy (%) | Cross‐validation consistency | χ2 | p‐value |
|---|---|---|---|---|---|
| rs2243250, rs2243283 | 55.5 | 50.5 | 5/10 | 5.223 | 0.022 |
| rs2243250, rs2227284, rs2243283 | 57.6 | 56.4 | 10/10 | 14.607 | p < 0.000 |
| rs2243250, rs2227284, rs2243283, rs2243289 | 58.0 | 56.7 | 9/10 | 16.652 | p < 0.000 |
MDR: Multifactor dimensionality reduction. The GenBank reference of IL‐4: NC_000005.10.
p‐values were calculated by Pearson χ2 test.
4. DISCUSSION
In the present hospital‐based case‐control study, we investigated the association between six single nucleotide polymorphisms in the IL‐4 gene and lung cancer risk in the Chinese male populations. Among these SNPs, rs2243250 and rs2243267 were associated with a decreased risk of lung cancer. In the genetic model analysis, rs2243250 decreased lung cancer risk under the log‐additive model, and the G/G genotype of rs2227284 conferred a protective effect on lung cancer susceptibility under the codominant and recessive models. Additionally, LD block was constructed with six SNPs in IL‐4, and we found that the CGGACA haplotype was related to a significantly decreased lung cancer susceptibility. In the result of MDR, the potential interactions among the three SNPs (rs2243250, rs2243284, rs2243283).
IL‐4 is a pleiotropic type Ⅱ cytokine which is encoded by the gene IL‐4 located within the cytokine gene cluster on chromosome 5q31.1 (Rosenwasser et al., 1995). Multiple biological processes are regulated by IL‐4, such as proliferation, differentiation, and apoptosis in various cell types (Nelms, Keegan, Zamorano, Ryan, & Paul, 1999). However, the role of IL‐4 in cancer is paradoxical. Okada et al. found that direct delivery of IL‐4 gene to a central nervous system tumor site prolongs the survival of animals by inhibiting tumor growth to some extent (Villa, 2001). Lee et al. found that IL‐4 exhibited a strong inhibitory function in the two critical steps of angiogenesis—migration of endothelial cells to the inflamed site and differentiation of migrated cells to the organized vessel structure (Lee et al., 2002). Based on the above evidences, it is suggested that IL‑4 has strong potential as a tumor therapy agent. But, there is still some evidence that IL‐4 is a tumor‑promoting molecule. For instance, Stremmel et al. found that IL‑4 knockout mice are more resistant to tumor challenge than IL‑4 competent mice, indicating that IL‐4 may play a critical role in abrogation of the antitumor immune response (Stremmel, Greenfield, Howard, Freeman, & Kuchroo, 1999). In addition, the expression of IL‐4 was up‐regulated in many cancers, such as colon cancer, NSCLC, and cervical cancer (Al‐Saleh et al., 1998; Asselin‐Paturel et al., 1998; Berghella et al., 1998). Therefore, further studies are required to clarify the real function of IL‐4 in cancers.
We found that rs2243250 was related to a lower risk of lung cancer. This was consistent with the previous studies. In 2012, Li et al. conducted a case‐control study in Chinese population and reported that frequencies of IL‐4 –590 genotype (TC and CC), and −590 C allele were significantly lower in patients with NSCLC than in healthy controls (Li et al., 2012). The same year, Gomes et al. reported that increased expression of IL‐4 associated with the TT genotype may contribute to immune surveillance during NSCLC development in Caucasian (Gomes et al., 2012). Besides, many studies showed that rs2243250 were associated with genetic susceptibility to other diseases, such as asthma, rheumatoid arthritis, and multiple sclerosis (Liu, Li, & Liu, 2012; Qiu et al., 2015). In our results, we revealed that rs2243267 and rs2227284, two less studied SNPs, were also found to be associated with lung cancer in this study. As well as, the relationship between rs2227284 and lung cancer risk has not been previously reported and we firstly reported it for the first time.
However, there were still some deficiencies in this study. Firstly, some bias in population selection should not be ignored. We observed the HWE in the controls, ORs, and 95% CIs on the basis of logistic regression analysis adjusted by age and gender to eliminate the bias to some extent. Secondly, the sample size was not larger and the reliability of the results needs to be further verified. Third, other risk factors (occupational exposures, physical activity etc.) were not included and should be considered to be assessed in the future and more comprehensive analyses about IL‐4 polymorphisms in lung cancer are needed to confirm the results.
In conclusion, we demonstrated that rs2243250, rs2243267, and rs2227284 in IL‐4 gene are associated with a decreased risk of lung cancer in northwestern Chinese males. These results may help improve the understanding of relationship between lung cancer and inflammation related gene IL‐4. On the other hand, the lung cancer‐associated molecular markers identified here might be useful as diagnostic and prognostic markers for lung cancer in future clinical studies.
CONFLICT OF INTERESTS
The authors declare no potential conflicts of interest.
ACKNOWLEDGEMENTS
The authors would like to express their thankfulness to everyone for assistance in the preparation of this manuscript and we also appreciate the support of first affiliated hospital of zhengzhou university, Northwest University, and Hainan General Hospital.
Tan N, Song J, Yan M, et al. Association between IL‐4 tagging single nucleotide polymorphisms and the risk of lung cancer in China. Mol Genet Genomic Med. 2019;7:e585 10.1002/mgg3.585
REFERENCE
- Adamec, C. (1964). Example of the use of the nonparametric test. Test X2 for comparison of 2 independent examples. Ceskoslovenske Zdravotnictvi, 12, 613–619. [PubMed] [Google Scholar]
- Al‐Saleh, W. , Giannini, S. L. , Jacobs, N. , Moutschen, M. , Doyen, J. , Boniver, J. , & Delvenne, P. (1998). Correlation of T‐helper secretory differentiation and types of antigen‐presenting cells in squamous intraepithelial lesions of the uterine cervix. Journal of Pathology, 184, 283–290. [DOI] [PubMed] [Google Scholar]
- Asselin‐Paturel, C. , Echchakir, H. , Carayol, G. , Gay, F. , Opolon, P. , Grunenwald, D. , … Mami‐Chouaib, F. (1998). Quantitative analysis of Th1, Th2 and TGF‐beta1 cytokine expression in tumor, TIL and PBL of non‐small cell lung cancer patients. International Journal of Cancer, 77, 7–12. [DOI] [PubMed] [Google Scholar]
- Barreiro, E. , Fermoselle, C. , Mateu‐Jimenez, M. , Sánchez‐Font, A. , Pijuan, L. , Gea, J. , & Curull, V. (2013). Oxidative stress and inflammation in the normal airways and blood of patients with lung cancer and COPD. Free Radical Biology & Medicine, 65, 859–871. 10.1016/j.freeradbiomed.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Barrett, J. C. , Fry, B. , Maller, J. , & Daly, M. J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Berghella, A. M. , Pellegrini, P. , Beato, T. D. , Marini, M. , Tomei, E. , Adorno, D. , & Casciani, C. U. (1998). The significance of an increase in soluble interleukin‐2 receptor level in colorectal cancer and its biological regulating role in the physiological switching of the immune response cytokine network from TH1 to TH2 and back. Cancer Immunology Immunotherapy, 45, 241–249. 10.1007/s002620050439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland, M. J. , & Altman, G. D. (2000). Statistics notes: The odds ratio. BMJ, 320, 1468 10.1136/bmj.320.7247.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Zheng, R. , Baade, P. D. , Zhang, S. , Zeng, H. , Bray, F. , … He, J. (2016). Cancer statistics in China, 2015. CA‐A Cancer Journal for Clinicians, 66, 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- Gabriel, S. , Ziaugra, L. , & Tabbaa, D. (2009). SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current Protocols in Human Genetics. Chapter 2: Unit 2.12. [DOI] [PubMed] [Google Scholar]
- Gomes, M. , Coelho, A. , Araújo, A. , Teixeira, A. L. , Catarino, R. , & Rui, M. (2012). Influence of functional genetic polymorphism (− 590C/T) in non‐small cell lung cancer (NSCLC) development: The paradoxal role of IL‐4. Gene, 504, 111–115. 10.1016/j.gene.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Gomes, M. , Teixeira, A. L. , Coelho, A. , Araújo, A. , & Rui, M. (2014). The role of inflammation in lung cancer. Advances in Experimental Medicine & Biology, 816, 1–23. [DOI] [PubMed] [Google Scholar]
- Granville, C. A. , & Dennis, P. A. (2005). An overview of lung cancer genomics and proteomics. American Journal of Respiratory Cell & Molecular Biology, 32, 169–176. 10.1165/rcmb.F290 [DOI] [PubMed] [Google Scholar]
- Gu, J. , Shen, Y. , & Zhang, Y. (2014). Association between interleukin‐4 polymorphisms and environment and nonsmall cell lung cancer in Chinese population. Journal of Cancer Research & Therapeutics, 10(Suppl.), C135–139. [DOI] [PubMed] [Google Scholar]
- Hong, Q. Y. , Wu, G. M. , Qian, G. S. , Hu, C. P. , Zhou, J. Y. , Chen, L. A. , … Wang, Q. (2015). Prevention and management of lung cancer in China. Cancer, 121(Suppl.), 3080–3088. 10.1002/cncr.29584 [DOI] [PubMed] [Google Scholar]
- Lee, I. Y. , Kim, J. , Ko, E. M. , Jeoung, E. J. , Kwon, Y. G. , & Choe, J. (2002). Interleukin‐4 inhibits the vascular endothelial growth factor‐ and basic fibroblast growth factor‐induced angiogenesis in vitro. Molecules & Cells, 14, 115–121. [PubMed] [Google Scholar]
- Leng, S. , Liu, Y. , Weissfeld, J. L. , Thomas, C. L. , Han, Y. , Picchi, M. A. , … Do, K. C. (2015). 15q12 Variants, sputum gene promoter hypermethylation, and lung cancer risk: A GWAS in smokers. Journal of the National Cancer Institute, 107 10.1093/jnci/djv035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Shi, W. , Yu, G. , Lin, L. , Yang, B. , Li, J. , … Gao, H. (2012). Interleukin‐4 ‐590T/C polymorphism influences the susceptibility to nonsmall cell lung cancer. DNA & Cell Biology, 31, 797–800. 10.1089/dna.2011.1425 [DOI] [PubMed] [Google Scholar]
- Lin, Z. , Su, Y. , Zhang, C. , Xing, M. , Ding, W. , Liao, L. , … Cui, D. (2013). The interaction of BDNF and NTRK2 gene increases the susceptibility of paranoid schizophrenia. PLoS ONE, 8, e74264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Li, T. , & Liu, J. (2012). Interleukin‐4 rs2243250 polymorphism is associated with asthma among Caucasians and related to atopic asthma. Cytokine, 59, 364–369. 10.1016/j.cyto.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Nelms, K. , Keegan, A. D. , Zamorano, J. , Ryan, J. J. , & Paul, W. E. (1999). The IL‐4 receptor: Signaling mechanisms and biologic functions. Annual Review of Immunology, 17, 701–738. 10.1146/annurev.immunol.17.1.701 [DOI] [PubMed] [Google Scholar]
- Qiu, L. J. , Ni, J. , Cen, H. , Wen, P. F. , Zhang, M. , Liang, Y. , … Ye, D. Q. (2015). Relationship between the IL‐4 gene promoter ‐590C/T (rs2243250) polymorphism and susceptibility to autoimmune diseases: A meta‐analysis. Journal of the European Academy of Dermatology & Venereology, 29, 48–55. 10.1111/jdv.12435 [DOI] [PubMed] [Google Scholar]
- Ritchie, M. D. , Hahn, L. W. , Roodi, N. , Bailey, L. R. , Dupont, W. D. , Parl, F. F. , & Moore, J. H. (2001). Multifactor‐dimensionality reduction reveals high‐order interactions among estrogen‐metabolism genes in sporadic breast cancer. American Journal of Human Genetics, 69, 138–147. 10.1086/321276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser, L. J. , Klemm, D. J. , Dresback, J. K. , Inamura, H. , Mascali, J. J. , Klinnert, M. , & Borish, L. (1995). Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clinical & Experimental Allergy, 25(Suppl.), 74–78. 10.1111/j.1365-2222.1995.tb00428.x [DOI] [PubMed] [Google Scholar]
- Shi, Y. Y. , & He, L. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research, 15, 97–98. 10.1038/sj.cr.7290272 [DOI] [PubMed] [Google Scholar]
- Sohal, S. S. , Mahmood, M. Q. , & Walters, E. H. (2014). Clinical significance of epithelial mesenchymal transition (EMT) in chronic obstructive pulmonary disease (COPD): Potential target for prevention of airway fibrosis and lung cancer. Clinical & Translational Medicine, 3, 33 10.1186/s40169-014-0033-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, B. , & Wild, C. (2014). World cancer report 2014. International Agency for Research on Cancer.
- Stremmel, C. , Greenfield, E. A. , Howard, E. , Freeman, G. J. , & Kuchroo, V. K. (1999). B7–2 expressed on EL4 lymphoma suppresses antitumor immunity by an interleukin 4–dependent mechanism. Journal of Experimental Medicine, 189, 919–930. 10.1084/jem.189.6.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, R. K. , Baker, A. C. , Debiasi, R. M. , Winckler, W. , Laframboise, T. , Lin, W. M. , … Garraway, L. A. (2007). High‐throughput oncogene mutation profiling in human cancer. Nature Genetics, 39, 347–351. 10.1038/ng1975 [DOI] [PubMed] [Google Scholar]
- Timofeeva, M. , Kropp, S. , Sauter, W. , Beckmann, L. , Rosenberger, A. , Illig, T. , … Wichmann, H.‐E. (2010). Genetic polymorphisms of MPO, GSTT1, GSTM1, GSTP1, EPHX1 and NQO1 as risk factors of early‐onset lung cancer. International Journal of Cancer, 127, 1547–1561. 10.1002/ijc.25175 [DOI] [PubMed] [Google Scholar]
- Toi, M. , Bicknell, R. , & Harris, A. L. (1992). Inhibition of colon and breast carcinoma cell growth by interleukin‐4. Cancer Research, 52, 275–279. [PubMed] [Google Scholar]
- Valls, J. , & Iniesta, R. (2006). SNPStats: A web tool for the analysis of association studies. Bioinformatics, 22, 1928–1929. 10.1093/bioinformatics/btl268 [DOI] [PubMed] [Google Scholar]
- Villa, L. (2001). Cytokine gene therapy of gliomas: Effective induction of therapeutic immunity to intracranial tumors by peripheral immunization with interleukin‐4 transduced glioma cells. Gene Therapy, 8, 1157–1166. 10.1038/sj.gt.3301496 [DOI] [PubMed] [Google Scholar]
- Yang, Q. , Guo, C. Y. , Cupples, L. A. , Levy, D. , Wilson, P. W. , & Fox, C. S. (2005). Genome‐wide search for genes affecting serum uric acid levels: The Framingham Heart Study. Metabolism Clinical & Experimental, 54, 1435–1441. 10.1016/j.metabol.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Yin, Z. , Cui, Z. , Ren, Y. , Xia, L. , Wang, Q. , Ying, Z. , … Zhou, B. (2016). Association between polymorphisms in pre‐miRNA genes and risk of lung cancer in a Chinese non‐smoking female population. Lung Cancer, 94, 15–21. 10.1016/j.lungcan.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Wang, M. , Tian, T. , Liu, K. , Liu, X. , Zhai, Y. , … Lu, J. (2017). Role of interleukin‐12 gene polymorphisms in the onset risk of cancer: A meta‐analysis. Oncotarget, 8, 29795–29807. 10.18632/oncotarget.16080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Zheng, Y. , Tian, T. , Liu, K. , Wang, M. , Lin, S. , … Dai, Z. (2018). Associations of interleukin‐6 gene polymorphisms with cancer risk: Evidence based on 49,408 cancer cases and 61,790 controls. Gene, 670, 136–147. 10.1016/j.gene.2018.05.104 [DOI] [PubMed] [Google Scholar]


