Abstract
Lutein, a hydroxylated carotenoid, is a pigment synthesised by plants and bacteria. Animals are unable to synthesise lutein, nevertheless, it is present in animal tissues, where its only source is dietary intake. Both in plants and animals, carotenoids are associated mainly with membranes where they carry out important physiological functions. Their trafficking to and insertion into membranes are not well recognised due to experimental difficulties. In this paper, a computational approach is used to elucidate details of the dynamics and energetics of lutein intercalation from the water to the phospholipid bilayer phase. The dynamics is studied using molecular dynamics simulation, and the energetics using umbrella sampling. Lutein spontaneous insertion into the bilayer and translocation across it proceed via formation of hydrogen bonds between its hydroxyl groups and water and/or phospholipid oxygen atoms as well as desolvation of its polyene chain. As lutein molecule is asymmetric, its bilayer intercalation is also asymmetric. The course of events and timescale of the intercalation are different from those of helical peptides. The time of full lutein intercalation ranges from 20 to 100 ns and its final orientation is predominately vertical. Nevertheless, some lutein molecules are in the final horizontal position and some aggregate in the water phase and remain there for the whole simulation time. The highest energy barrier for the intercalation process is ~2.2 kcal/mol and the energy gain is ~18 kcal/mol. The results obtained for lutein can be applied to other xanthophylls and molecules of a similar structure.
Keywords: Xanthophyll, Energy barrier, Hydrogen bond, Hydrophobic effect, Molecular dynamics, Umbrella sampling
Abbreviations: MD, Molecular dynamics; US, Umbrella sampling; PC, Phosphatidylcholine; Palmitoyl-oleoyl PC, POPC; OH-LUT, Lutein hydroxyl group; Op, Oe, Og, Oc collective names for the non-esterified phosphate, esterified phosphate, glycerol, and carbonyl oxygen atoms, respectively; Ow, Water oxygen atom
Graphical Abstract
Highlights
-
•
Lutein as an amphiphilic molecule readily intercalates into a lipid bilayer.
-
•
MD simulations with dense sampling revealed details of the intercalation process.
-
•
Time of full lutein membrane intercalation ranges from 20 to 100 ns.
-
•
Lutein intercalation from the ε ring end is less probable than from the β ring end.
-
•
Horizontal position of lutein in the bilayer is less probable than vertical.
1. Introduction
Xanthophylls (polar carotenoids) are natural pigments, synthesised mainly by plants and bacteria. In photosynthetic organisms, their main functions are energy collection and protection of the photosynthetic apparatus against photodamage [1]. The main xanthophylls involved in these processes are lutein, violaxanthin and neoxanthin. Lutein, the object of the present study, plays a key role in quenching triplet chlorophyll states in the major light-harvesting complex of higher plants [2,3].
Even though man is not capable of synthesising lutein, it has an important impact on human health [4] and must be supplied in the diet. In humans, lutein is present in blood serum and skin but especially high concentration of lutein can be found in the retina of the eye [5,6]. Lutein effectively protects the eye and skin from blue light and from oxygen free radical-induced damage [6,7] as well as from oxidative stress [8,9].
Theses protective roles that lutein plays in various organisms, are strictly related to and stem from its specific structural features. The molecule (C40H56O2) consists of a nonpolar polyene chain and two terminal polar monohydroxylated in C3 and C3′ positions ionone rings (Fig. 1a). The two rings are not equivalent (β and ε rings, Fig. 1a) as they differ in the position of the double bond. The chain comprises eight isoprene units with nine conjugated double bonds (Fig. 1a) which are responsible for the blue light absorbance and quenching of reactive oxygen intermediates [3,6,7]. Lutein, with polar and nonpolar groups, is an amphiphilic molecule and in the cell its final location are mainly membranes. In the membrane, the isoprenoid chain is positioned in the hydrophobic membrane core and the two hydroxylated rings are placed in the membrane polar interfacial region.
Fig. 1.
Chemical structures of (a) lutein and (b) POPC. In (a), the ionone rings and atoms that are used in analyses are indicated. In (b), the head group atoms are numbered in accordance with Sundaralingam [18]. The numbers of the acyl chains atoms, the chemical symbols for carbon atoms, C, and hydrogen atoms in the CH3, CH2, and CH groups have been omitted.
As it was mentioned above, in humans lutein is supplied in the diet. From small intestine it has to pass into the bloodstream where it is transported in some carriers [10] to selectively intercalate into the membranes of the eye retina or other tissues cells [11]. Also in plants, lutein that is synthesised in the envelope membrane has to be transported to the thylakoids, where it accumulates [12]; this process also involves the membrane intercalation phase.
Spontaneous transfer of a nonpolar molecule from a polar into a nonpolar environment like a lipid bilayer is common in nature and is driven by the free energy gain [13]. Nevertheless, the exact molecular mechanisms of a nonpolar molecule insertion into the bilayer are not well explained [10,14,15]. It is because they involve short-lived interatomic interactions e.g. [14,16]; besides, the molecules aggregate in the water phase e.g. [17]. This makes membrane insertion difficult to observe with currently available experimental methods. In contrast, molecular dynamics (MD) simulation having a femtosecond time and an atomic space resolution is particularly predisposed to investigate the process, so, it has been used in such studies.
In Ref. [19] the location of lutein molecules spontaneously intercalated into the phosphatidylcholine (PC) bilayer was analysed using computational methods. The study predicted two preferential lutein orientations in the bilayer and indicated basic interactions stabilising them. It also revealed that spontaneous lutein intercalation into the bilayer is fast and can complete even within <20 ns of MD simulation. However, the intercalation process itself was analysed neither in that study nor in any other. In contrast, peptide insertion into the lipid bilayer has been widely studied using experimental and computational methods e.g. [[20], [21], [22], [23], [24]].
In this computational study, the process of lutein membrane intercalation is analysed with sub-picosecond time resolution and one of the results is again the prediction of two main orientations of lutein in the bilayer – vertical and horizontal. These two preferential orientations were first suggested based on experimental results of the Gruszecki group e.g. [25]. In a recent experimental and computational study, Grudzinski et al. [26] determined that the location of a lutein molecule in the dimyristoyl-PC bilayer is trans-bilayer and the most probable orientation of the lutein polyene chain relative to the normal is 25° whereas the average orientation is ~37°. The most probable orientation was obtained from the distribution of lutein orientations and from the calculated free energy profile for rotation of a lutein molecule in the bilayer from the vertical to horizontal.
The liquid crystalline bilayer used here to study spontaneous lutein intercalation consists of 200 PC molecules. Such a bilayer size is large enough to enable minimisation of the stress put on the system during lutein intercalation. PC is chosen because it is a major lipid component of cell membranes of various organisms and PC bilayers have been used in relevant experimental e.g. [25,26] and MD simulation e.g. [26,27] studies. In addition to investigating the spontaneous process with unbiased classical MD simulation (no force pulling the molecule towards and through the bilayer hydrophobic core), umbrella sampling (US) simulation is performed to calculate free energy barriers for this process. The two approaches enable us to elucidate both dynamic and energetic aspects of the intercalation process of lutein which represents a class of molecules of certain structural features, particularly xanthophylls.
By full membrane intercalation of lutein we understand the state resulting from lutein spontaneous entering the bilayer from the water phase, in which its both ionone rings are anchored in the bilayer interface and its polyene chain is oriented either vertically or horizontally relative to the bilayer surface.
The key result of this study is that lutein intercalates into the bilayer preferentially with its β-ring. This finding is in perfect coincidence with the results of structural docking experiments which revealed that the ε-ring fits into the active site of the enzyme that catalyses the conversion of lutein to meso-zeaxanthin [28] and also into the asymmetric cavity of the lutein binding protein [29]. In both proteins, the β-ring binds outside the cavity.
2. Materials and Methods
In this study, both the dynamics and the energetics of the lutein intercalation into the bilayer are analysed in details. To investigate the dynamics, classical molecular modelling with atomic resolution, mainly MD simulation is used. To assess energetics, umbrella sampling (US) method is used.
2.1. MD Simulation
2.1.1. System Construction and Force Field Parameters
The basic system used in this MD simulation study consisted of a hydrated liquid crystalline 3-palmitoyl-2-oleoyl-1-phosphatidylcholine (POPC, Fig. 1b) bilayer, and six lutein molecules (Fig. 1a). The POPC bilayer containing 200 POPC (100 in each leaflet) and 6000 water molecules was built, equilibrated and validated in Ref. [30]. For POPC and lutein, all-atom optimised potentials for liquid simulations (OPLS-AA) force field [31] was used, and for water, TIP3P model [32] was used. The initial structure of the (3R, 3′R, 6′R) lutein molecule was created as described in Ref. [19]. In brief, the structure was built using the Avogadro program [33], and optimised using the Dreiding [34] force field with the Gasteiger-Marsili charges [35]. Afterwards, the structure was fully optimised at B3LYP/6-31G(d) level of theory using the Gaussian 09 program [36]. A rotational profile for the C6′-C7′ dihedral angle in the lutein molecule (Fig. 1a), calculated for the molecule parameterised in the OPLS-AA force field, was positively verified against that calculated at the B3LYP/6-31G(d) level of theory for twelve rotamers [19] (Fig. S1, SI) and that calculated by Landrum et al. [37]. Distributions of conformations of torsion C5-C6-C7-C8, β-ring, and torsion C5′-C6′-C7′-C8′, ε-ring, (Fig. S2, SI) follow strictly the quantum mechanical energy profiles for rotation for these torsions calculated in Ref. [37] and that for the C6′-C7′ torsion angle presented in Fig. S1 (SI). Time profiles of conformations of the torsion angles generated during MD simulations are shown in Fig. S3 (SI); they also follow the energy minima and the energy barriers of the quantum mechanical rotational profiles.
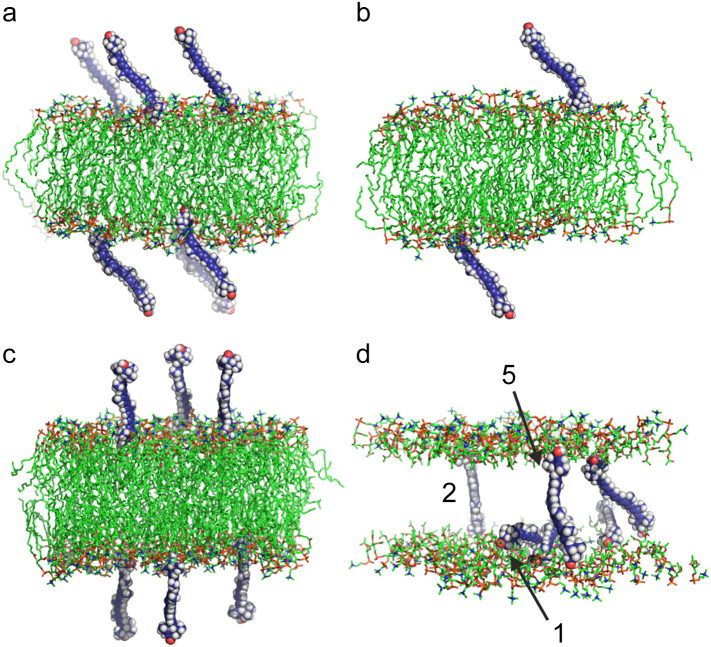
To construct the initial structures of the investigated systems, the water molecules were removed from the POPC bilayer [30], and six lutein molecules were placed near its surface (Fig. 2a). In total, four systems were MD simulated. Two of them included lutein molecules facing the bilayer with their β rings (system1 and system3) and two included molecules facing the bilayer with their ε rings (system2 and system4). Then, each system was rehydrated with over 40 water molecules per lipid (in total, ~10,000 water molecules were added to each system). The initial structure of one system is shown in Fig. 2a, b, c and its final structure in Fig. 2d. The initial and final positions of the rings of all lutein molecules that intercalated into the bilayer are schematically shown in Fig. S4 (SI). As can be seen from this figure, the entering lutein rings are initially placed within the regions of the phosphate groups and above the regions of the carbonyl groups. In the final states, both rings are placed in the regions of the carbonyl groups (Fig. S4, SI). Snapshots of the final positions of the lutein molecules in the bilayers are also shown the in Fig. S4 (SI).
Fig. 2.
(a, b, c) Initial and (d) final (after 40 ns of MD simulation) structures of the POPC-lutein bilayer. Initially, six lutein molecules are placed close to the bilayer surface. (a) View along the y-axis; (b) vertical cross section through the bilayer, to show the location of the lutein rings relative to the bilayer surface better; (c) view along the x-axis; in (d) only the POPC head groups are shown and the lutein molecules that are analysed are numbered as in the text. The atoms are represented in standard colours, except for the lutein carbon atoms which are dark blue. Water and lipid nonpolar hydrogen atoms are removed to show the details of the systems better.
2.1.2. Simulation Parameters
MD simulations of the bilayers were carried out in the NPT ensemble at the physiological temperature of 310 K (37 °C) and pressure of 1 atm with the GROMACS software package [38]. The phase transition temperature of the POPC bilayer is −2 °C thus at 37 °C the bilayer was in the liquid-crystalline phase (Lα). The temperatures of the solute and solvent were controlled independently by the Nosé-Hoover method [39,40] with a relaxation time of 0.6 ps. Pressure was controlled anisotropically by the Parrinello-Rahman method [41] with a relaxation time of 1.0 ps. The linear constraint solver (LINCS) algorithm [42] was used to preserve the length of any covalent bond with a hydrogen atom, and the time step was set to 2 fs. The van der Waals interactions were cut off at 1.2 nm. The long-range electrostatic interactions were evaluated using the particle-mesh Ewald summation method with a β-spline interpolation order of 5 and direct sum tolerance of 10−6 [43]. For the real space, a cut-off of 1.2 nm, three-dimensional periodic boundary conditions, and the usual minimum image convention were used [43]. The list of nonbonded pairs was updated every five time steps.
The system1 and 3 were MD simulated for 40 and 100 ns, respectively, and the systems 2 and 4 for 20 ns. The simulation time depended on the effectiveness of lutein full intercalation into the bilayer—each simulation was carried out until the intercalation criterion was satisfied by each of the lutein molecules in the system (cf. Introduction) but not shorter than 20 ns. In systems 1 and 2, the initial 1.5-ns trajectories were recorded every 2 fs and the remaining fragments of the trajectories (1.5–40 ns, and 1.5–20 ns), every 100 fs. The whole trajectory of the system3 was recorded every 100 fs. In the system4 none of the lutein molecules entered the bilayer so it was not analysed further. Dense trajectory samplings were necessary to capture formation of relatively short-lived hydrogen bonds (H-bond) between the hydroxyl group of the lutein ionone ring and POPC polar groups as well as water molecules in the early stages of lutein intercalation into the bilayer, while conserving the reasonable resolution during the whole in silico experiment.
2.2. Umbrella Sampling
To calculate the energy profile for a lutein molecule translocation from the water phase into the bilayer and across it, the umbrella sampling (US) method [[44], [45], [46]] was used. From the profile, the heights of the energy barriers for the process and the free energy gain of transferring the molecule from the water to the lipid phase were read. The system for US calculations consisted of the hydrated POPC bilayer built of 128 molecules hydrated with 45 H2O/POPC and a single lutein molecule. Initially, the molecule was placed in the bilayer water phase perpendicular to the bilayer surface.
To determine the starting configurations (windows) for US simulations, first the lutein molecule was pulled towards the bilayer interface using steered MD simulation [47]. The spring, with the spring constant of 250 kJ/(mol·nm2), was applied to the centre-of-mass (CM) of the lutein (Fig. 1a). The molecule was pulled along the bilayer normal (z-axis) with either its β or ε ring facing the bilayer. The pulling rate was 0.0001 nm/ps (1 nm/10 ns). When the CM of the lutein ring reached the POPC phosphate group region, to determine the remaining starting windows unbiased MD simulation was used where lutein intercalation into the bilayer proceeded spontaneously.
From the trajectories generated in MD simulations (steered and unbiased), 14 snapshots for lutein intercalation from the β ring side (β-intercalation) and 15 snapshots for the intercalation from the ε ring side (ε-intercalation) were taken at different bilayer depths as starting windows for US simulations. As histograms in Fig. S5 lower panel (SI) indicate the starting windows were chosen such as to uniformly cover all bilayer depths. The first window was located in the water phase. In each window, 300-ns restrained MD simulation was carried out. The spring constant of 100 kJ/(mol·nm2) restricted the vertical motion of the lutein molecule but did not restrain its lateral and rotational motion. In the case of the ε-intercalation, to improve sampling in the window at the depth of about 2.5 nm (near the barrier), a much higher spring constant of 1000 kJ/(mol·nm2) was used. The initial 10 ns of each restricted MD simulation were treated as the equilibration period and excluded from energy analyses.
The weighted histogram analysis method (WHAM) [48] provided by GROMACS gmx wham tool was used to confirm the correctness of sampling and to analyse the free energy change associated with lutein intercalation into the bilayer. Bayesian bootstrap analysis and 500 bootstrap profiles (Fig. S5 upper panel, SI) were used to calculate the average energy profile. Error bars are represented as mean square displacement of the obtained bootstrap profiles.
3. Results
3.1. Classic Molecular Dynamics Simulation – Spontaneous Lutein Intercalation
As it is described in Methods, four POPC bilayer systems, each containing six lutein molecules, were built (Fig. 2a) and MD simulated for 20–100 ns depending on the time of lutein full intercalation into the bilayer. Of the 24 lutein molecules initially placed near the surfaces of the bilayers, twelve faced the bilayer with their β rings and twelve with their ε rings. Altogether, twelve molecules either fully or partially spontaneously inserted into the bilayer (50%) – two from the ε ring side (~17%) and ten from the β ring side (~80%). The remaining molecules almost instantly aggregated in the water phase and stayed there as aggregates during the whole MD simulation time (Fig. S6, SI). Aggregation of lutein molecules in the water phase was demonstrated experimentally [17]. Of the three aggregates formed in this study one, containing 3 molecules, is of an H type (parallel packing), one, containing 2 molecules, is of a J type (antiparallel packing) and one, containing 3 molecules, is of a mixed type. The names of the aggregate types are in accordance with [17]. Of those molecules which intercalated fully, five (four from the β ring side and one from the ε ring side, Fig. 1a) were analysed in details to reveal subsequent steps of the intercalation process.
The initial surface area per POPC in the bilayer without lutein molecules was 62.89 ± 0.36 Å2. The experimentally determined values of the surface area/POPC are in the range between 63 and 66 Å2, e.g. [49,50]. The final surface area per lipid in the bilayer with one lutein molecule was 62.03 ± 0.41 Å2. The final area is slightly smaller than the initial one—this could be attributed the lutein condensing effect similar to that of cholesterol, e.g. [51], however, the effect is too small to really indicate condensation.
3.1.1. Hydrogen Bonds of the Monohydroxylated Ionone Ring in the First Bilayer Leaflet
In this section, H-bonds formed between the lutein hydroxyl group (OH-LUT) of the inserting ionone ring and POPC acceptor groups as well as water molecules during the initial 1.5 ns of the insertion process are analysed. To identify H-bonds, simple geometrical criteria are used where an H-bond donor (D) and acceptor (A) form an H-bond (Fig. 4) when the D–A distance is ≤0.35 nm and the angle between the D–A vector and the D-H bond is ≤30°. The limiting values for the distance and the angle are obtained from the relevant distributions. Time profiles of the H-bonds made by two lutein molecules of the system1 (mol1 and mol2, β-intercalation; 40 ns MD simulation), and one lutein molecule of the system2 (mol3, ε-intercalation; 20 ns MD simulation) with water and/or POPC oxygen atoms are shown in Fig. 3. In the subsequent steps of the intercalation process, the OH-LUT group of an inserting lutein molecule can be H-bonded in different configurations with one, two or three and occasionally with four water molecules and some of the POPC oxygen atoms (Figs. 3 and 4). As can be seen in Fig. 3, the OH-LUT⋯POPC H-bonds are made predominantly with the non-esterified (Op, a collective name for O13 and O14) but also with the esterified (Oe, a collective name for O11 and O12) phosphate oxygen atoms (Fig. 1b) as well as the glycerol backbone (Og, a collective name for O21 and O31) and the carbonyl (Oc, a collective name for O22 and O32) oxygen atoms (Fig. 1b); but there is no clear pattern in the selection of the H-bonding partner or time when such a bond is formed or broken. Most of OH-LUT⋯POPC direct H-bonds (Fig. 4) are short-lived (<10 ps) however, some of their lifetimes are longer than 100 ps. One has to bear in mind that OH-LUT⋯POPC and OH-LUT⋯H2O H-bonds are formed transiently by a moving lutein molecule, so by nature, they cannot be long-lived.
Fig. 4.

Snapshots of the time course of direct OH-LUT⋯POPC H-bonds. The atoms are represented in standard colours and the nonpolar hydrogen atoms have been omitted. In these H-bonds, the POPC Op, Oe, and Og atoms are engaged. The time in the upper left corner indicates the staring time of the course, the times in the lower left corners indicate the lapse time.
Fig. 3.
Time profiles of H-bonds formation of the intercalating rings of lutein molecules 1, 2 and 3 with water (water) and with the POPC oxygen atoms (Op, Oe, Og, Oc; for symbol explanation, see text) in the first bilayer leaflet (violet) during the initial 1500 ps of MD simulation with dense sampling (2- or 100-fs steps). The columns 1 and 2 represent intercalation of a lutein molecule with its β ring; the column 3 with its ε ring. H and V label the final position of the lutein molecule – H stands for horizontal, V for vertical.
The diagrams in Fig. 3 indicate that OH-LUT makes more H-bonds with water molecules than with POPC oxygen atoms, however; a POPC oxygen atom and a OH-LUT can be bridged by a water molecule [52] that is simultaneously H-bonded to both, as is illustrated in Fig. 5.
Fig. 5.

Snapshots of the time course of direct and water mediated OH-LUT⋯POPC H-bonds (water bridges). The atoms are represented in standard colours and the nonpolar hydrogen atoms have been omitted. In the water mediated H-bonds, the POPC Op and Oc atoms are engaged. The time in the upper left corner indicates the staring time of the course, the times in the lower left corners indicate the lapse time.
Time profiles of H-bonds formed by the entering lutein ring with water and POPC oxygen atoms during the whole intercalation processes of the analysed five lutein molecules are shown in Fig. 6. Due to the increasing depth of the ring insertion into the bilayer core, H-bonds made between OH-LUT and water and/or POPC in the first bilayer leaflet get broken (Fig. S7, SI). The time after which all these H-bonds are definitely broken ranges between ~2 and ~40 ns (Fig. 6), except for mol1, which inserts into the bilayer in the horizontal orientation and remains H-bonded to water molecules during the whole simulation time.
Fig. 6.
Time profiles of H-bonds formation of the intercalating rings of lutein molecules 1–5 with water (water) and with the POPC oxygen atoms (Op, Oe, Og, Oc) in both bilayer leaflets during the whole intercalation process (lasting from 20 to 100 ns). H-bonds made in the first leaflet are marked violet and those in the second leaflet are marked red. Four lutein molecules intercalated with their β rings and one with its ε ring. H and V label the final position of the lutein molecule – H stands for horizontal, V for vertical.
3.1.2. Desolvation of the Isoprenoid Chain
Initially, the lutein molecules are placed in the bilayer water phase. Since the lutein isoprenoid chain is nonpolar, water molecules in its neighbourhood form a clathrate around it (Fig. S8, SI). With increasing depth of lutein insertion into the bilayer, the water molecules forming the clathrate gradually dissociate concurrently with the change of the chain's environment from polar to nonpolar (Fig. 7). The chain desolvation process is depicted in Fig. 7 and in movie (movie, SI) as the diminishing and eventual disappearance of the first and second peaks in the radial distribution function (RDF) of the water oxygen atoms (Ow) relative to the lutein central chain atom, C15 (Fig. 1a). When C15 reaches the PC phosphate groups region the number of water molecules in the first and the second hydration zones of C15 decreases from 1.5 to ~1. When C15 reaches the PC carbonyl groups region the number of water molecules decreases to ~0.7. With deeper insertion of C15, the number of hydrating water molecules decreases to zero. Fig. 7 and the second column of Figs. 3 and 6 relate to the same lutein molecule – Figs. 3 and 6 show the history of H-bonding of its polar group and Fig. 7 shows desolvation of its nonpolar chain.
Fig. 7.
Desolvation of the lutein C15 atom, illustrated by the diminishing and eventual disappearance of the first and second peaks in the Ow-C15 RDF, concomitant with lutein intercalation into the bilayer. Snapshots of the intercalation process are on the left and RDFs are on the right. Only the POPC head groups are shown. The atoms are represented in standard colours, C15 atom is marked red, the water layer is shadow blue, and the hydrophobic core is shadow yellow. The nonpolar hydrogen atoms have been omitted. The whole time course is shown in movie (movie, SI).
3.1.3. Bilayer Lipids Rearrangement and Final Lutein Orientation
Spontaneous intercalation of a lutein molecule into the lipid bilayer is facilitated by perpetual motion of all bilayer components and by the geometrical features of the molecule that is relatively slim, rigid and long. Nevertheless, the intercalating molecule must enforce some rearrangement of the bilayer lipids to create enough space to move across the bilayer. However, since the intercalation process is spontaneous it causes only a small deformation of the bilayer surface. Even though the statistics is poor (time average over the last 5 ns of MD simulation), this deformation can be seen as a slight down-shift of POPC molecules close to the intercalated lutein molecule (Fig. S9, lower panel, SI). A POPC molecule is considered close when at least one atom of its head group is not further than 0.35 nm from any of the lutein atoms. Eight POPC molecules fulfilled this criterion for system shown in Fig. S9 (SI), so the ensemble average was over only eight molecules.
In general, lutein molecules translocate through the bilayer slightly tilted relative to the normal (Fig. 7, and movie, SI), and in general, their final positions are transmembrane but not strictly vertical (Fig. 2d and Fig. S4, SI) as they are tilted 31–45° relative to the normal (Fig. S10, SI). The tilting may result from nonlinearity of the lutein molecule as its polyene chain is distorted and assumes an S-shape [53,54]. It may also result from local mismatch between the width of the hydrophobic core of the POPC bilayer and an effective length of the lutein molecule. The average bilayer width measured as the distance between the average positions of the glycerol backbones in both bilayer leaflets is 3.03 ± 0.05 nm; the average apparent length of the lutein molecule (C3-C3′ distance) in the bilayer is 2.83 ± 0.05 nm, however, locally and temporarily the bilayer width may be smaller than the lutein effective length.
Snapshots of the lutein final positions in the bilayers are presented in Fig. S4 and also in Figs. S9, S11, S14 (SI). As shown, not all lutein molecules are in the perpendicular position. One out of five lutein molecules (mol1) analysed in this study orients itself horizontally at the first bilayer interface and remains in this orientation for the whole simulation time. Final horizontal orientation of a lutein molecule in PC bilayers was observed experimentally [25,55] and in our previous MD simulation study [19].
3.1.4. Hydrogen Bonds of the Monohydroxylated Ionone Ring in the Second Bilayer Leaflet
When the inserting ring reaches the depth corresponding to the middle of the acyl chain region of the first bilayer leaflet, all its H-bonds with water molecules and POPC acceptor groups formed in the first bilayer interfacial region are already broken (Fig. S7, SI). As the polar ring in the nonpolar environment and a water-exposed portion of the isoprenoid chain are in energetically unfavourable states, the lutein molecule proceeds further into the bilayer to counteract this. On entering the other bilayer leaflet, the lutein molecule starts rearranging PC molecules to create space enabling itself to move forward. This extra space facilitates diffusion of water molecules deeper into the leaflet. These water molecules are attracted by OH-LUT and form H-bonds with it. This is apparent in Fig. S11 (SI) where chains of mutually H-bonded water molecules anchor the lutein rings in the bilayer interfacial regions. The effect of a deeper local penetration of the bilayer by water is too small to be visible in the Op-Ow RDF presented in Fig. S9 (SI). OH-LUT also makes H-bonds with POPC oxygen atoms (Fig. 6), however, as Fig. 6 indicates, H-bonds with water molecules are made generally sooner due to higher mobility and higher motional freedom of water molecules compared to than with POPC head groups.
The time after which OH-LUT starts forming H-bonds in the opposite bilayer leaflet, ranges between ~7.5–15 and ~95 ns (Fig. 6), thus ~10 ns might be considered a minimum time needed for the lutein entering ring to cross the bilayer hydrophobic core.
3.1.5. Hydrogen Bonds of the Second Monohydroxylated Ionone Ring
For most of the MD simulation time, the second lutein ionone ring is fully hydrated and its OH-LUT group forms multiple H-bonds with water molecules (Fig. 8). About the time when the first (entering) ring starts forming H-bonds in the second bilayer leaflet or even slightly earlier, the second ring starts making H-bonds with POPC oxygen atoms in the first leaflet; this time ranges between ~7.5 and ~40 ns (Fig. 8). Once both lutein OH groups make H-bonds with water and POPC oxygen atoms in one or both bilayer interfacial regions, the lutein molecule is judged to be fully intercalated into the bilayer. The lifetimes of the second ionone ring OH-LUT⋯H2O and OH-LUT⋯POPC H-bonds range from <10 ps to >100 ps, as in the case of the first ring. There is no significant difference between the β and the ε ring in formation H-bonds with POPC and water.
Fig. 8.
Time profiles of H-bonds formation of the second (not-intercalating) rings of lutein molecules 1–5 with water (water) and with the POPC oxygen atoms (Op, Oe, Og, Oc) in the first bilayer leaflet (violet) during the whole intercalation process (lasting from 20 to 100 ns). Four lutein molecules intercalated with their β rings and one with its ε ring (first, intercalating rings). H and V label the final position of the lutein molecule – H stands for horizontal, V for vertical.
3.2. Energy Calculation
3.2.1. Umbrella Sampling
Profiles of the free energy of transfer of a whole lutein molecule from the water phase into the bilayer as a function of the relative distance between the lutein centre-of-mass (CM) and the bilayer centre, obtained from umbrella sampling, US, simulation are presented in Fig. 9. Two energy profiles are calculated – for the molecule intercalating from the β ring side (β-intercalation) and from the ε ring side (ε-intercalation). The free energies of transfer calculated from the profiles are −19.5 ± 1.0 kcal/mol for β-intercalation and −17.9 ± 1.1 kcal/mol for ε-intercalation (Table 1).
Fig. 9.
Schematic presentation of the initial state of US simulation (upper panel), free energy profiles obtained from US simulations (central panels), and the front-on view of the lutein molecules from the side of their penetrating rings (bottom panel). The arrow in the upper panel indicates the direction of the lutein trans-membrane motion and its length indicates the relative distance between the lutein centre-of-mass (CM) and the bilayer centre (Distance). As the molecule moves across the bilayer, the distance decreases and is zero when the lutein CM reaches the bilayer centre. The zero of the x-axes in the central panels corresponds to such a position of the lutein CM. With the decreasing distance, the energy of the system decreases and its minimum is set to zero, thus the profiles should be read from right to left to follow the direction of the arrow. The energy error bars are mean square displacement calculated by the WHAM method [48]. The plateau and the shoulder in the profiles do not represent barriers (for explanation, see text).
Table 1.
Positions and heights of the energy barriers to transfer a lutein molecule from the water phase into the POPC bilayer and across it.
| Location [nm] | Height [kcal/mol] | Energy gain [kcal/mol] | |
|---|---|---|---|
| β ring | ~2.9 | 0.2 ± 1.4 | −19.5 ± 1.0 |
| ε ring | ~2.5 | 2.2 ± 1.6 | −17.9 ± 1.1 |
The lutein molecule intercalated the bilayer from the β ring side (β ring) and the ε ring side (ε ring). The location of the barrier is determined for the lutein CM relative to the bilayer centre (Location). (Height) denotes the relative height of the barrier. The energy of the system reaches minimum on full cross-bilayer lutein intercalation (Energy gain).
Each of the profiles has one maximum and one plateau or shoulder. The maximum indicates a barrier to the translocation process, the plateau (shoulder) arises from rotation of the lutein molecule inside some of the US windows. At some bilayer depth (certain US windows), the molecule rotates about its CM and transiently orients horizontally in the hydrophobic bilayer core (Fig. S12, SI). Such an orientation is energetically disadvantageous so, for that depth of lutein insertion the energy of the system does not decrease.
For β-intercalation, the maximum in the profile is flat (Fig. 9) indicating a low energy barrier to the translocation process. The barrier is encountered by the intercalating ring; however, the barrier position in the profile (Fig. 9) is given for the lutein CM relative to the centre of the bilayer. The position of the barrier and its height are given in Table 1. The barrier coincides with the POPC glycerol backbones region of the first bilayer leaflet. This can be seen in Fig. S13 β-ring (SI) where time profiles of the positions along the bilayer normal of the lutein C3 atom and CM together with the average positions of the POPC phosphate and glycerol moieties (Fig. 1) for the maximum in the profile in Fig. 9 β-ring, are shown. The position of the barrier can also be anticipated from the movie (SI) where the inward movement of the lutein ring is impeded at the border between the polar interface and the hydrophobic core of the bilayer, for a considerable time.
The errors in the barrier heights are significant (Table 1), nevertheless it is apparent that the energy barrier to the translocation process for ε-intercalation is ~2 kcal/mol higher than for β-intercalation (Table 1). The barrier also coincides with the region of the POPC glycerol backbones which can be seen in Fig. S13 ε-ring (SI).
4. Discussion
Based on the chemical structure of the lutein molecule (Fig. 1a), one can expect that it readily partitions into the membrane and its final location there should be such that its both monohydroxylated ionone rings are placed at the bilayer interface and the isoprenoid chain is located in the bilayer nonpolar core. One can also predict that the structure of the bilayer imposes some obstacles to lutein cross-bilayer translocation. However, neither the heights of the energy barriers imposed by the bilayer on lutein insertion nor the timescale of the insertion process can be predicted from the structure.
This MD simulation study fully confirms rational expectations and predictions concerning partitioning and location of lutein in the membrane. But what is more important, it provides information about the dynamics and energetics of the intercalation process that is beyond predictions.
The lutein membrane insertion begins with random approaching the bilayer interface by one of the lutein ionone rings followed by formation of semi-stable direct and water mediated H-bonds of its OH group with POPC acceptor groups and water molecules at the bilayer interface. The main driving force that pushes the ring further into the bilayer core are unfavourable interactions between polar solvent and the large nonpolar part of the lutein molecule. However, further advancement of lutein into the bilayer core creates inferior conditions because of necessary dehydration of the OH-LUT group, unfavourable interactions between OH-LUT and the hydrophobic bilayer core, and local rearrangement of the relatively stiff POPC glycerol backbones. All these contribute to the energy barrier to lutein insertion. The barriers for β- and ε-intercalations are placed in the same bilayer region but that for ε-intercalation is significantly higher than that for β-intercalation (Fig. 9, Table 1). Nevertheless, both barriers can be freely overcome under physiological temperatures due to thermal activation. Thus, lutein intercalation from water into the lipid bilayer is spontaneous and the system gains ~18 kcal/mol on full intercalation. A higher barrier for the ε than β ring insertion makes the lutein ε-intercalation less likely, which is in agreement with other results of the present MD simulations indicating that spontaneous lutein β-intercalation is four times more probable than ε-intercalation.
Different heights of the barriers for β- and ε-intercalation are evidently caused by different preferential orientations of the β and ε rings resulting from sp2 hybridisation of the C6 carbon atom and sp3 hybridisation of the C6′ carbon atom (Fig. 1a). In the energy optimised lutein structure, the rings are oriented ~90° relative to each other [37]. The difference in the orientations of the rings is seen in Fig. 9 lower panel – the β ring is almost in the plane of the rigid polyene chain whereas the ε ring is almost perpendicular to the chain. Indeed, the average angle between the ring plane and the C6–C7 bond in lutein structures generated in MD simulations is 174° for the β ring and 112° for the ε ring. Moreover, the positions of the methyl groups on the ε ring (Fig. 9, lower panel) enhance the effect of the perpendicular ring orientation by making the cross-sectional area of the molecule larger. The ε ring is thus a greater obstacle to lutein penetration across any rigid region of the bilayer than the β ring.
The rapid decrease of the system's energy after passing the barrier implies that POPC acyl chains do not impose much resistance to the lutein motion which might be attributed to their great flexibility. Thus, the energy barrier to the lutein translocation across the bilayer is at the border of the polar interfacial region and the bilayer nonpolar core of the first leaflet. In US simulation, the lutein intercalating ring is not forced to move across the glycerol backbones region of the second leaflet because once it reaches the region, it forms H-bonds with carbonyl oxygen atoms and water molecules that stabilise its position there and lowers energy of the system. Thus, lutein US simulation is completed when its both monohydroxylated ionone rings make H-bonds with POPC oxygen atoms and water molecules in the bilayer interfacial regions as is shown in Fig. S11 (SI).
In four out of five cases of lutein spontaneous insertion into the bilayer analysed in detail, the final position of the molecule is cross-bilayer (vertical orientation) (Figs. S4 and S11, SI). The mutual arrangement of POPC and lutein in the final state is shown in Fig. S9 upper left (SI). In one case, the molecule is oriented horizontally below the interfacial region of the first leaflet with both rings located in the same bilayer interface. Such a position is stabilised by H-bonds involving both lutein OH-LUT groups, POPC oxygen atoms and water molecules, as is shown in Fig. S14 (SI). Our computer simulation [19] and experimental [8,25,26,55,56] studies also indicated that the horizontal lutein orientation is possible but that the cross-bilayer one is more favourable.
Whereas peptide insertion into the lipid bilayer has been widely studied using experimental and computational methods e.g. [[20], [21], [22], [23], [24]], there have been no such studies for a relatively rigid, long and mainly nonpolar molecule with two terminal polar groups as is lutein. This study fills the gap and demonstrates that the course and timescale of lutein insertion and translocation across the bilayer are vastly different from those of peptides. Membrane partitioning of transmembrane helical peptides generally requires partitioning-folding coupling as energetic cost of partitioning a peptide bond into the bilayer is high [16]. Peptide partitioning is mainly a three-stage process in which the peptide adsorbs at the membrane interface, folds into helix, and inserts to span the bilayer [20] however, peptide adsorption and folding at the interface are not precondition for insertion [23]. In the adsorption and folding pathway, insertion of the peptide requires change of its orientation from horizontal to tilted or vertical [22]. This is a very slow process undergoing on μs to ms timescale [20,22]. The barriers for insertion-rotation are ~11 to ~15 kcal/mol for 16–27 aa helical peptides [15,20,22,57]. Thus the insertion time and barrier for peptides are much greater than for lutein. These might be attributed among others to peptides larger sizes, various polarities of the amino acids in their sequences, local unfolding, and internal flexibility. Compared to peptides, insertion of lutein is much more straightforward as it does not involve folding and, in most cases, rotation. Energetics associated with lutein rotation in the bilayer was analysed in Ref. [26].
The results obtained in this study (positions of the barriers, their heights, intercalation timescales) are consistent with predictions and with experimental results showing that the octanol/water partition coefficient of lutein is very high and between 6.84 and 14.8 [[58], [59], [60]]. Thus lutein, having a high affinity for lipophilic environment is supposed to spontaneously and fast intercalate into the lipid bilayer. As lutein is a member of the xanthophyll family, the details revealed in this study are most likely applicable to other xanthophylls as well as to molecules of a similar chemical structure.
Based on the result of this study that lutein ε-intercalation into the membrane is less probable than β-intercalation, one can draw an interesting and rather unexpected conclusion that the β and ε rings can be found mainly in different leaflets of the membrane. This might have far reaching biological consequences as lutein is widely present in membranes of both plant and animal kingdoms. In humans, lutein plays a significant protective role against photoinduced damage to the lens and the retina [61]. Also in plants, lutein plays a vital photoprotective role [2]. But similar roles are played by another xanthophyll, zeaxanthin, which has two β rings and not the β and the ε ring, as lutein has. However, before one could speculate on the possible biological role of the ε ring position in the biomembrane, the predicted non-equivalent positions of the lutein β and ε rings in the bilayer have to be experimentally evidenced.
A great support to our results of the preferential lutein β-intercalation into the bilayer is very recent docking computations which revealed that the lutein ε-ring fits much better than the β-ring into the active site of the enzyme that catalyses the conversion of lutein to meso-zeaxanthin [28] and also into the asymmetric cavity of the lutein binding protein [29]. In both proteins, the β-ring binds outside the cavity. The isomerohydrolase enzyme RPE65 which putatively converts lutein to meso-zeaxanthin is a membrane associated protein in which reactants and products likely enter and leave the active site through a hydrophobic tunnel which is thought to open into the lipid membrane [62]. Thus, the lutein ε-ring that is located on a different side of the membrane than the β-ring is readily accessible to the enzyme.
Acknowledgments
Acknowledgments
This work was inspired by discussions with K. Strzalka. MPG thanks W. Gruszecki for a constructive discussion. Faculty of Biochemistry, Biophysics and Biotechnology of Jagiellonian University is a partner of the Leading National Research Centre (KNOW) supported by the Ministry of Science and Higher Education. This research was supported in part by PL-Grid Infrastructure.
Declarations of Interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.04.001.
Appendix A. Supplementary data
Desolvation of the lutein C15 atom concomitant with lutein intercalation into the POPC bilayer. Desolvation is illustrated with the diminishing and eventual disappearance of the maxima in the radial distribution function (RDF) of the water molecules relative to the C15 lutein atom (displayed on the left hand side) as the water molecules are not shown in the movie for clarity. The course of lutein intercalation into the bilayer is displayed in the lower panel on the right hand side. The actual position of the C15 lutein atom is displayed in the upper panel on the right hand side. The pink horizontal lines in the panel indicate the average positions of the POPC phosphorous atoms in both bilayer leaflets at a given time.
Supplementary material
References
- 1.Moise A.R., Al-Babili S., Wurtzel E.T. Mechanistic aspects of carotenoid biosynthesis. Chem Rev. 2014;114:164–193. doi: 10.1021/cr400106y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballottari M., Mozzo M., Girardon J., Hienerwadel R., Bassi R. Chlorophyll triplet quenching and photoprotection in the higher plant monomeric antenna protein Lhcb5. J Phys Chem B. 2013;117:11337–11348. doi: 10.1021/jp402977y. [DOI] [PubMed] [Google Scholar]
- 3.Dall'Osto L., Lico C., Alric J., Giuliano G., Havaux M., Bassi R. Lutein is needed for efficient chlorophyll triplet quenching in the major LHCII antenna complex of higher plants and effective photoprotection in vivo under strong light. BMC Plant Biol. 2006;6:32. doi: 10.1186/1471-2229-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granado F., Olmedilla B., Blanco I. Nutritional and clinical relevance of lutein in human health. Br J Nutr. 2003;90:487–503. doi: 10.1079/bjn2003927. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein P.S., Khachik F., Carvalho L.S., Muir G.J., Zhao D.Y., Katz N.B. Identification and quantitation of carotenoids and their metabolites in the tissues of the human eye. Exp Eye Res. 2001;72:215–223. doi: 10.1006/exer.2000.0954. [DOI] [PubMed] [Google Scholar]
- 6.Kijlstra A., Tian Y., Kelly E.R., Berendschot T.T.J.M. Lutein: more than just a filter for blue light. Prog Retin Eye Res. 2012;31:303–315. doi: 10.1016/j.preteyeres.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Koushan K., Rusovici R., Li W.H., Ferguson L.R., Chalam K.V. The role of lutein in eye-related disease. Nutrients. 2013;5:1823–1839. doi: 10.3390/nu5051823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sujak A., Gabrielska J., Grudzinski W., Borc R., Mazurek P., Gruszecki W.I. Lutein and zeaxanthin as protectors of lipid membranes against oxidative damage: the structural aspects. Arch Biochem Biophys. 1999;371:301–307. doi: 10.1006/abbi.1999.1437. [DOI] [PubMed] [Google Scholar]
- 9.Sindhu E.R., Preethi K.C., Kuttan R. Antioxidant activity of carotenoid lutein in vitro and in vivo. Indian J Exp Biol. 2010;48:843–848. [PubMed] [Google Scholar]
- 10.Lancrajan I., Diehl H.A., Socaciu C., Engelke M., Zorn-Kruppa M. Carotenoid incorporation into natural membranes from artificial carriers: liposomes and beta-cyclodextrins. Chem Phys Lipids. 2001;112:1–10. doi: 10.1016/s0009-3084(01)00138-4. [DOI] [PubMed] [Google Scholar]
- 11.Landrum J.T., Bone R.A., Moore L.L., Gomez C.M. Analysis of zeaxanthin distribution within individual human retinas. Methods Enzymol. 1999;299:457–467. doi: 10.1016/s0076-6879(99)99043-2. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Sola M.A., Rodríguez-Concepcion M. The arabidopsis book. American Society of Plant Biologists; 2012. Carotenoid biosynthesis in arabidopsis: a colorful pathway. (pp e0158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Holde K.E., Johnson W.C., Ho P.S. Pearson/Prentice Hall; 2006. Principles of Physical Biochemistry. [Google Scholar]
- 14.Shinoda W. Permeability across lipid membranes. Biochim Biophys Acta-Biomem. 2016;1858:2254–2265. doi: 10.1016/j.bbamem.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 15.Pourmousa M., Wong-ekkabut J., Patra M., Karttunen M. Molecular dynamic studies of transportan interacting with a DPPC lipid bilayer. J Phys Chem B. 2013;117:230–241. doi: 10.1021/jp310255r. [DOI] [PubMed] [Google Scholar]
- 16.White S.H. How hydrogen bonds shape membrane protein structure. Adv Protein Chem. 2005;72:157–172. doi: 10.1016/S0065-3233(05)72006-4. [DOI] [PubMed] [Google Scholar]
- 17.Polyakov N.E., Magyar A., Kispert L.D. Photochemical and optical properties of water-soluble xanthophyll antioxidants: aggregation vs complexation. J Phys Chem B. 2013;117:10173–10182. doi: 10.1021/jp4062708. [DOI] [PubMed] [Google Scholar]
- 18.Sundaralingam M. Discussion paper: molecular structures and conformations of the phospholipids and sphingomyelins. Ann N Y Acad Sci. 1972;195:324–355. [PubMed] [Google Scholar]
- 19.Pasenkiewicz-Gierula M., Baczynski K., Murzyn K., Markiewicz M. Orientation of lutein in a lipid bilayer - revisited. Acta Biochim Pol. 2012;59:115–118. [PubMed] [Google Scholar]
- 20.Ulmschneider M.B., Doux J.P.F., Killian J.A., Smith J.C., Ulmschneider J.P. Mechanism and kinetics of peptide partitioning into membranes from all-atom simulations of thermostable peptides. J Am Chem Soc. 2010;132:3452–3460. doi: 10.1021/ja909347x. [DOI] [PubMed] [Google Scholar]
- 21.Yue T.T., Sun M.B., Zhang S., Ren H., Ge B.S., Huang F. How transmembrane peptides insert and orientate in biomembranes: a combined experimental and simulation study. Phys Chem Chem Phys. 2016;18:17483–17494. doi: 10.1039/c6cp01133k. [DOI] [PubMed] [Google Scholar]
- 22.Irudayam S.J., Pobandt T., Berkowitz M.L. Free energy barrier for melittin reorientation from a membrane-bound state to a transmembrane state. J Phys Chem B. 2013;117:13457–13463. doi: 10.1021/jp406328d. [DOI] [PubMed] [Google Scholar]
- 23.Nymeyer H., Woolf T.B., Garcia A.E. Folding is not required for bilayer insertion: replica exchange simulations of an alpha-helical peptide with an explicit lipid bilayer. Proteins. 2005;59:783–790. doi: 10.1002/prot.20460. [DOI] [PubMed] [Google Scholar]
- 24.Babakhani A., Gorfe A.A., Kim J.E., McCammon J.A. Thermodynamics of peptide insertion and aggregation in a lipid bilayer. J Phys Chem B. 2008;112:10528–10534. doi: 10.1021/jp804710v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruszecki W.I., Strzalka K. Carotenoids as modulators of lipid membrane physical properties. Biochim Biophys Acta-Mol Basis Dis. 2005;1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Grudzinski W., Nierzwicki L., Welc R., Reszczynska E., Luchowski R., Czub J. Localization and orientation of xanthophylls in a lipid bilayer. Sci Rep. 2017;7:9619. doi: 10.1038/s41598-017-10183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson Q.R., Mostofian B., Gomez G.F., Smith J.C., Cheng X.L. Effects of carotenoids on lipid bilayers. Phys Chem Chem Phys. 2018;20:3795–3804. doi: 10.1039/c7cp07126d. [DOI] [PubMed] [Google Scholar]
- 28.Shyam R., Gorusupudi A., Nelson K., Horvath M.P., Bernstein P.S. RPE65 has an additional function as the lutein to meso-zeaxanthin isomerase in the vertebrate eye. P Natl Acad Sci USA. 2017;114:10882–10887. doi: 10.1073/pnas.1706332114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath M.P., George E.W., Tran Q.T., Baumgardner K., Zharov G., Lee S. Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein. Acta Crystallogr Sect F. 2016;72:609–618. doi: 10.1107/S2053230X16010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plesnar E., Subczynski W.K., Pasenkiewicz-Gierula M. Saturation with cholesterol increases vertical order and smoothes the surface of the phosphatidylcholine bilayer: a molecular simulation study. Biochim Biophys Acta-Biomem. 2012;1818:520–529. doi: 10.1016/j.bbamem.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 32.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 33.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Chem. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayo S.L., Olafson B.D., Goddard W.A. Dreiding - a generic force-field for molecular simulations. J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 35.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity - a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 36.Frisch M.J., Trucks G.W., Schlegel H.B. Gaussian, Inc; Wallingford CT: 2009. Gaussian 09, Revision B.01. [Google Scholar]
- 37.Landrum J.T., Chatfield D.C., Mebel A.M., Alvarez-Calderon F., Fernandez M.V. The conformation of end-groups is one determinant of carotenoid topology suitable for high fidelity molecular recognition: a study of beta- and epsilon-end-groups. Arch Biochem Biophys. 2010;493:169–174. doi: 10.1016/j.abb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 39.Nose S. A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys. 1984;81:511–519. [Google Scholar]
- 40.Hoover W. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 41.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182. [Google Scholar]
- 42.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 43.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577. [Google Scholar]
- 44.Patey G.N., Valleau J.P. Free energy of spheres with dipoles - Monte-Carlo with multistage sampling. Chem Phys Lett. 1973;21:297–300. [Google Scholar]
- 45.Torrie G.M., Valleau J.P. Monte Carlo free energy estimates using non-Boltzmann sampling: application to the sub-critical Lennard-Jones fluid. Chem Phys Lett. 1974;28:578–581. [Google Scholar]
- 46.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 47.Stepaniants S., Izrailev S., Schulten K. Extraction of lipids from phospholipid membranes by steered molecular dynamics. J Mol Model. 1997;3:473–475. [Google Scholar]
- 48.Kumar S., Bouzida D., Swendsen R.H., Kollman P.A., Rosenberg J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 49.Smaby J.M., Momsen M.M., Brockman H.L., Brown R.E. Phosphatidylcholine acyl unsaturation modulates the decrease in interfacial elasticity induced by cholesterol. Biophys J. 1997;73:1492–1505. doi: 10.1016/S0006-3495(97)78181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kucerka N., Nieh M.P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta-Biomem. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 51.Yin J.J., Subczynski W.K. Effects of lutein and cholesterol on alkyl chain bending in lipid bilayers: a pulse electron spin resonance spin labeling study. Biophys J. 1996;71:832–839. doi: 10.1016/S0006-3495(96)79284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasenkiewicz-Gierula M., Takaoka Y., Miyagawa H., Kitamura K., Kusumi A. Hydrogen bonding of water to phosphatidylcholine in the membrane as studied by a molecular dynamics simulation: location, geometry, and lipid−lipid bridging via hydrogen-bonded water. J Phys Chem A. 1997;101:3677–3691. [Google Scholar]
- 53.Qian P., Mizoguchi T., Fujii R., Hara K. Conformation analysis of carotenoids in the purple bacterium Rhodobium marinum based on NMR spectroscopy and AM1 calculation. J Chem Inf Comput Sci. 2002;42:1311–1319. doi: 10.1021/ci0255230. [DOI] [PubMed] [Google Scholar]
- 54.Fiedor L., Pilch M. Side methyl groups control the conformation and contribute to symmetry breaking of isoprenoid chromophores. Angew Chem Int Ed. 2018;57:6501–6506. doi: 10.1002/anie.201802094. [DOI] [PubMed] [Google Scholar]
- 55.Tan C., Xia S.Q., Xue J., Xie J.H., Feng B.A., Zhang X.M. Liposomes as vehicles for lutein: preparation, stability, liposomal membrane dynamics, and structure. J Agric Food Chem. 2013;61:8175–8184. doi: 10.1021/jf402085f. [DOI] [PubMed] [Google Scholar]
- 56.Sujak A., Mazurek P., Gruszecki W.I. Xanthophyll pigments lutein and zeaxanthin in lipid multibilayers formed with dimyristoylphosphatidylcholine. J Photochem Photobiol B. 2002;68:39–44. doi: 10.1016/s1011-1344(02)00330-5. [DOI] [PubMed] [Google Scholar]
- 57.Ulmschneider J.P., Smith J.C., White S.H., Ulmschneider M.B. In silico partitioning and Transmembrane insertion of hydrophobic peptides under equilibrium conditions. J Am Chem Soc. 2011;133:15487–15495. doi: 10.1021/ja204042f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dima S., Dima C. Protection of bioactive compounds. In: Apetrei C., editor. Frontiers in bioactive compounds. Bentham Science Publishers; 2016. pp. 255–301. [Google Scholar]
- 59.Reboul E., Abou L., Mikail C., Ghiringhelli O., Andre M., Portugal H. Lutein transport by Caco-2 TC-7 cells occurs partly by a facilitated process involving the scavenger receptor class B type I (SR-BI) Biochem J. 2005;387:455–461. doi: 10.1042/BJ20040554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.EPA Chemistry dashboard. 2018. https://comptox.epa.gov/dashboard/
- 61.Roberts J.E., Dennison J. The photobiology of lutein and zeaxanthin in the eye. Aust J Ophthalmol. 2015;2015 doi: 10.1155/2015/687173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiser P.D., Zhang J.Y., Badiee M., Li Q.J., Shi W.X., Sui X.W. Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nat Chem Biol. 2015;11:409. doi: 10.1038/nchembio.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Desolvation of the lutein C15 atom concomitant with lutein intercalation into the POPC bilayer. Desolvation is illustrated with the diminishing and eventual disappearance of the maxima in the radial distribution function (RDF) of the water molecules relative to the C15 lutein atom (displayed on the left hand side) as the water molecules are not shown in the movie for clarity. The course of lutein intercalation into the bilayer is displayed in the lower panel on the right hand side. The actual position of the C15 lutein atom is displayed in the upper panel on the right hand side. The pink horizontal lines in the panel indicate the average positions of the POPC phosphorous atoms in both bilayer leaflets at a given time.
Supplementary material