Abstract
The field of proteomics is rapidly gaining territory as a promising alternative to genomic approaches in the efforts to unravel the complex molecular mechanisms underlying schizophrenia and other psychiatric disorders. X-aptamer technology has emerged as a novel proteomic approach for high-sensitivity analyses, and we hypothesized that this technology would identify unique molecular signatures in plasma samples from schizophrenia patients (n = 60) compared to controls (n = 20). Using a combinatorial library of X-aptamer beads, we developed a two-color flow cytometer-based approach to identify specific X-aptamers that bound with high specificity to each target group. Based on this, we synthesized two unique X-aptamer sequences, and specific proteins pulled down from the patient and control groups by these X-aptamers were identified by mass spectrometry. We identified two protein biomarkers, complement component C4A and ApoB, upregulated in plasma samples from schizophrenia patients. ELISA validation suggested that the observed differences in C4 levels in patients are likely due to the presence of the illness itself, while ApoB may be a marker of antipsychotic-induced alterations. These studies highlight the utility of the X-aptamer technology in the identification of biomarkers for schizophrenia that will advance our understanding of the pathophysiological mechanisms of this disorder.
Key Words: Schizophrenia, Proteomics, X-aptamer, C4, ApoB
Introduction
Schizophrenia is a severe mental illness characterized by symptoms of psychosis, cognitive abnormalities, and blunted volition. The mode of schizophrenia transmission is complex and the phenotypic expression of the wide range of symptoms varies extensively between individuals [1]. While numerous genes of weak individual effect, interacting with environmental factors, are thought to be involved in the development and expression of schizophrenia, the precise molecular mechanisms that underlie schizophrenia pathophysiology are not well understood. To date, genomic techniques such as genome-wide association studies (GWAS) and analysis of gene expression have been most commonly used in attempts to identify biological markers underlying schizophrenia susceptibility, with recent GWAS identifying credible susceptibility loci [2]. However, the final output of gene expression, the protein, is regulated by a number of different cellular processes and posttranslational modifications – crucial for proper protein function – that are not detected by genomic techniques. High-throughput proteomic approaches have been recently gaining territory as a promising alternative to identifying molecular signatures of disease in schizophrenia and other psychiatric disorders [3, 4], and they are widely used to study illnesses such as Alzheimer's disease and cancer [5, 6].
X-aptamer technology [7, 8] combined with mass spectrometric techniques [9] has emerged as a novel proteomic approach for high-sensitivity analyses in clinical diagnosis and research. X-aptamers are chemically modified DNA affinity agents that employ a wide range of chemical modifications such as amino acid-like side chains, phosphorodithioates, and small molecules, which allow X-aptamers to bind to target proteins with high specificity and affinity. As novel elements for molecular recognition, X-aptamers possess specific recognition properties toward low-molecular-weight substrates or macromolecules [10]. Due to their affinity and specificity for target molecules, high stability, and facility of synthesis and modification, aptamers are feasible and practical alternatives to antibodies for many applications including research, diagnostics, and therapeutics [7, 11, 12]. Importantly, aptamers bind to target molecules in their native state, creating a true molecular profile of disease. The aptamer technology has been most successfully applied in the field of cancer biology, where aptamers have been able to identify phenotypic variations between cell types, representing differences in molecular signatures [13, 14]. We have shown that aptamers can safely target and reduce cancer metastasis [15, 16, 17], can be easily selected against tissues using a laser microdissection method [9], and enhance binding of liposomes and nanoparticles [17, 18, 19, 20].
Based on these studies, we hypothesized that the X-aptamer technology would identify unique molecular signatures in plasma samples from schizophrenia patients compared to controls. A convenient bead-based combinatorial library format enabled us to develop a two-color flow cytometer-based approach to identify specific biomarkers from patient/control sample sets. Using this combinatorial library of X-aptamer beads, we identified two protein biomarkers, complement C4 and ApoB, upregulated in plasma samples from schizophrenia patients.
Subjects and Methods
Subjects
Sixty patients with schizophrenia (n = 60), recruited from local community mental health centers, met the DSM-IV-TR criteria for schizophrenia by the Structured Clinical Interview for DSM-IV (SCID). The patients had been on the same antipsychotic treatment for at least 3 months prior to enrollment, and the prescribed doses of all the antipsychotics were within the clinically recommended range, e.g., olanzapine 10–20 mg/day, risperidone 1–5 mg/day, and ziprasidone 20–80 mg/day.
Twenty healthy controls (n = 20), matched with the patients for gender, ethnicity, age, and years of education, were screened for DSM-IV axis I disorders using the SCID nonpatient version. Subjects with current or past DSM-IV axis I psychiatric disorders, or with first-degree relatives with any axis I psychiatric disorder, were excluded.
Weight and BMI were obtained from all subjects. Subjects with a history of diabetes or dyslipidemia were excluded.
Processing of Blood Samples
Fasting blood was drawn by venipuncture and collected into heparin-containing tubes. The blood was immediately processed by centrifugation at 3,400 rpm for 10 min, and plasma was collected, aliquoted, and stored at −80°C until the experiments were performed. Twenty-five microliters of plasma from each subject per group were pooled for the initial X-aptamer screening.
Fluorescent Labeling
Approximately 150 μL of total pooled sample from each group was subjected to albumin depletion (Thermo Fisher Scientific, Waltham, MA, USA) according to instructions, and then buffer exchanged to 50 mM sodium borate buffer (pH 8.5) using ZebaTM spin columns (Thermo Fisher). Protein concentrations were estimated using a NanoDrop 2000TM (NanoDrop Technologies, Wilmington, DE, USA). Fifty microliters of sample from each group were used for fluorescent labeling, and another 50 μL were used for biotinylation. Schizophrenia plasma proteins were labeled with Alexa Fluor® 488 (green) (NHS chemistry; Thermo Fisher) and labeled proteome was purified through a spin column (Thermo Fisher). Control samples were labeled with Alexa Fluor® 647 (red) and purified. The volume of sample solution before and after labeling did not change appreciably (∼50 μL), and therefore the originally estimated protein concentrations were considered for the X-aptamer screening process. Biotinylation of proteome from the remaining 50 μL of sample per group was carried out using sulfosuccinimidyl-6-(biotinamido)hexanoate (EZ-linkTM Sulfo-NHS-LC-Biotin; Thermo Fisher). Biotinylated proteins were purified through ZebaTM spin columns (Thermo Fisher).
Two-Color Flow Sorting of X-Aptamer Beads
The X-aptamer bead library was purchased from AM Biotechnologies, LLC (Houston, TX, USA) and split into three parts (∼3 million beads each) [8]. The split bead library was individually incubated with 2 µg of fluorescently labeled proteome from the pooled patient group and the pooled control group. After 1 h of incubation, a two-color flow sort was performed through a BD FACS ARIA II flow cytometer (BD Biosciences, San Jose, CA, USA). After sorting, green beads (patient proteome) and red beads (control proteome) were separately collected.
Solution Phase Screen to Recapture Proteome-Specific X-Aptamers
Each sorted bead contains many copies of a single unique X-aptamer sequence attached to the bead surface by a single RNA base that can be cleaved by alkaline hydrolysis, releasing X-aptamers to the solution phase. Alkaline hydrolysis was performed using 0.2 M sodium hydroxide at 65°C for 30 min. The supernatant solution was separated from the beads by centrifugation, and sodium hydroxide was removed by dilution and filtering through 3-kDa cutoff spin filters. Biotinylated proteome (50 μL) from each sample group was individually loaded onto 1 mg of streptavidin-coated DynabeadsTM M-280 (Invitrogen, Carlsbad, CA, USA) and incubated with the corresponding sorted X-aptamer solution phase to recapture proteome-specific X-aptamers. Following incubation, the DynabeadsTM were used for PCR amplification of captured X-aptamers and next-generation sequencing (NGS). Barcoded primers were used during PCR reactions to distinguish between the X-aptamer sequences from the patient and control groups. PCR products were purified and pooled at equal concentrations for NGS (PrimBio Research Institute, Exton, PA, USA).
Analysis of NGS Data and Identification of X-Aptamers Specific to Patients
The NGS data were analyzed using Aptaligner software [21], specifically designed for the analysis of NGS data from bead-based X-aptamer selection projects. The data file contained sequence information and frequencies for the patient and control pools, for both the bead-based selection (flow sorting) and the solution phase selection (biotinylation). Potentially selective aptamers were chosen based on relatively high NGS frequencies (∼binding) in both phases of the selection process in patients or controls, while having lower frequencies in the opposite experiment group. Thus, in Table 2, SC-XA1 is the sequence that occurred the most in the experiments using patient blood samples, and Ctrl-XA1 (which is not a control in the usual sense) is the sequence that occurred the most in the experiments using control patient blood samples. Both sequences arose from the same XA library and thus have identical primer sequences (except for bar codes) as well as ∼50% identity in the random region.
Table 2.
Top 5 X-aptamer frequencies found for each selection experiment
| X-aptamer sequence | Risk group |
|
|---|---|---|
| control | SC | |
| SC-XA1 | 0 | 190 |
| SC-XA2 | 0 | 97 |
| SC-XA3 | 0 | 88 |
| SC-XA4 | 0 | 60 |
| SC-XA5 | 0 | 30 |
| Ctrl-XA1 | 1,870 | 0 |
| Ctrl-XA2 | 205 | 0 |
| Ctrl-XA3 | 105 | 0 |
| Ctrl-XA4 | 45 | 0 |
| Ctrl-XA5 | 43 | 0 |
XA, X-aptamer; Ctrl, control; SC, schizophrenia.
Generation of Unique X-Aptamers
Potentially selective X-aptamers were synthesized on an Expedite 8909 Oligo Synthesizer (Applied Biosystems, Foster City, CA, USA) at the 1-µmole scale, using specialized X-phosphoramidites (AM Biotechnologies), standard phosphoramidites, and DNA Synthesis Reagents (Glen Research, Sterling, VA, USA). The X-aptamers were deprotected under basic conditions following recommended methods (AM Biotechnologies) and purified on an AKTA 10 (GE Healthcare Life Sciences, Pittsburgh, PA, USA) purifier over a semipreparative Hamilton PRP-1 reverse-phase column using a 100-mM triethylamine acetate (pH 8.4) loading buffer and an increasing gradient (1/3%/min) of acetonitrile from 0 to 40% with a 2 mL/min flow rate.
Mass Spectrometry
Biotinylated X-aptamers individually synthesized (1 µmole) were loaded on 1 mg of DynabeadsTM and incubated with 5 µg of albumin-depleted patient or control pooled plasma samples – in duplicate, separate, individual reactions – at room temperature for 1 h. After incubation, the DynabeadsTM were washed with PBS and suspended in 1× Laemmli buffer, and clear supernatant (20 µL) was electrophoresed on 10% denaturing polyacrylamide gel for 10 min. Within gel, trypsin digestion was performed, followed by mass spectrometric analysis (Orbitrap Fusion ETD; Thermo Fisher), as previously described [19].
The raw data files were processed to generate a Mascot Generic Format file with Mascot Distiller and searched against the IPI_human_3.87.fastq database using the Mascot search engine v2.3.02 (www.matrixscience.com/search_form_select.html). Mascot provides tentative identifications of proteins by identifying peptides expected to arise from each protein in the queried database. It calculates a score for each tentatively identified protein based on the number of observed peptides compared to the expected number of peptides. For proteins at high concentration, all or most of the theoretical peptides will be observed, resulting in a high score (high confidence of protein assignment). Low-abundance proteins do not provide as many detected peptides, and thus the confidence (score) in the protein assignment is lower. If a protein concentration is high in two samples, but 5 times higher in one of them, both samples can yield the same score because the same number of peptides (not concentration) was detected in each mass spectrometry run. The proteins are then listed (ranked) in order of decreasing score, with the most confident assignment being ranked 1. Top-ranked proteins (ranks 1–3) typically have scores well over 1,000 (online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000492331) and proteins with scores less than 70 are considered noise. Potential protein biomarkers should exhibit a significant rise or drop in rank in patients compared to controls.
Enzyme-Linked Immunosorbent Assays
Complement C4 (C4) and apolipoprotein B (ApoB) levels were measured by ELISA (ab108824 and ab190806; Abcam, Cambridge, UK) in plasma from each individual patient according to manufacturer's instructions. Samples were diluted 1: 100 for measurement of C4 and 1: 4,000 for ApoB.
Statistical Analyses
Statistical analyses for the ELISA experiments were completed using GraphPad Prism version 5 (GraphPad Software, La Jolla, CA, USA). After log10 transformation, the data were found to follow a normal distribution (Kolmogorov-Smirnov test). Student's t test was used to compare controls against each patient group, separately. Data are presented as the mean ± standard error of the mean (SEM). p values < 0.05 were considered statistically significant.
Results
Demographics
No differences in gender, ethnicity, years of education, BMI, weight, or age were found between the groups (Table 1).
Table 1.
Characteristics of the subjects
| Variable | SC patients (n = 60) | Controls (n = 20) |
|---|---|---|
| Age, years | 42.5±1.4 | 41.1±2.6 |
| Sex (male/female),n | 46/14 | 14/6 |
| Ethnicity (C/AA/H), n | 9/12/39 | 4/3/13 |
| BMI | 32.13±1.8 | 28.55±1.5 |
| Weight, lb | 204.8±10.38 | 188.4±9.4 |
| Years of education | 11.56±0.37 | 12.6±0.2 |
| Aripiprazole, ziprasidone | 1, 10 | NA |
| Quetiapine, risperidone | 5, 22 | NA |
| Clozapine, olanzapine | 2, 20 | NA |
BMI, weight, age, and years of education are indicated as the mean ± SEM. SC, schizophrenia; C, Caucasian; AA, African American; H, Hispanic; NA, not applicable.
Identifying Putative X-Aptamers for Biomarker Detection
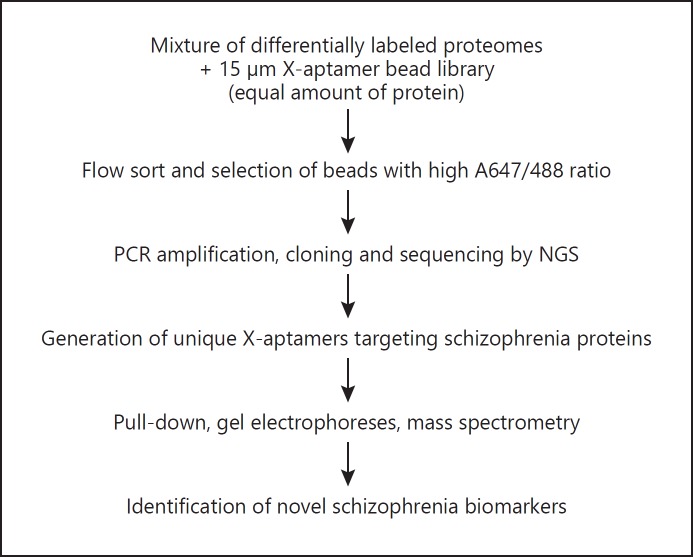
The overall approach for X-aptamer selection and biomarker identification is shown in Figure 1. First, the X-aptamer bead libraries were incubated with 150 μL of pooled plasma samples from the patient and control groups, separately, to identify which X-aptamers would bind with a differential preference to plasma from schizophrenia patients relative to control plasma (Table 2). For each of the groups, a number of X-aptamers bound with high specificity. The top 5 sequences (by frequency) are listed along with the frequency counts. For the patient population, X-aptamer SC-XA1 was observed 190 times, but it was found 0 times in the control experiment. One would then expect this aptamer to bind to a protein upregulated in the schizophrenia group. For the control group, X-aptamer Ctrl-XA1 was detected 1,870 times, and it was not observed in the schizophrenia group.
Fig. 1.
X-aptamer analysis of schizophrenia plasma samples to identify novel biomarkers. NGS, next-generation sequencing.
C4A and ApoB Identified by Two Unique X-Aptamers in the Schizophrenia Patient Group
Based on the results in Table 2, two X-aptamer sequences, SC-XA1 and Ctrl-XA1, were synthesized in their biotinylated form and bound to streptavidin on beads. Each bead-bound X-aptamer was then utilized for protein pull-down experiments against the group of pooled samples from patients and the pooled samples from controls. The proteins were removed, run on a gel, and subjected to mass spectrometric analysis (online suppl. Table 1), from which proteins were ranked based on the individual scores and unique spectra (Table 3). From the pull-down experiments, two proteins, ApoB and C4A, had very different rankings in plasma from patients compared to controls, thus suggesting that they might be useful biomarkers for schizophrenia. ApoB, which ranked as the 3rd highest protein for both the SC-XA1 and Ctrl-XA1 pull-down experiments in the schizophrenia group, was only ranked as the 13th highest protein in the control plasma for binding to SC-XA1 (∼5 times fewer ions as shown by the emPAI score) and was not detected at all in the Ctrl-XA1 binding experiment with control plasma. Showing similar behavior, protein IPI00643 (C4A), while ranked as the 6th highest protein in the schizophrenia group in binding experiments against both SC-XA1 and Ctrl-XA1, only ranked as the 11th highest protein (∼50% smaller emPAI score) in the control plasma pool for binding with SC-XA1 and was not detected in the binding experiment using Ctrl-XA1. These differences in ApoB and C4A suggest they may be biomarkers for schizophrenia. The other highly ranked proteins exhibited substantially similar rankings across the groups and were thus not considered as possible biomarkers. It should be noted that due to the sensitivity of the spectrometer, environmental proteins such as keratin and keratin fragments are always detected.
Table 3.
Mascot rank of proteins detected by mass spectrometry from X-aptamer bead pull-down experiments
| Protein ID | Description | SC-XA1 ranking | Ctrl-XA1 ranking | ||
|---|---|---|---|---|---|
| SC | control | SC | control | ||
| IPI00783987 | Complement C3 (fragment) | 1 | 2 | 1 | 5 |
| IPI00220327 | KRT1 keratin | 2 | 1 | 2 | 1 |
| IPI00022229 | APOB apolipoprotein B-100 | 3 | 13 | 3 | ND |
| IPI00009865 | KRT10 keratin | 4 | 3 | 4 | 2 |
| IPI00550731 | Putative uncharacterized protein | 5 | 4 | 5 | 3 |
| IPI00643 | C4A uncharacterized protein | 6 | 11 | 6 | ND |
| IPI00478003 | Alpha-2-macroglobulin | 7 | 5 | 7 | 10 |
| IPI00745872 | ALB isoform 1 of serum albumin | 8 | 6 | 8 | 4 |
| IPI00896380 | IGHM isoform 2 of Ig mu chain C | 9 | 12 | 11 | 16 |
| IPI00021841 | APOA1 apolipoprotein A-I | 10 | 7 | 10 | 7 |
Proteins in bold are considered potential biomarkers for schizophrenia. SC, schizophrenia; Ctrl, control; ND, not detected.
Of interest, C4A has been identified as a potential schizophrenia susceptibility gene in a recent study following a GWAS signal [22].
Validation in Individual Plasma Samples
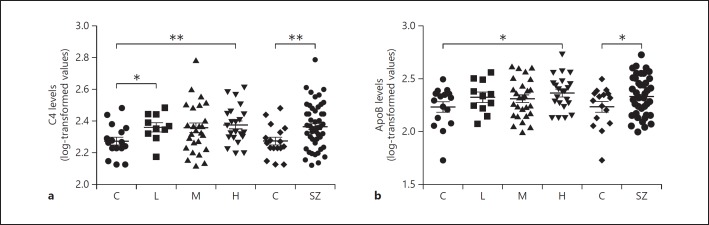
ELISAs specific for C4 (which recognize all C4 isoforms) and ApoB, considered the top 2 most significant proteins identified by the ranking analyses, were performed on the individual schizophrenia (n = 60) and control samples (n = 20) to validate the X-aptamer results obtained from the pooled samples. We observed increased levels of C4 and ApoB in the schizophrenia group compared to the controls (p = 0.0097 and p = 0.047, respectively) (Fig. 2).
Fig. 2.
log-transformed relative data for levels of C4 (a) and ApoB (b) in controls (C; n = 20) and patients with schizophrenia (SZ; n = 60) undergoing treatment with antipsychotics conferring a low (L; n = 11), medium (M; n = 27), or high (H; n = 22) risk for developing metabolic syndrome. The data were assessed by Student's t test comparing the controls with each group separately. * p < 0.05, ** p < 0.01.
To at least partially assess whether the elevated levels of C4 and ApoB in the patients could be related to medication, the patients were subdivided into groups according to the type of second-generation antipsychotic (SGA) used, based on the known risk for metabolic syndrome conferred by these medications (low, medium, or high [23]), and each group was compared to the controls. Levels of C4 were significantly increased in patients using low- and high-risk SGAs as compared to controls (p = 0.0293 and p = 0.0072, respectively), with a trend also observed for the medium-risk group, suggesting that differences in levels between all patients and controls may be due to the presence of the schizophrenia illness itself, and not due to the type of SGA used (Fig. 2a). On the other hand, we observed significantly increased levels of ApoB only in patients under treatment with high-risk antipsychotics compared to controls (p < 0.0282), and not in patients under treatment with low- or medium-risk medications, suggesting that the initial finding of ApoB in the pooled samples may be due to the type of SGA used by the patients (Fig. 2b).
Discussion
The development of high-throughput protein isolation techniques, such as X-aptamers, has recently revolutionized modern approaches to proteomic analyses in clinical research [7]. To our knowledge this is the first study to use an X-aptamer library to identify potential biomarkers for schizophrenia. Using a different approach, SOMAmers and the SOMAscan platform also utilize modified DNA for biomarker detection, although this has not been used for schizophrenia [24]. For the initial screening, our strategy was to use pooled samples from healthy controls and schizophrenia patients. After incubation of fluorescently labeled pooled samples from patients and controls with the X-aptamer library, we identified two unique X-aptamer sequences by NGS, one overrepresented in patients and the other overrepresented in controls. After mass spectrometric analyses of pooled-down peptides, we identified C4A and ApoB as the two proteins exhibiting differential expression in patients compared to controls.
The gene coding for C4A is located within the major histocompatibility complex (MHC) locus on chromosome 6, a region found to be associated with schizophrenia in the largest GWAS of schizophrenia to date (36,989 cases and 113,075 controls), in which among the 108 loci that met the criteria for genome-wide significance, the locus on chromosome 6 harboring MHC genes was the most significant [2]. A follow-up study pinpointed the MHC association to alleles of the C4 gene [22] coding for C4A and C4B, and also found elevated levels of C4A in postmortem brains of schizophrenia patients compared to controls. C4 activates C3, marking the synapse for phagocytosis [25], and drives synaptic pruning by microglia [26]. The finding of C4 alleles associated with schizophrenia indicates an important genetic influence on the extent of synaptic pruning, which has been hypothesized to play an important role in schizophrenia development [27]. Recently, an RNA sequencing-based transcriptomic profiling study on lymphoblastoid cell lines from a sample of European ancestry (529 schizophrenia cases and 660 controls) found differential expression in components of the complement system (CR1, CR2, CD55, and C3), though not for C4A [28]. Our study demonstrates the potential of C4 as a biomarker of diagnostic value, as it can be identified and differentiated in easily accessible peripheral tissue. Our findings further suggest that the observed differences in C4 levels in schizophrenia patients are likely due to the presence of the illness itself and not alterations caused by SGAs, since significant increases were observed across different types of SGAs known to differ in their risk to induce metabolic alterations. However, the possibility exists that C4 levels are altered due to other medications or factors not evaluated in this study, and thus further studies are needed to completely rule out effects of medications on C4 expression.
ApoB is a member of the apolipoprotein family that interacts with the low-density lipoprotein receptor and is involved in the distribution of fat molecules to tissues [29]. High levels of ApoB are thought to be a primary cause of vascular complications such as atherosclerosis, heart disease, and stroke, and mutations in the APOB gene are associated with hypercholesterolemia [30]. Studies have suggested cholesterol and apolipoproteins as candidate psychiatric biomarkers [31]. We observed elevated levels of ApoB only in subjects under treatment with SGAs that cause a high risk for metabolic syndrome, suggesting this protein as a marker for antipsychotic-induced metabolic alterations. Using the same cohort of patients, we have previously demonstrated that high-risk SGAs are associated with the disruption of proteins involved in energy metabolism [22]. Similarly, Wang et al. [32] found that schizophrenia patients who are considered metabolically obese but of normal weight under treatment with olanzapine or risperidone have elevated levels of cholesterol, very-low-density lipoprotein, and ApoB compared with those considered metabolically non-obese, suggesting ApoB as a marker for hyperlipidemia and cardiovascular complications in subjects with schizophrenia. In addition, Ryoo and Park [33] demonstrated a significant association between elevated serum ApoB levels and future development of the metabolic syndrome among a large number of Korean men during a 5-year follow-up period. Future prospective studies should consider the role of ApoB in the development of metabolic complications in schizophrenic patients under treatment with clozapine or olanzapine.
Due to budget limitations, our pool-down and mass spectrometry experiments were performed using only two of the unique X-aptamer sequences identified to have differential binding in patients compared to controls (Table 2). Further studies are warranted to follow up on additional X-aptamers that exhibited preferential binding to the patient group, as these could lead to the identification of additional potential biomarkers for schizophrenia. Further studies to trim down the X-aptamers for optimal performance are also planned. As these X-aptamers have not yet been optimized, comparisons with the more established SOMAmer reagents, and comparisons of binding affinities and off-rates, are not yet warranted.
Our present results should be taken with caution given the lack of information related to potential confounding factors such as smoking and alcohol/drug abuse history. Further, the relatively small sample size and cross-sectional nature of the study necessitate further validation in larger longitudinal studies. In addition, in order to clearly determine the effect of SGAs and other medications on C4 and ApoB, future studies should be performed with patients before and after treatment or on medication- naïve patients.
In summary, we have used a novel X-aptamer proteomic approach to identify potential novel biomarkers for schizophrenia. We identified C4 as a potential diagnostic marker for schizophrenia and ApoB as a marker of antipsychotic-induced metabolic alterations. This study highlights the utility of the X-aptamer technology in the identification of biomarkers for schizophrenia that will advance our understanding of the pathophysiological mechanisms of this disorder.
Statement of Ethics
This study was performed with approval from the Institutional Review Boards in accordance with the Declaration of Helsinki.
Disclosure Statement
D.V.'s conflicts of interest are: advisory board and honoraria – Genentech, AbbVie, Amgen, and Otsuka Lundbeck; research grants – Genentech and Amgen; speakers bureau – Otsuka Lundbeck and Janssen. D.G.G. has a financial interest in AM Biotechnologies. All other authors declare no conflict of interest.
Supplementary Material
Supplementary data
Acknowledgements
This study was supported in part by the Center for Clinical and Translational Sciences (CCTS) (UL1TR000371) via a Pilot Project Award (C.W.-B.) and salary support for D.E.V. and Amy Hazen, U54CA151668 (D.G.G.), and the UTHSCSA Department of Psychiatry: Friends for Psychiatric Research (D.V.). We thank Dr. Amy Hazen and the CCTS-supported UTHealth Flow Cytometry Center and Nanochemistry Service Center for their assistance.
References
- 1.Henriksen MG, Nordgaard J, Jansson LB. Genetics of schizophrenia: overview of methodsfindings and limitations. Front Hum Neurosci. 2017;11:322. doi: 10.3389/fnhum.2017.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davalieva K, Maleva Kostovska I, Dwork AJ. Proteomics research in schizophrenia. Front Cell Neurosci. 2016;10:18. doi: 10.3389/fncel.2016.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nascimento JM, Garcia S, Saia-Cereda VM, Santana AG, Brandao-Teles C, Zuccoli GS, Junqueira DG, Reis-de-Oliveira G, Baldasso PA, Cassoli JS, Martins-de-Souza D. Proteomics and molecular tools for unveiling missing links in the biochemical understanding of schizophrenia. Proteomics Clin Appl. 2016;10:1148–1158. doi: 10.1002/prca.201600021. [DOI] [PubMed] [Google Scholar]
- 5.Veenstra TD. Neuroproteomic tools for battling Alzheimer's disease. Proteomics. 2016;16:2847–2853. doi: 10.1002/pmic.201600211. [DOI] [PubMed] [Google Scholar]
- 6.Latosinska A, Frantzi M, Vlahou A, Merseburger AS, Mischak H. Clinical proteomics for precision medicine: the bladder cancer case. Proteomics Clin Appl. 2018;12 doi: 10.1002/prca.201700074. prca.201700074. [DOI] [PubMed] [Google Scholar]
- 7.He W, Elizondo-Riojas MA, Li X, Lokesh GL, Somasunderam A, Thiviyanathan V, Volk DE, Durland RH, Englehardt J, Cavasotto CN, Gorenstein DG. X-aptamers: a bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry. 2012;51:8321–8323. doi: 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lokesh GL, Wang H, Lam CH, Thiviyanathan V, Ward N, Gorenstein DG, Volk DE. X-aptamer selection and validation. Methods Mol Biol. 2017;1632:151–174. doi: 10.1007/978-1-4939-7138-1_10. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Li X, Volk DE, Lokesh GL, Elizondo-Riojas MA, Li L, Nick AM, Sood AK, Rosenblatt KP, Gorenstein DG. Morph-X-Select: morphology-based tissue aptamer selection for ovarian cancer biomarker discovery. Biotechniques. 2016;61:249–259. doi: 10.2144/000114473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao T, Liu R, Ding X, Zhao J, Yu H, Wang L, Xu Q, Wang X, Lou X, He M, Xiao Y. Nanoprobe-enhanced, split aptamer-based electrochemical sandwich assay for ultrasensitive detection of small molecules. Anal Chem. 2015;87:7712–7719. doi: 10.1021/acs.analchem.5b01178. [DOI] [PubMed] [Google Scholar]
- 11.Ma C, Zhao C, Ge Y, Shi C. Aptameric molecular switch for cascade signal amplification. Clin Chem. 2012;58:384–390. doi: 10.1373/clinchem.2011.173195. [DOI] [PubMed] [Google Scholar]
- 12.Meng HM, Liu H, Kuai H, Peng R, Mo L, Zhang XB. Aptamer-integrated DNA nanostructures for biosensingbioimaging and cancer therapy. Chem Soc Rev. 2016;45:2583–2602. doi: 10.1039/c5cs00645g. [DOI] [PubMed] [Google Scholar]
- 13.Lyu Y, Chen G, Shangguan D, Zhang L, Wan S, Wu Y, Zhang H, Duan L, Liu C, You M, Wang J, et al. Generating cell targeting aptamers for nanotheranostics using cell-SELEX. Theranostics. 2016;6:1440–1452. doi: 10.7150/thno.15666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Identification of liver cancer-specific aptamers using whole live cells. Anal Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 15.Kang SA, Hasan N, Mann AP, Zheng W, Zhao L, Morris L, Zhu W, Zhao YD, Suh KS, Doo ley WC, Volk D, et al. Blocking the adhesion cascade at the premetastatic niche for prevention of breast cancer metastasis. Mol Ther. 2015;23:1044–1054. doi: 10.1038/mt.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang SA, Tsolmon B, Mann AP, Zheng W, Zhao L, Zhao YD, Volk DE, Lokesh GL, Morris L, Gupta V, Razaq W, et al. Safety evaluation of intravenously administered mono-thioated aptamer against E-selectin in mice. Toxicol Appl Pharmacol. 2015;287:86–92. doi: 10.1016/j.taap.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mai J, Huang Y, Mu C, Zhang G, Xu R, Guo X, Xia X, Volk DE, Lokesh GL, Thiviyanathan V, Gorenstein DG, et al. Bone marrow endothelium-targeted therapeutics for metastatic breast cancer. J Control Release. 2014;187:22–29. doi: 10.1016/j.jconrel.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann AP, Bhavane RC, Somasunderam A, Liz Montalvo-Ortiz B, Ghaghada KB, Volk D, Nieves-Alicea R, Suh KS, Ferrari M, Annapragada A, Gorenstein DG, et al. Thioaptamer conjugated liposomes for tumor vasculature targeting. Oncotarget. 2011;2:298–304. doi: 10.18632/oncotarget.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangala LS, Wang H, Jiang D, Wu SY, Somasunderam A, Volk DE, Lokesh GL, Li X, Pra deep S, Yang X, Haemmerle M, et al. Improving vascular maturation using noncoding RNAs increases antitumor effect of chemotherapy. JCI Insight. 2016;1:e87754. doi: 10.1172/jci.insight.87754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard F, Ha NP, Sule P, Alexander JF, Volk DE, Lokesh GLR, Liu X, Cirillo JD, Gorenstein DG, Yuan J, Chatterjee S, et al. Thioaptamer targeted discoidal microparticles increase self immunity and reduce <italic>Mycobacterium tuberculosis</italic> burden in mice. J Control Release. 2017;266:238–247. doi: 10.1016/j.jconrel.2017.09.038. [DOI] [PubMed] [Google Scholar]
- 21.Lu E, Elizondo-Riojas MA, Chang JT, Volk DE. Aptaligner: automated software for aligning pseudorandom DNA X-aptamers from next-generation sequencing data. Biochemistry. 2014;53:3523–3525. doi: 10.1021/bi500443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paredes RM, Quinones M, Marballi K, Gao X, Valdez C, Ahuja SS, Velligan D, Walss-Bass C. Metabolomic profiling of schizophrenia patients at risk for metabolic syndrome. Int J Neuropsychopharmacol. 2014;17:1139–1148. doi: 10.1017/S1461145714000157. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Z, Pan X, Parlayan C, Ojima H, Kondo T. Proteomic study of hepatocellular carcinoma using a novel modified aptamer-based array (SOMAscan) platform. Biochim Biophys Acta. 2017;1865:434–443. doi: 10.1016/j.bbapap.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Presumey J, Bialas AR, Carroll MC. Complement system in neural synapse elimination in development and disease. Adv Immunol. 2017;135:53–79. doi: 10.1016/bs.ai.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Sellgren CM, Sheridan SD, Gracias J, Xuan D, Fu T, Perlis RH. Patient-specific models of microglia-mediated engulfment of synapses and neural progenitors. Mol Psychiatry. 2017;22:170–177. doi: 10.1038/mp.2016.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders AR, Drigalenko EI, Duan J, Moy W, Freda J, Göring HHH, Gejman PV. Transcriptome sequencing study implicates immune-related genes differentially expressed in schizophrenia: new data and a meta-analysis. Transl Psychiatry. 2017;7:e1093. doi: 10.1038/tp.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linton MF, Yancey PG, Davies SS, Jerome WGJ, Linton EF, Vickers KC. The role of lipids and lipoproteins in atherosclerosis. In: De Groot LJ, Chrousos G, Dungan K, et al.Endotext, editors. South Dartmouth. MDText.com Inc 2000–2015, last update Dec 24, 2015. [Google Scholar]
- 30.Ramasamy I. Update on the molecular biology of dyslipidemias. Clin Chim Acta. 2016;454:143–185. doi: 10.1016/j.cca.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 31.Woods AG, Sokolowska I, Taurines R, Gerlach M, Dudley E, Thome J, Darie CC. Potential biomarkers in psychiatry: focus on the cholesterol system. J Cell Mol Med. 2012;16:1184–1195. doi: 10.1111/j.1582-4934.2012.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Liu EY, Freudenreich O, Goff D, Henderson DC, Fan X. Phenotypic characteristics in metabolically obese but normal weight non-diabetic patients with schizophrenia. Schizophr Res. 2010;124:49–53. doi: 10.1016/j.schres.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Ryoo JH, Park SK. Association of apolipoprotein B and incidence of metabolic syndrome in Korean men: a 5-years’ follow-up study. Atherosclerosis. 2013;226:496–501. doi: 10.1016/j.atherosclerosis.2012.11.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data