Abstract
Mitochondrial dysfunction has been associated with schizophrenia (SZ) and bipolar disorder (BD). This review examines recent publications and novel associations between mitochondrial genes and SZ and BD. Associations of nuclear-encoded mitochondrial variants with SZ were found using gene- and pathway-based approaches. Two control region mitochondrial DNA (mtDNA) SNPs, T16519C and T195C, both showed an association with SZ and BD. A review of 4 studies of A15218G located in the cytochrome B oxidase gene (CYTB, SZ = 11,311, control = 35,735) shows a moderate association with SZ (p = 2.15E-03). Another mtDNA allele A12308G was nominally associated with psychosis in BD type I subjects and SZ. The first published study testing the epistatic interaction between nuclear-encoded and mitochondria-encoded genes demonstrated evidence for potential interactions between mtDNA and the nuclear genome for BD. A similar analysis for the risk of SZ revealed significant joint effects (34 nuclear-mitochondria SNP pairs with joint effect p ≤ 5E-07) and significant enrichment of projection neurons. The mitochondria-encoded gene CYTB was found in both the epistatic interactions for SZ and BD and the single SNP association of SZ. Future efforts considering population stratification and polygenic risk scores will test the role of mitochondrial variants in psychiatric disorders.
Key Words: Mitochondrial DNA, Epistatic interactions, Genome-wide association, Pathway analysis, Schizophrenia, Bipolar disorder
Introduction
Schizophrenia (SZ) and bipolar disorder (BD) are psychiatric disorders with high heritability affecting at least 2% of the population and have a significant genetic correlation between the disorders. Mitochondrial dysfunctions in synaptic, energetic, and metabolic pathways have been associated with the neurobiology of both SZ and BD.
Until recently, the field of mitochondrial genetics in psychiatric disorders has been largely ignored in genome-wide association studies (GWAS). We summarize the recent genetic evidence of associations of mitochondrial-related genes with SZ and BD. This recent genetic evidence, while still subject to future analyses, is discussed in this review to provide an interim status on mitochondrial genetics studies.
In this discussion, the mitochondrial-related genes include both mitochondria-encoded genes and nuclear-encoded genes with mitochondrial function. Essentially, the nuclear genome contributes ∼1,500 genes and the mitochondrial DNA (mtDNA) encodes 37 genes together comprising the mitochondrial-related gene set. This overview is divided into three parts: (a) nuclear-encoded mitochondrial genes and SZ; (b) mtDNA-encoded variants in SZ and BD; and (c) epistatic interaction between nuclear and mtDNA variants in SZ and BD.
Recent reviews of psychiatric disorders and mitochondrial-related genes are available for background studies not included in this review [2, 3]. This paper presents novel associations between mitochondrial genes and these disorders as well as recently published work to inform the mitochondria hypothesis of SZ and BD.
Mitochondrial Genes in the Nuclear Genome and SZ
Nuclear-encoded mitochondrial genes have been implicated in the etiology and pathology of SZ in a number of published studies. Mitochondrial dysfunction in synaptic, energetic, and metabolic pathways has been associated with the neurobiology of both SZ and BD [4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34]. Table 1 is abridged from Hjelm et al. [10] to show genes with corroboratory evidence from 3 or more independent studies. Genes are ranked in Table 1 based on the number of independent associations with SZ and are then ranked based on Maestro Score (from MitoCarta) to display confidence in mitochondrial localization. In the full list of genes [10] there were 10 Complex I genes replicated in 2 or more independent studies. Studies evaluated in Table 1 include analyses of copy number variants (CNVs), rare and de novo mutations, GWAS, transcriptome alterations in SZ brains and in controls during adolescence (i.e., during stages of brain development physiologically relevant to the onset of SZ symptoms), and proteomics [34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46].
Table 1.
A subset of nuclear-encoded mitochondrial genes associated with SZ etiology or pathology in at least 3 independent studies
| Gene/studies | Study type [Ref.] |
|---|---|
| MDH2/4 | denovo CNV [37], CNV [36],↓ RNA (DLPFC pyramidal) [34],↓RNA(DG) [42] |
| NDUFS2/4 | GWAS (early onset) [34],↓ RNA (DLPFC pyramidal) [34], Peak Expression (15–25 y) [41],DER(teen>adult) [45] |
| ATP5A1/3 | Rare mutation [38],↓ RNA (PFC area 9) [43],↑ Protein (DLPFC/ACC) [46] |
| IDH3A/3 | ↓RNA (DLPFC pyramidal) [34],↓RNA (DG) [42],↓Protein (ACC)[46] |
| NDUFA2/3 | GWAS (PGC) [40],↓RNA (DLPFC pyramidal) [34],↑ Protein (DLPFC) [46] |
| BDH1/3 | denovo CNV [37], CNV [36],DER(teen> adult) [45] |
| CLPX/3 | denovo mutation [39], Peak Expression (15–25 y) [41],DER(teen>child) [45] |
| GOT2/3 | CNV [36],↓RNA (PFC area 9) [43], Peak Expression (15–25 y) [41] |
| MDH1/3 | ↓RNA (PFC area 9) [43],DER(teen>child) [45],↓Protein (DLPFC) [46] |
The study type(s) and type of genetic variants for each gene are shown as adapted from a published analysis [10]. CNV, copy number variant; DLPFC, dorsolateral prefrontal cortex; DG, dentate gyrus; GWAS, genome-wide association study; DER, differentially expressed regions; PFC, prefrontal cortex; ACC, anterior cingulate cortex; PGC, psychiatric genomics consortium; CC, corpus callosum. ↓↑ direction of change in mRNA or protein compared to controls.
A closer inquiry of CNVs was conducted in a Swedish SZ sample (n = 4,719 cases and 5,917 controls) [36]. This study reported the nuclear-encoded mitochondrial gene set was significantly enriched in large CNVs in subjects with SZ [36]. Utilizing a subset of the same sample [38], several SNPs were nominally associated (p < 1E-06) with the risk of SZ, and pathways related to mitochondrial dysfunction and immunity pathways were associated with risk of SZ (Table 2). The p values in Table 2 are calculated using a right-tailed Fisher's exact test based on the distinct sets of genes that are mapped to all variants that were associated with SZ (p < 1E-06), compared to those genes that are known to be associated with a given pathway and results are adapted from Ingenuity Variants Analysis (IVA) Pathway Report. The p values less than 0.01 indicate that there are significantly more genes with the annotation than expected by chance relative to all genes implicated in the annotation from the Ingenuity Knowledge Base. The total number of pathways tested in IVA is 670; in Table 2, the p values that remain highly significant when correcting for multiple pathway comparisons are shown.
Table 2.
Pathways from the Swedish Exome Study [36] (n = 2,536 SZ and 2,543 controls)
| p value of enrichment | Genes,n | |
|---|---|---|
| Hematopoiesis frompluripotentstem cells | 4.997E-19 | 9 |
| Communication, innate and adaptive immune cells | 1.951E-17 | 29 |
| Super pathway of cholesterol biosynthesis | 7.362E-17 | 13 |
| OX40 signaling pathway | 8.421E-12 | 29 |
| Altered T cell and B cell signaling in rheumatoid arthritis | 8.412E-11 | 37 |
| Systemic lupus erythematosus signaling | 2.022E-09 | 102 |
| Apoptosis of target cells | 4.453E-09 | 14 |
| Mitochondrial dysfunction | 4.067E-08 | 73 |
| Dolichyl-diphosphooligosaccharide biosynthesis | 5.236E-08 | 6 |
| Nur77 signaling in T lymphocytes | 1.123E-07 | 26 |
The evidence shown in Tables 1 and 2 supports an enrichment of SZ-associated variants in nuclear-encoded mitochondrial genes in existing studies.
The large GWAS of SZ conducted by the Schizophrenia Working Group of the PGC (n = 36,989 cases and 113,075 controls) [40] identified 108 genomic loci associated with SZ. These 108 loci encompass 350 genes – and an enrichment analysis demonstrated an overrepresentation of genes expressed in the brain, as well as those specific to glutamatergic signaling and immunity [12]. We further noted that these 108 loci contain 22 genes implicated in mitochondrial function: NDUFA6, HSPD1, PCCB, NDUFA2, SHMT2, ATPAF2, NDUFA13, IMMP2L, DDX28, EFHD1, COQ10B, USMG5, C2orf47, DUS2L, HSPA9, MARS2, SFXN2, HSPE1, IREB2, AS3MT, DRG2, and TMTC1 [10]. This list represents a significant enrichment of the mitochondrial network relative to all RefSeq genes (5,000 randomly selected lists of 350 RefSeq genes containing 22 or more MitoCarta genes, p = 0.012).
The PGC study was extended to a larger analysis in terms of subjects using a set of 818 nuclear-encoded mitochondrial genes (Table 3, from Goncalves [47]). Meta-analysis results of mitochondria function in a combined discovery and replication sample (total n = 106,226) found several mitochondrial genes were significantly associated with SZ after multiple testing correction (Table 3). Note that this analysis does not cover mtDNA encoded genes. The genes in Table 3 were further queried in the CommonMind Consortium (CMC) human dorsolateral prefrontal cortex RNA-Seq data (https://www.synapse.org/CMC [48]). Expression data from 514 subjects showed 3 mitochondrial genes (SMDT1, PCCB, and NDUFA6) that were found to have statistically significantly decreased expression in SZ cases compared to controls after adjusting for covariate effects of brain pH, age, PMI, RIN, institution, and the manner of death (Table 3, bolded genes). Two genes (SMDT1 and PCCB) were differentially expressed in BD in the CMC DLPFC samples compared to controls (Table 3) and were significant but in the opposite direction compared to SZ.
Table 3.
List of mitochondrial genes significantly associated with SZ risk through meta-analysis after the conservative Bonferroni correction for the 818 multiple hypothesis tests conducted
| Gene symbol | Chr | Start, bp | Discovery (n = 82,315) |
Replication (n = 23,911) |
Meta-analysis (n = 106,226) |
|
|---|---|---|---|---|---|---|
| p a | p a | metap | Bonferroni-correctedp a | |||
| HSPE1-MOB4 | 2 | 198364721 | 1.59E-08 | 0.7527 | 2.30E-07 | 0.0002 |
| C2orf47 | 2 | 200820040 | 6.65E-07 | 0.2897 | 3.17E-06 | 0.0026 |
| SMDT1↓↑ | 22 | 42475695 | 1.54E-05 | 0.0206 | 5.05E-06 | 0.0041 |
| ATPAF2 | 17 | 17921334 | 1.19E-06 | 0.3115 | 5.88E-06 | 0.0048 |
| PCCB↓↑ | 3 | 135969167 | 7.27E-07 | 0.9221 | 1.02E-05 | 0.0083 |
| NDUFA6↓ | 22 | 42481530 | 8.03E-06 | 0.1106 | 1.33E-05 | 0.0108 |
| HSPA9 | 5 | 137890571 | 2.22E-06 | 0.5973 | 1.93E-05 | 0.0158 |
| DNAJA3 | 16 | 4475806 | 5.87E-06 | 0.3596 | 2.97E-05 | 0.0243 |
| FOXO3 | 6 | 108881026 | 1.79E-05 | 0.2440 | 5.83E-05 | 0.0477 |
Data from Goncalves et al. [47]. PGC-SZ2 (discovery)n = 35,476 cases and 46,839 controls. iPSYCH-SZ [66] (replication) n = 2,290 cases and 21,621 controls. Bolded genes show nominal differential expression (p < 0.05) in CommonMind Consortium (CMC) human dorsolateral prefrontal cortex RNA-Seq data [48];↓, decreased expression in SZ cases compared to controls;↑, significantly increased expression in bipolar disorder in CMC compared to controls. a Gene-basedp values using the MAGMA tool [90].
In a recent cross-disorder analysis of gene expression, the mitochondrial and neuronal gene coexpression module was shared between BD and SZ cortical regions [49], and the shared genes were downregulated, independent of antipsychotic treatment [50]. Not all recent studies of mitochondrial-related pathways reported an association with SZ. For example, the careful work of PGC Cross-Disorder and Pathway Groups did not show highly ranked mitochondria pathways as being involved in major psychiatric disorders [51]. The mitochondria pathway involves large numbers of genes; one possibility is lack of mitochondrial “pathway” involvement indicates that it is likely that there is not a major disruption of large numbers of common genes with broad effects on mitochondria which give rise to a disease association. This leaves open the possibility, as we discuss below, that rare variants as well as complex interactions involving mitochondrial-related genes can be associated with psychiatric disorders [2]. However, this cross-disorder pathway study did find that the synaptic compartment is highly relevant to SZ etiology. Synaptic activity is closely linked to mitochondrial function [52, 53]; synaptic currents are stabilized most when mitochondria are present near or within the synapse terminal. This area of research will likely produce more data that link mitochondria functioning and transport in neurons to synaptic physiology as one of the key determinants of synaptic functions [54, 55].
The early formulation of the theory of mitochondria dysfunction by Dr. Tadafumi Kato [7] indicated that brain energy metabolism might be altered in BD subjects, and there might be increased large deletions and genetic polymorphisms for BD. Studies of mitochondria involvement in BD show evidence of mitochondria dysfunction in BD [28, 30, 31]. Genetic link to postmortem brain pH was found for mtDNA super haplogroup (U, K, UK), which had a significantly higher postmortem pH (7.00 ± 0.18 SD) compared to the remaining subjects’ pH (6.8 ± 0.18 SD) [1]. A small association study of BD showed an association with T195C [1] and also with a 5178C/10398A mtDNA haplotype [8]. A decreased mtDNA copy number in BD has been reported [10, 56, 57]. However, pathway analysis of GWAS of BD did not reveal an association of pathways and mitochondria dysfunction [58]. Of the 226 genes that passed FDR in 3 of 4 GWAS [58], only 8 of these genes had predicted mitochondria function (MTHFD1L, ACSS3, SIAH3, PAK7, GOLSYN, SLIT3, KCNMA1, NAV2) which did not reach significance for the enrichment of mitochondrial genes. Mitochondria evidence was uncovered by studying iPSC-induced neurons from subjects with BD that were lithium nonresponders compared to lithium responder subjects. Induced neurons from lithium nonresponders showed hyperexcitability and increased mtDNA expressed genes [14]. This information suggests that the nonresponder endophenotype of BD subjects might harbor mitochondria risk factors, as this endophenotypic evidence was captured in cell lines derived from BD patients.
In summary, using gene-based analysis of mitochondrial-related nuclear genes, we find evidence that mitochondrial genes are associated with the architecture (CNV, SNV, and functional expression at protein and mRNA level) of SZ [10]. Genes involved in mitochondria Complex I appeared to be of particular importance in the gene-based analysis for the risk of SZ. The genetic evidence, while not unequivocal, did pass meta-analysis association threshold for several mitochondrial-related genes using a combined study of over 106,226 subjects.
Although the mitochondria genetic evidence has not yet been as thoroughly tested for BD mitochondrial risk compared to the test for SZ, there is accumulating evidence supporting mitochondria involvement in BD risk. These findings, in turn, motivate inquiry of involvement of variants in the mtDNA genome and its interaction with the nuclear genome in SZ, as well as BD, as reviewed in the following sections.
mtDNA-Encoded Variants in SZ and BD
The role of evolution in creating the genetic landscape for risk of SZ and maintaining these genes has been described [13]. According to this view, the human brain has evolved into a high energy consumption tissue, thus invoking mitochondrial genes during this selection process. Compared to other primates whose brain consumes about 13% of total energy at rest the human brain consumes about 20% of total energy at rest [59]. The SZ subjects might represent brains which do not reach the necessary bioenergetics demand due to maladaptive mitochondria. The presence of recent selective pressures acting on mtDNA is still an open question, but there is compelling evidence for balanced purifying selection acting against highly deleterious mutations on this genome in germline cells [60].
To investigate the selection of mtDNA variants, the number of nonsynonymous substitutions (dN) is compared to the number of synonymous substitutions (dS). The synonymous substitution rate is generally greater than the nonsynonymous rate in mtDNA in humans and other primates [61]. The rate was compared in SZ, controls, BD, and major depressive disorder (Table 4). The differences between psychiatric groups in this rate were tested using pairwise comparisons. Table 4 shows the number of times the null hypothesis of strict neutrality (dN = dS) was accepted (column [B]) or rejected in favor of dS > dN (column [A]). Using pairwise comparisons, a significant increase was found in the ratio (column [A]/[A + B]) for SZ compared to controls. The rate of synonymous substitutions was 22% higher (p = 0.0017) in DLPFC of individuals with SZ compared to controls [1], and there was a trend toward a higher substitution rate in BP, albeit not significant, and likely due to lack of overall power in the study. There was also an elevated rate of synonymous substitution in the MT-ND5 gene in SZ [62] in blood-derived mtDNA.
Table 4.
Codon-based test of purifying selection for pairwise analysis between sequences previously published [1]
| Group | [A] dS > dN |
[B] dN = dS |
[A]/[A + B] | [A]/[A + B]/ control | χ2 | p value |
|---|---|---|---|---|---|---|
| BD | 193 | 239 | 0.44 | (1.11) | 2.42 | 0.119 |
| MDD | 202 | 296 | 0.40 | (1.01) | 0.03 | 0.85 |
| SZ | 247 | 257 | 0.49 | (1.22) | 9.82 | 0.0017 |
| Control | 297 | 445 | 0.40 | (1.00) | NA | NA |
This effect was calculated using the transitions/transversions bias calculation for mtDNA coding sequences as previously published [1] using samples of DLPFC. The number of subjects for each group: BD = 12, MDD = 15, SZ = 14, control = 36.
A further analysis of the overall transition/transversion bias (R) for all DLPFC mtDNA sequences was conducted for each of the four subject groups separately using only coding genes. The observed transition/transversion ratios were 2.25-fold higher in SZ compared to controls [1]. This suggested a higher number of substitutions in DLPFC in SZ compared to controls in the 13 mtDNA genes. Two caveats apply here: the overall sample size was small (n = 77) and the total number of transversions was modest (n = 22), which can inflate R. With these caveats, the number of substitutions in SZ pairwise comparisons exceeded the rate in control comparisons, and the overall R rate is higher in SZ than in C. As an example, the maximum R ratio (transition/transversion) for mitochondrial complex I was 10-fold higher in SZ compared to controls [1]. The trend for BD elevation of purifying selection could be tested with larger samples becoming available.
The rate of transitions in mitochondria can be elevated due to instability in the mtDNA genome due to oxidation and lack of histone protection. Factors such as oxidative stress, reactive oxygen, and lack of mtDNA repair mechanisms can facilitate transmission of substituted bases. Since there are no histone groups protecting mtDNA from potential oxidation by reactive oxygen species, elevated transition bias merits further investigation. However, the calculation of dN and dS in a single population at a single time can lead to a violation of assumptions of purifying selection due to lack of independence within a single species. Although there is evidence reported of purifying selection pressure on mtDNA genomes from subjects with SZ, this finding needs to be replicated in a large case-control analysis of subjects from major ethnic haplogroups, to exclude occult population stratification.
A transition occurring in the hypervariable region in mtDNA was also associated with SZ and BD at positions T195C and T16519C [63]. The common T16519C transition is a recurring sporadic T>C mutation, that was shown to be elevated in three of the four populations studied. The original “C” ancestral allele was protective (Table 5). These positions in the hypervariable region are somewhat unstable in the mtDNA genome, are sometimes heteroplasmic, and occur independently in multiple haplogroups from different ethnic origins. As a side note, although T16519C is a heteroplasmic locus, homozygous calls are often forced for haploid chromosomes such as the mitochondria, or the allele is eliminated from analysis due to low confidence in homozygous calls.
Table 5.
T16519C association in schizophrenia was significant (p = 0.015)
| Study | Cases |
Controls |
n | p value | OR | ||
|---|---|---|---|---|---|---|---|
| 16519C | 16519T | 16519C | 16519T | ||||
| Mosquera [92] | 149 | 305 | 212 | 404 | 1,070 | 0.585 | 0.93 |
| Cardiff | 193 | 428 | 239 | 406 | 1,266 | 0.024 | 0.76 |
| Gain-WTCCC2 [63] | 329 | 805 | 1,285 | 2,620 | 5,039 | 0.013 | 0.83 |
| Costa Rica-Whole Exome | 425 | 304 | 243 | 212 | 1,184 | 0.093 | 1.22 |
| Pooled OR (Mantel-Haenszel) | 95% CI upper – lower: 0.80–0.98 | 8,559 | Two-tailedp = 0.027 | 0.894 | |||
OR, odds ratio. A Mantel-Haenszelχ2 test [91] was used to analyze the significance of the association across four combined samples of the mtDNA hypervariable T16519C locus. Note: ancestral allele 16519C is protective.
Common Haplogroup Variants in mtDNA and Association with SZ and BD
The mtDNA 195C allele was protective for BD and SZ analyzed together compared to controls (total n = 5,889) [63]. When the SZ group was analyzed separately, this effect was also significant. To test whether an ethnic-specific mtDNA SNP A12308G which defines the super-haplogroup U, K, UK matrilineages affected this association, a Cochran-Mantel-Haenszel test remained significant when adjusting for this major haplogroup, the association remained nominally significant (p = 0.0011). Thus, the risk decreases for both psychiatric disorders when the mtDNA control region alleles 16519C and 195C are found in different ethnic haplogroups, most likely as a back mutation.
For BD, an increased risk for psychosis was demonstrated in haplogroup U-K [64] at A12308G (OR = 2.48, p = 0.006). Recent data from a large case-control study of Swedish subjects [3] showed a nonsignificant association at A12308G (OR: 0.97, p = 0.27) with similar G allele frequency in SZ (0.26) and controls (0.25). Further analysis is required to clarify this association, as possibly due to differences in the haplogroup background of the subjects.
In addition to the A12308G variant, the mtDNA allele A15218G was significantly associated with risk of SZ. Two studies showed similar results unadjusted for potential population stratification (Table 6). An increased risk for SZ at A15218G was shown in the results of the WTCCC2 study [65]. This WTCCC2 study examined 45 mtDNA SNPs using subjects (n = 3,058 SZ cases and 5,033 controls) mainly of Irish descent. A study of Danish subjects using n > 25,000 subjects also showed modest association (p < 0.05) of A15218G for this ethnic SNP [66]. In a third published dataset using array genotype data, in the Swedish population [3] the risk allele A15218G was protective for risk of SZ (OR = 0.80, p = 0.02).
Table 6.
Association of an mtDNA variant (A15218G) shows an increased risk for G allele in a meta-analysis of four independent studies (total subjects n = 47,451)
| Study population [Ref.] | Schizophrenia, n | Controls,n | OR | p value |
|---|---|---|---|---|
| Swedish [3] | 4,579 | 5,610 | 0.8 | 0.02 |
| UK (WTCCC2) [65] | 3,058 | 5,033 | 2.87 | 4.99E-08 |
| Danish [66] | 2,538 | 2,3743 | 1.25 | 0.021 |
| USA (dB GAIN) [63] | 1,136 | 1,349 | 1.11 | 0.665 |
| Total SZ (n = 11,311) | Total C (n = 35,735) | 1.20 pooled Lower–upper 95%: 1.07–1.35 | 2.15E-03 | |
Combining these studies showed a modest but not genome-wide significant value (2.15E-03) for the association of the A15218G variant for risk of SZ with a pooled OR of 1.2 (Table 6). This evidence across different populations could be attributable to the enrichment of varying haplogroups carrying the 15218G allele (Table 6). This suggests that the mtDNA genetic backgrounds of each population are highly dependent on the haplogroups sampled [66], which adds complexity to the interpretation of the result. There has also been the suggestion that nSNPs and mtSNPs are in some linkage disequilibrium (LD) which will be addressed by using a joint and interaction analyses of the entire genome and stratifying cases and controls into subgroups while accounting for effects of multidimensional covariates [63]. It would provide a better understanding if the randomization method for each study could be run to test for association with stratification by study; unfortunately, the genotypes at the subject level data are not available for this analysis for all 4 studies of A15218G.
The A15218G variant occurs in multiple haplogroup branches at Mitomap [67] (A, B, D, F, H, HV, J, K, L, M, T, U). Due to the low frequency observed in ancestral populations, e.g. L2 and L3 branches, it is likely that the mutation gained prevalence later in the haplogroups diverging into Europe, such as the HV1a, HV1b, HV1c haplogroups, where 100% of sequenced individuals carry the variant. From Phylotree Build 17 [68], there are 5 haplogroups with A15218G as a defining branch SNP: M7a1a2, M10a, HV1a′b′c, H13a2c, U5a1. Overall, this variant is at 1.6% prevalence in 45,494 individuals worldwide. The widespread distribution of this variant suggests an independent mutation that arises independently worldwide. This variant is located in Complex III gene CYTB causing a nonsynonymous Thr = >Ala with the Thr being conserved across 27 primate species.
A recent paper [3] summarized additional relevant mtDNA haplogroup association studies across Asian and European populations (the reader is referred to a compilation). Most studies are small by GWAS parameters; however, given that many individuals in the HV haplogroup are carriers of the A15218G variant, a study of Israeli-Arab SZ and controls [69] is of interest. This study showed an OR of 1.99 based on sample size of n = 202 (SZ) and n = 202 (controls). Spanish samples purportedly showed a lack of association of HV subjects with SZ [70]. However, this does not rule out a role for A15218G, as it is present in only deeper branches of H and HV.
In summary, a risk allele in mtDNA present in multiple haplogroups (A15218G) shows a modest association in a meta-analysis of 4 independent studies. Not all of these combined studies of mtDNA and SZ (n = 47,046 subjects, Table 6) were corrected for potential population stratification effects [66] using principal components (PC) analysis, which could potentially account for the signal detection at this locus. Further, large GWAS of the control region variants T195C and T16519C have suggested that the ancestral allele is protective. At the time of this writing, we are not aware of any mtDNA association studies that have reached GWA significance levels and that have been corrected for population stratification. However, this does not imply that common variants or potentially ethnic-specific variants will not carry the association signal, but this will require careful attention to latent differences in cases and controls for ethnic haplogroup differences. The genotype array data do not provide adequate information for imputation of rare and private mutations of mtDNA variants.
Rare mtDNA Mutations Associated with SZ
Deep sequencing of the genome can provide rare mutations at the gene level to unravel the complex relationship among multiple rare mtDNA variants that might not be found with traditional haplogroup analysis [71]. Applying SKAT [72] to a sample of Swedes (n = 10, 771) and analyzing a set of rare SNPs with minor allele frequency (MAF) < 0.01, a nominal rare variant association with SZ (p = 0.007) was reported [3]. This is the largest study to date of mtDNA for analysis of rare coding region SNPs with MAF ≤1%. Multiple centers around the world are conducting deep sequencing studies to ultimately associate rare variants in mitochondrial genes with neuropsychiatric disorders. For analysis of rare variants, there are studies of whole-genome and whole-exome sequencing that can be combined: Danish (Dr. Michael Christiansen, n = 3,000 SZ and 20,000 controls), the Whole Genome Sequencing Consortium of Psychiatric Disorders (WGSPD) [73] (Dr. Stephan Sanders, n = 1,670 SZ, 2,067 BD, 4,378 controls), and Cardiff (Prof. Michael O'Donovan, n = 8,000 SZ, 8,000 BD, 16,000 controls). The total samples are n = 12,670 (SZ), n = 10,067 (BD), and n = 40,378 (controls). These three projects and others that make available mtDNA variants will be useful for conducting the rare mtDNA variant analysis in SZ and BD. It is anticipated that future aggregation of sequencing data will include the release of mtDNA variants, but at this time large centers have not yet adopted this plan especially in view of discovery of novel and rare variants in mtDNA. For example, the Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org/gene/ENSG00 000198899) indicates that mtDNA has no coverage for the exomes in the database. gnomAD is a coalition of investigators seeking to aggregate and harmonize exome and genome sequencing data from a variety of large-scale sequencing projects. The dataset provided on this website spans 123,136 exomes and 15,496 genomes from unrelated individuals sequenced as part of various disease-specific and population genetic studies [74]. In our experience, we have found highly concordant variant calls between Sanger-, exome-, and whole-genome sequencing of mtDNA [Vawter et al., unpubl. data] such that either WES or WGS usually give highly reliable mtDNA NGS results.
Interaction Analysis of Mitochondrial and Nuclear-Encoded Genes
The concept that mtDNA can direct the mode of energy production of the cell was supported by recent cybrid studies conducted by Dr. C. Kenney's group [75, 76]. Cybrids are created by removing mitochondria from a cell line by passaging in presence of ethidium bromide. These cells are then fused with platelets (containing mitochondrial but not nuclear DNA [nDNA]) from varying haplogroups. The resulting cybrid cells contain identical nDNA genes, but varying mitochondrial genes. Thus, the cross talk between nuclear and mitochondrial genes can be disrupted or enhanced and tested in vitro. Using a cybrid model with the same nuclear background, Dr. Kenney's group measured the expression of the mtDNA-encoded genes in respiratory complexes I, III, IV, and V of the OXPHOS pathway and found that J cybrids had significantly lower expression levels in 7 mitochondrial genes compared to H cybrids (Complex I: MT-ND1, MT-ND2, MT-ND3, MT-ND4/ND4L; Complex IV: MT-CO2, MT-CO3; Complex V: MT-ATP6. Further, these same J cybrids showed lower OXPHOS utilization compared to H cybrids.
The concept that mtDNA haplogroups can mediate the energy production levels is supported by another study which compares H (most common European) versus L (African maternal origin) haplogroups within cybrids from a retinal pigment epithelial cell line [76]. The L cybrids had lower mtDNA copy numbers but showed 9 mtDNA genes with significantly increased expression (1.57- to 2.11-fold) compared with H cybrids. In addition, the L cybrids had significantly decreased ATP turnover and spare respiratory capacity [76]. These patterns across both studies by Dr. Kenney's group showed L cybrids having the highest respiratory activities, the H cybrids intermediate and the J cybrids showing the lowest activities. This indicates that each of the cybrid haplogroups (L vs. H vs. J) had a unique bioenergetics profile. This strongly supports the hypothesis that cellular bioenergetics are greatly affected by the mtDNA haplogroups within a cell and moderated by nuclear background.
A mitochondria transfer to patient's cells with SZ showed the cells receiving mitochondria transfer from controls had a higher mitochondria membrane potential compared to the same cells that did not receive a mitochondria transfer [77]. Mitochondria membrane potential is maintained at sufficiently high levels to promote ADP to ATP conversion. This work suggests that the mitochondria organelle itself can be transplanted with therapeutic effects, given it is from a healthy donor. This therapeutic transfer of mitochondria has been translated into clinical practice in the UK through three parents mixing technique involving a pronuclear transfer [78] to correct mtDNA mutations.
mtSNP × nSNP Interactions
To further understand nuclear and mitochondria interactions, we recently published the first GWAS to investigate mtSNP × nSNP epistatic analysis [79]. This study utilized publicly available GWAS data (dbGAP Whole Genome Association Study of Bipolar Disorder (dbGaP Study Accession phs000017.v3.p1) from 2,035 European American subjects (BD = 1,001, controls = 1,034) originally published [80]. In brief, the epistatic method, restated verbatim from Ryu et al. [79], involved 1df and 2df association tests. For each association test, logistic regression models were used to assess the joint effects of the nSNP main effect and the interaction effect between nSNP and mtSNP, for each pair of mtSNP and nSNP, using 2df likelihood ratio tests. The 2df test jointly evaluates the effect of the last 2 terms (i.e., the nSNP effect and the nSNP × mtSNP interaction effect) from the following logistic regression model: Logit (p) = a0 + a1 × mtSNP + a2 × nSNP + a3 × nSNP × mtSNP, where p is the probability of the outcome (e.g., SZ vs. control), and a0/a1/a2/a3 are regression parameters for terms included in the model. The 2df analysis jointly assesses the effect of the nSNP main effect and/or the nSNP × mtSNP interaction term (i.e., testing if both a2 = 0 and a3 = 0), thereby testing the nSNP effect while allowing for modification of the effect by the mtSNP genotype. In addition, 1df likelihood ratio tests of the nSNP × mtSNP interaction terms were also performed, in this case assessing whether a3 = 0.
Interestingly, the top mtSNP rs3088309 located in mitochondria cytochrome B (CYTB) showed joint effects of several nSNPs and their interactions with mtSNP rs3088309 for risk of BD (2df test, Table 7). The mtSNP rs3088309 is a missense variant that causes an amino acid change from leucine (major allele) to isoleucine (minor allele). Being essential for electron transport chain, the presence of CYTB rs3088309 might leave the neurons more vulnerable to gene × environment risk factors for BD. The mtSNP × nSNP interaction between rs3088309 and rs1880924 in the MGAM gene showed suggestive association with BD risk (Table 7; interaction p = 1.4E-04), while the 2 df test signal was stronger (p = 8.2E-08), suggesting the presence of both main and interaction effects. We are careful not to overinterpret these comparisons of the main effects of nSNPs and the interactions with mtSNPs. Interestingly, the top nuclear-mitochondria pairs were not overrepresented in mitochondrial genes.
Table 7.
BD interaction analysis
| MtSNP |
Nuclear SNP |
Association results BD risk |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID#/HGVS | gene | MA | MAF | SNP | Chr: BP | gene | MA | MAF | 2 dfp | interactionp | ORa | ORb |
| rs3088309 | ||||||||||||
| m.15452C>A | CYTB | A | 0.21 | rs1880924 | 7:141717225 | MGAM | A | 0.14 | 8.2E-08 | 1.4E-04 | 1.27 | 3.42 |
| rs12733666 | 1: 77927820 | AK5 | C | 0.18 | 2.9E-06 | 7.1E-07 | 1.14 | 0.38 | ||||
| rs7782502 | 7: 105695500 | CDHR3 | G | 0.40 | 6.6E-06 | 3.6E-05 | 0.72 | 1.45 | ||||
In presence of a major mtSNP allele, the odds ratio for adding one copy of nSNP minor allele based on interaction analysis.
In presence of a minor mtSNP allele, the odds ratio for adding one copy of nSNP minor allele based on interaction analysis.
Applying a similar epistatic analysis approach, we assessed the association between the risk of SZ and mtSNP × nSNP interactions using logistic regression models with and without adjusting for the first PC based on nSNPs using the Irish subjects (n = 1,560 SZ cases and 1,745 controls). The results were very similar regardless of the PC adjustment. Two separate analyses were performed, 1 df for testing the mtSNP × nSNP interaction and 2 df tests for main effect and interaction effect jointly. The top association results implied that 2 nuclear genes FAM155A and ADNP2 may interact with mtSNPs for risk of SZ, although they have yet to be independently replicated. In total, 80 unique potential epistatic pairs had 1 df or 2 df p values < 1E-07, involving 21 nuclear genes and 10 mitochondrial genes. Three nuclear genes SOX5, TLE4, and ANK3 in these top 21 nuclear genes have already been implicated in SZ or BD GWAS, whose main effects were genome-wide significant as published in the GWAS catalog (http://www.ebi.ac.uk/gwas/docs/file-downloads). These 3 genes and FAM155A have medium to high relative gene expression in the brain compared to other tissues (GTEx; https://www.gtexportal.org/home/). For SOX5, the association of nSNP rs41369145 in the large PGC dataset (n ∼ 150,000) was p = 0.023, while modeling mtSNP rs2853513 with the same nSNP rs41369145 showed a considerably stronger interaction result (p = 8.0E-08) in the present sample (n = 2,508). Six of 10 mtDNA genes in the top epistatic pairs were located in the functional unit of Complex I of the mitochondria electron transport chain. There were 34 nuclear-mitochondria SNP pairs (joint effect p ≤ 5E-07).
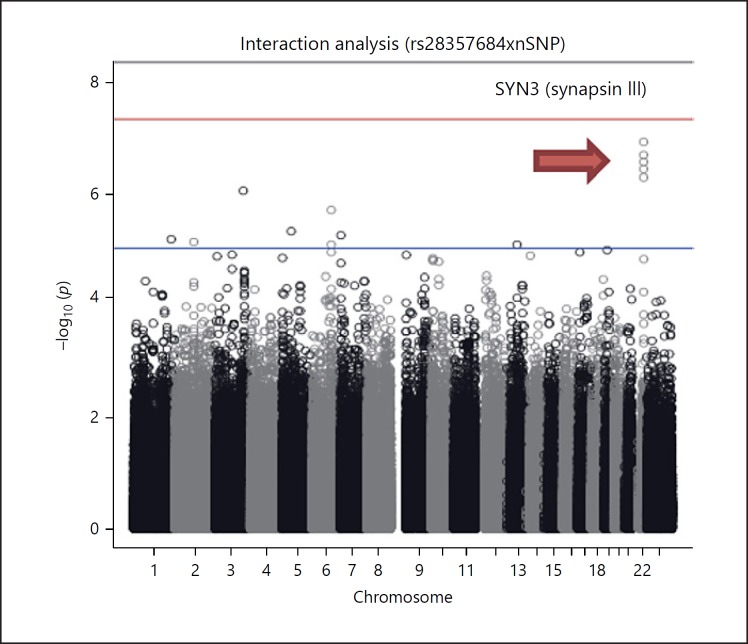
A nuclear-encoded mitochondria pair in the epistatic analysis shows a signal at the SYN3-CYTB gene pair (p = 1.26E-07) in the SZ analysis. The Manhattan plot for this 1 df mtSNP × nSNP interaction is shown (Fig. 1).
Fig. 1.
The Manhattan plot for the mtSNP × nSNP interaction result in SZ shows an interesting signal at the SYN3 gene, the significance threshold is red line.
SYN3 is a member of the synapsin gene family which is implicated in synaptogenesis and the modulation of neurotransmitter release, suggesting a potential role in several neuropsychiatric diseases. The SYN3 example is shown as there have been positive linkage studies of SZ at chr22q12.3 [81].
Although we have shown potential mtDNA × nDNA interaction effects of SNPs in 5 nuclear genes (Tables 7, 8), none of those nSNPs had strong main effects on risk of SZ or BD. In general, the working hypothesis is that the effect of some nSNPs depends on a person's mtSNP composition, and the contribution of nSNPs may be detected only through its interaction with mtSNP, not as the main effects alone. For example, the OR for adding one minor nSNP allele was 3.42 among carriers of minor allele C of the mtSNP rs3088309, while the lower OR of 1.27 was found in carriers of common allele A of the mtSNP. These results for BD and SZ, while nearing statistical significance at the genome-wide level, show that epistatic analysis can potentially locate relevant genes (p value 1.58E-08 for SZ; n = 1,560 SZ cases and 1,745 controls), and with larger studies the power will increase for signal detection.
Table 8.
Interaction analysis in schizophrenia and top epistatic pairs
| nSNP | nMAF | nA2 | CHR | POS | nGene | mtSNP | mtGene | mtA2 | mtMAF | 1 df | 2 df |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs8093433 | 0.49 | A | 18 | 7.59E+07 | ADNP2 | rs3135030 | ND6 | T | 0.020 | 9.44E-07 | 2.07E-08 |
| rs7317862 | 0.07 | A | 13 | 1.07E+08 | FAM155A | rs28357684 | CYTB | G | 0.046 | 1.58E-08 | 8.69E-08 |
| rs7317862 | 0.07 | A | 13 | 1.07E+08 | FAM155A | rs28358275 | ND3 | T | 0.042 | 3.46E-08 | 1.90E-07 |
The DAVID database for the enrichment of pathways was queried (https://david.ncifcrf.gov/) by submitting nuclear-mtDNA SNP pairs meeting a nominal association threshold of p < 0.0001 for both joint and interaction effects. There were 530 unique genes, and those genes were enriched in neuron projection development and morphogenesis, from the results of the epistatic interactions. These results, shown in Table 9, are Bonferroni corrected and were found in a single category that showed a significant association. This analysis supports a recent study showing involvement preferentially of projection neurons as altered in SZ transcriptomic wide association study involvement [82]. As shown above for BD epistatic analysis, mitochondrial genes were not overenriched in the 530 genes analyzed.
Table 9.
Pathway analysis for the interaction pairs of SNPs in SZ epistatic analysis
| Category enrichment score: 6.57 | Count | Bonferroni |
|---|---|---|
| Neuron projection morphogenesis | 37 | 7.60E-06 |
| Neuron projection development | 45 | 6.60E-05 |
| Cell morphogenesis involved in neuron differentiation | ||
| 32 | 4.20E-04 | |
| Neuron development | 47 | 1.10E-03 |
| Neurogenesis | 61 | 1.80E-03 |
| Generation of neurons | 58 | 2.50E-03 |
| Cell projection morphogenesis | 40 | 5.40E-03 |
| Cell part morphogenesis | 40 | 9.80E-03 |
| Neuron differentiation | 52 | 1.40E-02 |
| Cell projection organization | 54 | 1.60E-02 |
| Excitatory synapse | 15 | 4.80E-02 |
These results for BD and SZ show that epistatic analysis can potentially identify additional genes associated with BD and/or SZ, especially larger studies with better statistical power. These results also support the recent cross-disorder analysis of gene expression where the mitochondrial and neuronal gene coexpression module was shared between BD and SZ cortical regions [49], and the shared genes were downregulated, independent of antipsychotic treatment [50].
As mentioned in the common variant section, there is slight LD between the mitochondrial and nuclear genomes to take into account in interaction analysis. While the average r2 = 0.004 globally is low across the mtDNA and the remaining nuclear genomes, it is not different than the within nuclear genome LD (r2 = 0.004) [83]. The mean D′ is also similar between the mitochondrial and nuclear genomes globally. For future interaction analyses, the potential effect of local genotype-geographic influences [66] will need to be assessed.
Others have investigated gene-environment interactions by testing for association between complex traits (e.g., depression) with interactions between polygenic risk scores (PRS; derived from SNP main effects on the trait), and environmental factors (e.g., stress) [84, 85, 86, 87]. We propose a similar approach to test whether the effect of mitochondrial SNPs on the risk of BD or SZ is modified by a person's polygenic risk from nuclear SNPs. We will then analyze potential improvement of using the mtSNP × nSNP PRS interaction to predict BD or SZ over prediction based solely on the nSNP PRS. In this proposed method, disease-specific nSNP PRSs would be constructed in the replication cohorts from LD-clumped SNPs for a range of p value thresholds in the discovery cohorts by summing over nSNP risk alleles weighted by their effect sizes observed in the discovery cohorts, using PRSice [88]. This will then permit an evaluation of the association of the disease with the PRS, and the interaction between the PRS and each mtSNP, in the replication cohort using Nagelkerke's R2 or alternatively R2 on the liability scale [89].
Summary
The reviewed analyses of variants in the mitochondrial genome shows genes in both nDNA and mtDNA potentially carry the risk of SZ and BD. A meta-analysis of A15218G (SZ = 11,311, control = 35,735) shows a moderate association with SZ (p = 2.15E-03, OR = 1.2, 95% CI 1.07–1.35) with 3 of the 4 studies showing the same direction of risk. The A15218G variant is located in the cytochrome B oxidase gene (CYTB), and conserved across 27 primate species. Another mtDNA allele A12308G was nominally associated with psychosis in BD type I subjects and SZ. The compiled results of the variant association of A15218G in 47,046 subjects indicate that a more detailed analysis is required to rule out an occult population stratification. A preliminary meta-analysis for the mtDNA A15218G nonsynonymous variant shows an association of SZ which is to be interpreted with caution as further analysis is required to rigorously control for haplogroup origin in testing for association. Furthermore, genotypes captured from array data do not yield or provide adequate information for imputation of rare and private mutations, nor do they adequately control for specific sub-haplogroup assignment. However, the current legacy databases do not generally carry information on mtDNA variants, nor do the large psychiatric genetics consortia. The WES and WGS pipelines have mtDNA variants but often do not include this in data releases.
In addition to variants in mtDNA, we also reviewed the association of nuclear-encoded mitochondrial variants with SZ. Different gene-based and pathway-based approaches for nuclear-encoded mitochondrial-related genes showed significant associations with SZ. Several nuclear-encoded genes in the electron chain forming Complex I appear to be of particular importance in informing the risk of SZ. Meanwhile, in-depth analyses have yet to be conducted for BD notwithstanding multiple reports of mitochondria involvement in BD. For example, a novel analysis testing the epistatic interaction between nuclear-encoded and mitochondria-encoded genes demonstrated evidence for potential interactions between mtDNA and the nuclear genome for BD. The first published study (BD = 1,001 and controls = 1,034) showed that the odds ratios for BD risk decreased from 1.1 to 0.38 by adding an mtSNP minor allele compared to the mtSNP major allele for the nuclear gene AK5 and the CYTB mitochondria-encoded gene. AK5 (adenylate kinase 5) is a brain-specific gene in the synthesis of ADP, a precursor to ATP. A similar analysis for the risk of SZ (n = 2,508 subjects) also revealed a highly significant joint effect (collective effect of nuclear SNP and its interaction with mtSNP; p ≤ 1E-07). Three of the top signals in the epistatic analysis involved genes with established genome-wide significant associations with SZ and enriched brain expression (ANK3, SOX5, TLE4) and the mitochondria-encoded Complex III gene CYTB. This suggests that both genomes jointly determine the amount of cellular energy production requirements, the impairment of which is associated with the risk of BD and SZ.
Further analysis of the interactions in nDNA and mtDNA variants might uncover more information about the genetic architecture and patient risk strata with this approach. Multiple variants in Complex I genes suggest that this may be an important site for genetic interaction analysis that will identify subjects with high risk as well as pathogenic variants. By carefully controlling for geographic stratification and other potential confounders, it will be possible to test the hypothesis that rare and common variants in mitochondrial genes are associated with SZ and BD. Analysis of PRS by addition of mtDNA SNPs might be a possible method to determine whether stratification by mtDNA SNPs actually increases the risk score.
Conclusion
In this review, the control region showed protective associations with SZ and BD, primarily with the ancestral alleles. It is of interest that the mitochondria-encoded gene CYTB was found in both the epistatic interactions for SZ and BD and the single SNP association of SZ. These interactions may ultimately transform the genetic landscape of these disorders far better than single SNP associations. Analysis of epistatic interactions when stratifying a relatively small GWAS of SZ by mtDNA SNPs produced a projection neuron single cell category that was also found in 2 much larger SZ GWAS of PGC and CLOZUK (n > 250,000 subjects), which was highly significant [82]. With large volumes of data being generated from the sequencing of mtDNA, further effort is required to compile and evaluate these studies to completely understand the role of rare and common mitochondrial variants in psychiatric disorders.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgments
Larry and Judy Foundation (V.G.); Danish National Biobank, Novo Nordisk Foundation, iPSYCH, Lundbeck Foundation (M.C.); Assurex Health, Mayo Foundation, Myriad, National Institute of Alcohol Abuse and Alcoholism (NIAAA), National Institute of Mental Health (NIMH), Pfizer (M.A.F.); Pritzker Neuropsychiatric Research Fund, NIMH R01MH085801 (M.P.V.).
Funding support for the Whole Genome Association Study of Bipolar Disorder was provided by the National Institute of Mental Health (NIMH), and the genotyping of samples was provided through the Genetic Association Information Network (GAIN). The datasets used for the analyses described in this article were obtained from the database of Genotypes and Phenotypes (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession No. phs000017.v3.p1. Samples and associated phenotype data for the Collaborative Genomic Study of Bipolar Disorder were provided by the NIMH Genetics Initiative for Bipolar Disorder. Data and biomaterials were collected in four projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1991 to 1998, the principal investigators and coinvestigators were: Indiana University, Indianapolis, IN, U01 MH46282, John Nurnberger, MD, PhD, Marvin Miller, MD, and Elizabeth Bowman, MD; Washington University, St. Louis, MO, U01 MH46280, Theodore Reich, MD, Allison Goate, PhD, and John Rice, PhD; Johns Hopkins University, Baltimore, MD, U01 MH46274, J. Raymond DePaulo, Jr., MD, Sylvia Simpson, MD, MPH, and Colin Stine, PhD; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, Elliot Gershon, MD, Diane Kazuba, BA, and Elizabeth Maxwell, MSW. Data and biomaterials were collected as part of ten projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1999 to 2003, the principal investigators and coinvestigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, MD, PhD, Marvin J. Miller, MD, Elizabeth S. Bowman, MD, N. Leela Rau, MD, P. Ryan Moe, MD, Nalini Samavedy, MD, Rif El-Mallakh, MD (at University of Louisville), Husseini Manji, MD (at Wayne State University), Debra A. Glitz, MD (at Wayne State University), Eric T. Meyer, MS, Carrie Smiley, RN, Tatiana Foroud, PhD, Leah Flury, MS, Danielle M. Dick, PhD, Howard Edenberg, PhD; Washington University, St. Louis, MO, R01 MH059534, John Rice, PhD, Theodore Reich, MD, Allison Goate, PhD, Laura Bierut, MD; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis, MD, J. Raymond DePaulo, Jr., MD, Dean F. MacKinnon, MD, Francis M. Mondimore, MD, James B. Potash, MD, Peter P. Zandi, PhD, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini MD, PhD; University of California at Irvine, CA, R01 MH60068, William Byerley, MD, and Mark Vawter PhD; University of Iowa, IA, R01 MH059548, William Coryell, MD, and Raymond Crowe, MD; University of Chicago, IL, R01 MH59535, Elliot Gershon, MD, Judith Badner, PhD, Francis McMahon, MD, Chunyu Liu, PhD, Alan Sanders, MD, Maria Caserta, Steven Dinwiddie, MD, Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, MD, Rebecca McKinney, BA; Rush University, IL, R01 MH059556, William Scheftner, MD, Howard M. Kravitz, DO, MPH, Diana Marta, BS, Annette Vaughn-Brown, MSN, RN, and Laurie Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, MD, Layla Kassem, PsyD, Sevilla Detera-Wadleigh, PhD, Lisa Austin, PhD, Dennis L. Murphy, MD.
References
- 1.Rollins B, Martin MV, Sequeira PA, Moon EA, Morgan LZ, Watson SJ, et al. Mitochondrial variants in schizophreniabipolar disorder and major depressive disorder. PLoS One. 2009;4((3)):e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasahara T, Kato T. What Can Mitochondrial DNA Analysis Tell Us About Mood Disorders? Biol Psychiatry. 2018 May;83((9)):731–8. doi: 10.1016/j.biopsych.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Gonçalves V.F., Giamberardino S.N., Crowley J.J., Vawter M.P., Saxena R., Bulik C.M., Yilmaz Z., Hultman C.M., Sklar P., Kennedy J.L., Sullivan P.F., Knight J. Examining the role of common and rare mitochondrial variants in schizophrenia. PLoS ONE. 2018 doi: 10.1371/journal.pone.0191153. 13(1 e0191153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato T. Neurobiological basis of bipolar disorder: mitochondrial dysfunction hypothesis and beyond. Schizophr Res. 2017 Sep;187:62–6. doi: 10.1016/j.schres.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 5.Kato T. DNA polymorphisms and bipolar disorder. Am J Psychiatry. 2001 Jul;158((7)):1169–70. doi: 10.1176/appi.ajp.158.7.1169-b. [DOI] [PubMed] [Google Scholar]
- 6.Kato T. The otherforgotten genomemitochondrial DNA and mental disorders. Mol Psychiatry. 2001 Nov;6((6)):625–33. doi: 10.1038/sj.mp.4000926. [DOI] [PubMed] [Google Scholar]
- 7.Kato T, Kato N. Mitochondrial dysfunction in bipolar disorder. Bipolar Disord. 2000 Sep;2((3 Pt 1)):180–90. doi: 10.1034/j.1399-5618.2000.020305.x. [DOI] [PubMed] [Google Scholar]
- 8.Kato T, Kunugi H, Nanko S, Kato N. Mitochondrial DNA polymorphisms in bipolar disorder. J Affect Disord. 2001 Feb;62((3)):151–64. doi: 10.1016/s0165-0327(99)00173-1. [DOI] [PubMed] [Google Scholar]
- 9.Kato T, Murashita J, Kamiya A, Shioiri T, Kato N, Inubushi T. Decreased brain intracellular pH measured by 31P-MRS in bipolar disorder: a confirmation in drug-free patients and correlation with white matter hyperintensity. Eur Arch Psychiatry Clin Neurosci. 1998;248((6)):301–6. doi: 10.1007/s004060050054. [DOI] [PubMed] [Google Scholar]
- 10.Hjelm BE, Rollins B, Mamdani F, Lauterborn JC, Kirov G, Lynch G, et al. Evidence of Mitochondrial Dysfunction within the Complex Genetic Etiology of Schizophrenia. Mol Neuropsychiatry. 2015 Dec;1((4)):201–19. doi: 10.1159/000441252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rollins BL, Morgan L, Hjelm BE, Sequeira A, Schatzberg AF, Barchas JD, et al. Mitochondrial Complex I Deficiency in Schizophrenia and Bipolar Disorder and Medication Influence. Mol Neuropsychiatry. 2018 Feb;3((3)):157–69. doi: 10.1159/000484348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjelm B.E., Rollins B., Morgan L., Sequeira A., Mamdani F., Pereira F., Damas J., Schatzberg A.F., Barchas J.D., Lee F.S., Akil H., Watson S.J., Myers R.M., Thompson P.M., Bunney W.E., Vawter M.P. Splice-Break: Exploiting an RNA-seq splice junction algorithm to discover mitochondrial. DNA deletion breakpoints Nucleic Acids Research in submission. doi: 10.1093/nar/gkz164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goncalves VF, Andreazza AC, Kennedy JL. Mitochondrial dysfunction in schizophrenia: an evolutionary perspective. Hum Genet. 2014 doi: 10.1007/s00439-014-1491-8. [DOI] [PubMed] [Google Scholar]
- 14.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, et al. Pharmacogenomics of Bipolar Disorder Study Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015 Nov;527((7576)):95–9. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Shachar D, Zuk R, Gazawi H, Reshef A, Sheinkman A, Klein E. Increased mitochondrial complex I activity in platelets of schizophrenic patients. Int J Neuropsychopharmacol. 1999 Dec;2((4)):245–53. doi: 10.1017/S1461145799001649. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Shachar D. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J Neurochem. 2002 Dec;83((6)):1241–51. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- 17.Dror N, Klein E, Karry R, Sheinkman A, Kirsh Z, Mazor M, et al. State-dependent alterations in mitochondrial complex I activity in platelets: a potential peripheral marker for schizophrenia. Mol Psychiatry. 2002;7((9)):995–1001. doi: 10.1038/sj.mp.4001116. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Shachar D, Laifenfeld D. Mitochondria, synaptic plasticityand schizophrenia. Int Rev Neurobiol. 2004;59:273–96. doi: 10.1016/S0074-7742(04)59011-6. [DOI] [PubMed] [Google Scholar]
- 19.Karry R, Klein E, Ben Shachar D. Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biol Psychiatry. 2004 Apr;55((7)):676–84. doi: 10.1016/j.biopsych.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Shachar D, Karry R. Neuroanatomical pattern of mitochondrial complex I pathology varies between schizophreniabipolar disorder and major depression. PLoS One. 2008;3((11)):e3676. doi: 10.1371/journal.pone.0003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner-Lavie H, Klein E, Ben-Shachar D. Mitochondrial complex I as a novel target for intraneuronal DA: modulation of respiration in intact cells. Biochem Pharmacol. 2009 Jul;78((1)):85–95. doi: 10.1016/j.bcp.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Craddock N, O'Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005 Mar;42((3)):193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamshere ML, Bennett P, Williams N, Segurado R, Cardno A, Norton N, et al. Genomewide linkage scan in schizoaffective disorder: significant evidence for linkage at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19p13. Arch Gen Psychiatry. 2005 Oct;62((10)):1081–8. doi: 10.1001/archpsyc.62.10.1081. [DOI] [PubMed] [Google Scholar]
- 24.Weinberger D.R. Genetic mechanisms of psychosis: in vivo and postmortem genomics. Clinical therapeutics, 2005;27(Suppl A):p. S8–15. doi: 10.1016/j.clinthera.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006 Jan;32((1)):9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park C, Park SK. Molecular links between mitochondrial dysfunctions and schizophrenia. Mol Cells. 2012 Feb;33((2)):105–10. doi: 10.1007/s10059-012-2284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014 Nov;515((7527)):414–8. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004 Mar;61((3)):300–8. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 29.Naydenov AV, MacDonald ML, Ongur D, Konradi C. Differences in lymphocyte electron transport gene expression levels between subjects with bipolar disorder and normal controls in response to glucose deprivation stress. Arch Gen Psychiatry. 2007 May;64((5)):555–64. doi: 10.1001/archpsyc.64.5.555. [DOI] [PubMed] [Google Scholar]
- 30.Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2011 May;29((3)):311–24. doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konradi C, Sillivan SE, Clay HB. Mitochondria, oligodendrocytes and inflammation in bipolar disorder: evidence from transcriptome studies points to intriguing parallels with multiple sclerosis. Neurobiol Dis. 2012 Jan;45((1)):37–47. doi: 10.1016/j.nbd.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000 Oct;28((1)):53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 33.Sequeira PA, Martin MV, Vawter MP. The first decade and beyond of transcriptional profiling in schizophrenia. Neurobiol Dis. 2011 doi: 10.1016/j.nbd.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. 2015 Nov;20((11)):1397–405. doi: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010 Jun;11((6)):402–16. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014 Jul;19((7)):762–73. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012 Feb;17((2)):142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014 Feb;506((7487)):185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014 Feb;506((7487)):179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014 Jul;511((7510)):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Zhang W, Tang J, Tan L, Luo XJ, Chen X, et al. Do nuclear-encoded core subunits of mitochondrial complex I confer genetic susceptibility to schizophrenia in Han Chinese populations? Sci Rep. 2015 Jun;5((1)):11076. doi: 10.1038/srep11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, et al. Deficient hippocampal neuron expression of proteasomeubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005 Jul;58((2)):85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 43.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002 Apr;22((7)):2718–29. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris LW, Lockstone HE, Khaitovich P, Weickert CS, Webster MJ, Bahn S. Gene expression in the prefrontal cortex during adolescence: implications for the onset of schizophrenia. BMC Med Genomics. 2009 May;2((1)):28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2015 Jan;18((1)):154–61. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.English JA, Pennington K, Dunn MJ, Cotter DR. The neuroproteomics of schizophrenia. Biol Psychiatry. 2011 Jan;69((2)):163–72. doi: 10.1016/j.biopsych.2010.06.031. [DOI] [PubMed] [Google Scholar]
- 47.Goncalves VF. A comprehensive analysis of mitochondrial genetic dysfunction in schizophrenia. Biol Psychiatry. Forthcoming. [Google Scholar]
- 48.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016 Nov;19((11)):1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. CommonMind Consortium PsychENCODE Consortium iPSYCH-BROAD Working Group Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018 Feb;359((6376)):693–7. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin MV, Mirnics K, Nisenbaum LK, Vawter MP. Olanzapine Reversed Brain Gene Expression Changes Induced by Phencyclidine Treatment in Non-Human Primates. Mol Neuropsychiatry. 2015 Jul;1((2)):82–93. doi: 10.1159/000430786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronalimmune and histone pathways. Nat Neurosci. 2015 Feb;18((2)):199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin MY, Sheng ZH. Regulation of mitochondrial transport in neurons. Exp Cell Res. 2015 May;334((1)):35–44. doi: 10.1016/j.yexcr.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou B, Lin MY, Sun T, Knight AL, Sheng ZH. Characterization of mitochondrial transport in neurons. Methods Enzymol. 2014;547:75–96. doi: 10.1016/B978-0-12-801415-8.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kann O, Kovács R. Mitochondria and neuronal activity. Am J Physiol Cell Physiol. 2007 Feb;292((2)):C641–57. doi: 10.1152/ajpcell.00222.2006. [DOI] [PubMed] [Google Scholar]
- 55.Sheng ZH. Mitochondrial trafficking and anchoring in neurons: new insight and implications. J Cell Biol. 2014 Mar;204((7)):1087–98. doi: 10.1083/jcb.201312123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang CC, Jou SH, Lin TT, Liu CS. Mitochondrial DNA variation and increased oxidative damage in euthymic patients with bipolar disorder. Psychiatry Clin Neurosci. 2014 Jul;68((7)):551–7. doi: 10.1111/pcn.12163. [DOI] [PubMed] [Google Scholar]
- 57.Wang D, Li Z, Liu W, Zhou J, Ma X, Tang J, et al. Differential mitochondrial DNA copy number in three mood states of bipolar disorder. BMC Psychiatry. 2018 May;18((1)):149. doi: 10.1186/s12888-018-1717-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nurnberger JI, Jr, Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, et al. Psychiatric Genomics Consortium Bipolar Group Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 2014 Jun;71((6)):657–64. doi: 10.1001/jamapsychiatry.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mink JW, Blumenschine RJ, Adams DB. Ratio of central nervous system to body metabolism in vertebrates: its constancy and functional basis. Am J Physiol. 1981 Sep;241((3)):R203–12. doi: 10.1152/ajpregu.1981.241.3.R203. [DOI] [PubMed] [Google Scholar]
- 60.Burr SP, Pezet M, Chinnery PF. Mitochondrial DNA Heteroplasmy and Purifying Selection in the Mammalian Female Germ Line. Dev Growth Differ. 2018 Jan;60((1)):21–32. doi: 10.1111/dgd.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nachman MW, Brown WM, Stoneking M, Aquadro CF. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 1996 Mar;142((3)):953–63. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bamne MN, Talkowski ME, Moraes CT, Manuck SB, Ferrell RE, Chowdari KV, et al. Systematic association studies of mitochondrial DNA variations in schizophrenia: focus on the ND5 gene. Schizophr Bull. 2008 May;34((3)):458–65. doi: 10.1093/schbul/sbm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sequeira A, Martin MV, Rollins B, Moon EA, Bunney WE, Macciardi F, et al. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet. 2012 Jun;3:103. doi: 10.3389/fgene.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frye MA, Ryu E, Nassan M, Jenkins GD, Andreazza AC, Evans JM, et al. Mitochondrial DNA sequence data reveals association of haplogroup U with psychosis in bipolar disorder. J Psychiatr Res. 2017 Jan;84:221–6. doi: 10.1016/j.jpsychires.2016.09.027. [DOI] [PubMed] [Google Scholar]
- 65.Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 2014 May;10((5)):e1004369. doi: 10.1371/journal.pgen.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hagen CM, Goncalves VF, Hedley PL, Bybjerg-Grauholm J, Baekvad-Hansen M, Hansen CS, et al. SNPs associated with Schizophrenia exhibit Highly Variable Inter-allelic Haplogroup Affiliation and Nuclear Genogeographic Affinity: Bi-Genomic Linkage Disequilibrium raises Major Concerns for Link to Disease. bioRxiv. 2017 [Google Scholar]
- 67.Lott M.T., Leipzig J.N., Derbeneva O., Xie H.M., Chalkia D., Sarmady M., Procaccio V., Wallace D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Current protocols in bioinformatics, 2013;44:p. 1 23–1-26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009 Feb;30((2)):E386–94. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 69.Amar S, Shamir A, Ovadia O, Blanaru M, Reshef A, Kremer I, et al. Mitochondrial DNA HV lineage increases the susceptibility to schizophrenia among Israeli Arabs. Schizophr Res. 2007 Aug;94((1-3)):354–8. doi: 10.1016/j.schres.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 70.Torrell H, Salas A, Abasolo N, Morén C, Garrabou G, Valero J, et al. Mitochondrial DNA (mtDNA) variants in the European haplogroups HVJT, and U do not have a major role in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2014 Oct;165B((7)):607–17. doi: 10.1002/ajmg.b.32264. [DOI] [PubMed] [Google Scholar]
- 71.Raule N, Sevini F, Li S, Barbieri A, Tallaro F, Lomartire L, et al. The co-occurrence of mtDNA mutations on different oxidative phosphorylation subunitsnot detected by haplogroup analysisaffects human longevity and is population specific. Aging Cell. 2014 Jun;13((3)):401–7. doi: 10.1111/acel.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011 Jul;89((1)):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders SJ, Neale BM, Huang H, Werling DM, An JY, Dong S, et al. Whole Genome Sequencing for Psychiatric Disorders (WGSPD) Whole genome sequencing in psychiatric disorders: the WGSPD consortium. Nat Neurosci. 2017 Dec;20((12)):1661–8. doi: 10.1038/s41593-017-0017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 Aug;536((7616)):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, et al. Inherited mitochondrial DNA variants can affect complementinflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet. 2014 Jul;23((13)):3537–51. doi: 10.1093/hmg/ddu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim Biophys Acta. 2014 Feb;1842((2)):208–19. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robicsek O, Ene HM, Karry R, Ytzhaki O, Asor E, McPhie D, et al. Isolated Mitochondria Transfer Improves Neuronal Differentiation of Schizophrenia-Derived Induced Pluripotent Stem Cells and Rescues Deficits in a Rat Model of the Disorder. Schizophr Bull. 2018 Feb;44((2)):432–42. doi: 10.1093/schbul/sbx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NM, Fragouli E, et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature. 2016 Jun;534((7607)):383–6. doi: 10.1038/nature18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryu E, Nassan M, Jenkins GD, Armasu SM, Andreazza A, McElroy SL, et al. A Genome-Wide Search for Bipolar Disorder Risk Loci Modified by Mitochondrial Genome Variation. Mol Neuropsychiatry. 2018 Feb;3((3)):125–34. doi: 10.1159/000464444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009 Aug;14((8)):755–63. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gill M, Vallada H, Collier D, Sham P, Holmans P, Murray R, et al. A combined analysis of D22S278 marker alleles in affected sib-pairs: support for a susceptibility locus for schizophrenia at chromosome 22q12. Schizophrenia Collaborative Linkage Group (Chromosome 22) Am J Med Genet. 1996 Feb;67((1)):40–5. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 82.Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, et al. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genetic identification of brain cell types underlying schizophrenia. Nat Genet. 2018 Jun;50((6)):825–33. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sloan DB, Fields PD, Havird JC. Mitonuclear linkage disequilibrium in human populations. Proc Biol Sci. 2015 Sep;282((1815)):282. doi: 10.1098/rspb.2015.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agerbo E, Sullivan PF, Vilhjálmsson BJ, Pedersen CB, Mors O, Børglum AD, et al. Polygenic Risk ScoreParental Socioeconomic StatusFamily History of Psychiatric Disorders and the Risk for Schizophrenia: A Danish Population-Based Study and Meta-analysis. JAMA Psychiatry. 2015 Jul;72((7)):635–41. doi: 10.1001/jamapsychiatry.2015.0346. [DOI] [PubMed] [Google Scholar]
- 85.Mullins N, Power RA, Fisher HL, Hanscombe KB, Euesden J, Iniesta R, et al. Polygenic interactions with environmental adversity in the aetiology of major depressive disorder. Psychol Med. 2016 Mar;46((4)):759–70. doi: 10.1017/S0033291715002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Musliner KL, Seifuddin F, Judy JA, Pirooznia M, Goes FS, Zandi PP. Polygenic riskstressful life events and depressive symptoms in older adults: a polygenic score analysis. Psychol Med. 2015 Jun;45((8)):1709–20. doi: 10.1017/S0033291714002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peyrot WJ, Van der Auwera S, Milaneschi Y, Dolan CV, Madden PA, Sullivan PF, et al. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Does Childhood Trauma Moderate Polygenic Risk for Depression? A Meta-analysis of 5765 Subjects From the Psychiatric Genomics Consortium. Biol Psychiatry. 2017 Sep;:S0006-3223(17)31993-5. doi: 10.1016/j.biopsych.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Euesden J, Lewis CM, O'Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015 May;31((9)):1466–8. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36((3)):214–24. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
- 90.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput Biol. 2015 Apr;11((4)):e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959 Apr;22((4)):719–48. [PubMed] [Google Scholar]
- 92.Mosquera-Miguel A., Torrell H., Abasolo N., Arrojo M., Paz E., Ramos-Rios R., Agra S., Paramo M., Brenlla J., Martinez S., Vilella E., Valero J., Gutierrez-Zotes A., Martorell L., Costas J., Salas A. No evidence that major mtDNA European haplogroups confer risk to schizophrenia. American journal of medical genetics Part B Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics, 2012;159B((4)):p. 414–421. doi: 10.1002/ajmg.b.32044. [DOI] [PubMed] [Google Scholar]