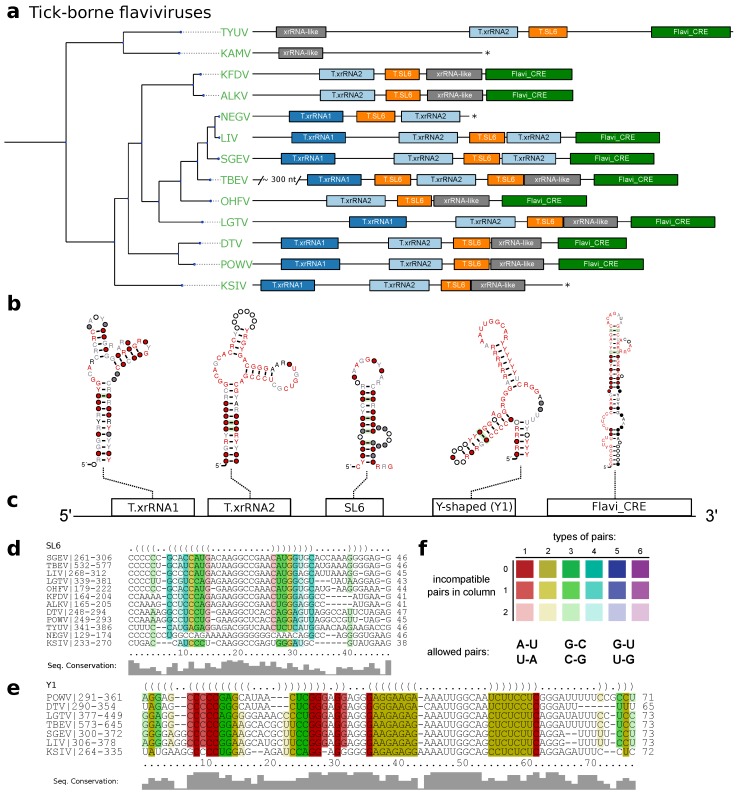
Figure 3.
(a) Annotated 3UTRs of TBFVs. The phylogenetic tree on the left has been computed from complete coding sequence nucleotide alignments and corresponds to the TBFV subtree in Figure 1. For each species with available 3UTR sequence a sketch of the 3UTR is drawn to scale next to the leaves of the tree. Colored boxes represent conserved RNA structural elements. Identifiers within the boxes indicate the CM which was used to infer homology at this position. Asterisks indicate incomplete 3UTR sequences. Species without available 3UTR are not shown. (b) Consensus structure plots of CM hits as calculated by mlocarna. (c) Schematic depiction of the common structural architecture of TBFV 3UTRs. (d,e) Structural alignments of elements SL6 and Y1. (f) RNAalifold coloring scheme for paired columns in alignments. Colors indicate the number of basepair combinations found in pair of columns. Fainter colors indicate that some sequences cannot form a base pair.