Abstract
Cardiovascular diseases (CVDs) have become the leading cause of disability and death worldwide, particularly in low- and middle-income countries. Hypertension, a major cause of CVD progression, is widely attributable to genetic, behavioral, and environmental risk factors. Among the genetic reasons, angiotensin II enzyme, produced as a result of abnormal functioning of the renin–angiotensin system, is reported as the foremost cause of hypertension. A cascade of genes, including those encoding for WNK kinases (WNK1 and WNK4), Bp1, Bp2, angiotensinogen, and other enzymes, is involved in the conversion of angiotensin I to angiotensin II. However, the angiotensin-converting enzyme (ACE) plays a crucial role in this pathway. Therefore, ACE could be a potential therapeutic target in regulating the conversion of angiotensin I to angiotensin II and eventually controlling hypertension. In this study, a molecular docking-based approach was utilized for identifying and evaluating potential inhibitors of ACE present in herbs, other natural sources, and synthetic sources, on the basis of these compounds’ binding affinities and other physicochemical features. In addition, the suitability of these inhibitors as drugs for biological systems, considering their adsorption, distribution, metabolism, and excretion (ADME), was predicted using Lipinski’s rule. In conclusion, our study provides a novel and clearer insight into the interaction properties of known putative inhibitors of ACE.
Keywords: angiotensin-converting enzyme, ligands, hypertension, molecular docking, drug designing
1. Introduction
Cardiovascular diseases (CVDs) refer to disorders of the heart and blood vessels, including coronary artery diseases (CAD) such as angina and myocardial infarction, that are very common and represent a major challenge to sustainable human development nowadays [1]. With the turn of the century, these diseases have become the leading cause of disability and death around the globe, particularly in developing countries. Global status reports have shown that approximately 12.3 million deaths (25.8% of all deaths) globally were caused by CVDs in 1990 and increased to 17.9 million deaths (32.1% of all deaths) in 2015 [2]. In the United States, 11%, 37%, 71%, and 85% of people are affected by CVDs between 20 and 40, 40 and 60, 60 and 80, and over 80 years of age, respectively [3]. Risk factors of CVDs include genetic, behavioral, and environmental factors, such as high blood pressure, smoking, diabetes, physical inactivity, obesity/overweight, high blood cholesterol, low socioeconomic status, and excessive alcohol. Among these, high blood pressure is the main risk factor for CVD deaths (13%), followed by the tobacco (9%), diabetes (6%), lack of exercise (6%), and obesity (5%), and the prevalence of risk factors fluctuates in various regions [4].
Hypertensive heart disease mostly known as hypertension, refers to a group of disorders including heart failure, ischemic heart disease, and left ventricular hypertrophy and is becoming a major cause of death associated with high blood pressure worldwide [5]. Several genes including WNK1, WNK4, Bp1, Bp2, AGT, and ACE were reported to be involved in hypertension [6]. Mutation in WNK1 and WNK4 genes could cause disturbances in the homeostasis of K+, salts, and pH level [7], whereas a mutation in AGT genes present on chromosome 1 results in an imbalance of angiotensinogen production, ultimately leading to hypertension [8]. It was revealed that ACE plays a key role in hypertension, its dysfunction being the most frequent cause of hypertension [9]. The most comon biological reason behind hypertension is the production of angiotensin II enzyme, which is produced from the conversion of angiotensin I by the action of a series of the enzymes [10]. Therefore, regulating the conversion of angiotensin I to angiotensin II could be an effective strategy to control hypertension.
Angiotensin-converting enzyme (ACE) is considered crucial in this pathway and has received considerable attention as a therapeutic target for controlling hypertension. Repressing ACE expression has been proved as an effective strategy in controlling hypertension, as its downregulation will inhibit the conversion of angiotensin I to angiotensin II [11]. In this study, the binding affinities of various natural, synthetic, and herbal inhibitors for active sites of ACE were predicted using the molecular docking approach, which is becoming an extremely important tool in drug design. Molecular docking is playing a major role in structure-based molecular biology and computer-based drug design. The molecular docking methodology can be utilized to demonstrate the cooperation between a small molecule and a protein at the nanoscale, which empowers us to describe the behavior of small particles in the binding site of the proteins and explain key biochemical processes. Furthermore, drug-likeness and compatibility with gastrointestinal and brain absorption were computed for all the inhibitors tested to evaluate their suitability as potential therapeutic agents and orally active drugs for the treatment of hypertension.
2. Materials and Methods
2.1. Physiochemical Properties
The physiochemical properties of human ACE were predicted using Protparam [12]. The Protparam tool works on the basis of the Edelhoch method [13], determining the weight value of instability with respect to 400 different dipeptides (DIWV) and the hydropathy values for extinction coefficients, instability index (II), and GRAVY value (grand average of hydropathy value).
2.2. Secondary Structure Predictions
The number of helix turns and coils was calculated using “Psipred” [14]. Psipred used two feed-forward neural networks which perform an analysis of output obtained from PSI–BLAST (Position-Specific Iterated–BLAST) for secondary structure prediction.
2.3. Domain and Motif Analysis
Domains of human ACE were retrieved from the NCBI conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?), and motifs were predicted using the MEME [15] software. To overcome the limitation of finding gapped motifs, an efficient algorithm of MEME motif discovery is integrated with the algorithm of GLAM2 [16] which enables users to discover novel motifs having gaps.
2.4. Selection of Ligand, Receptor, and Active Site Prediction
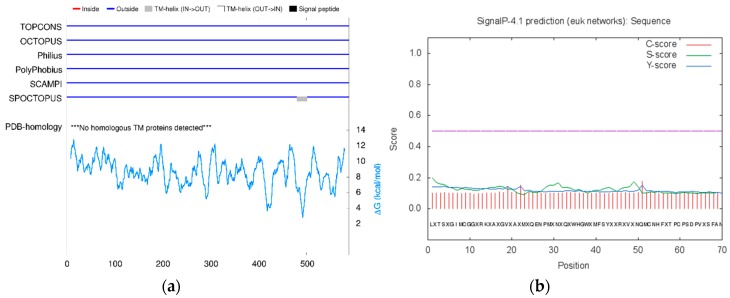
The structures of ligand molecules were downloaded from PubChem that selects compounds based on their chemical formula and physiochemical properties [17]. To explore the binding sites of ligands (inhibitors) on the ACE structure, its 3D-structure was retrieved from RCSB-PDB (Figure 1). Active sites of human ACE were retrieved from InterPro (https://www.ebi.ac.uk/interpro/).
Figure 1.
Results from (a) TOPCONS and (b) Signalp4 showed that ACE is present outside of the cell membrane and has no signal peptide.
2.5. Preparation of Ligands and Receptor
Ligands and receptor were prepared for docking by minimizing their energy and then 3D protonating in MOE 2009.10 [18] by removing solvent molecules (water) and other sites on ACE, facilitating the interaction of only inhibitors or ligands with the selected receptor.
2.6. Molecular Docking
The scope of therapeutic targets in drug design and discovery has broadened with the completion of the human genome project. At the same time, advanced strategies such as excessive-throughput crystallography, protein purification, and nuclear magnetic resonance (NMR) spectroscopy have been providing structural information of protein–ligand and protein complexes [19,20]. All these advancements have resulted in the development of computer-aided drug design, also known as molecular docking. Molecular docking consists of structure-based and ligand-based methods. The ligand-based methods such as QSAR (quantitative structure–activity relationship) are preferred when information about a ligand is large; otherwise, structure-based docking methods are used [21]. In this study, molecular docking of reported synthetic and natural inhibitors of ACE was performed using diverse computational tools, with the aim to discover the optimum inhibitor, which ultimately would provide the basis for designing drugs against hypertension by inhibiting ACE. The structure-based docking method was used because structure-based Computer Aided Drug Designing (CADD) relies on the knowledge of the target protein structure to calculate interaction energies for all compounds tested, whereas ligand-based CADD exploits the knowledge of known active and inactive molecules through chemical similarity searches or construction of predictive, QSAR models [22]. Structure-based CADD is generally preferred where high-resolution structural data of the target protein are available, i.e., for soluble proteins that can readily be crystallized. These advancements in research allow computational techniques to analyze all factors involved in drug design and discovery.
A plethora of computational tools have been developed and are widely used for protein-ligand docking, particularly for discovering drugs based on small well-structured molecules. These tools include Glide, Gold, Auto dock, PyRx, Surflex, ICM, FITTED, MOE (Molecular Operating Environment). MOE, which helps to visualize, characterize, and evaluate protein interactions with other proteins or ligands, was utilized in this study. Proteins and small molecules can be designed using modern in silico design applications, and structure–activity relationships (SAR) in micro-molecules can be developed. It works on the basis of high-throughput screening, docking, energy determination, combining biology, chemistry, and information technology [23]. A list of widely used docking tools and their description are presented in Table 1.
Table 1.
A list of widely used tools for docking.
| No. | Software/Tools | Algorithm | Scoring Term | Advantages | Ref. |
|---|---|---|---|---|---|
| 1. | Molecular Operating Environment (MOE) | High-Speed α shapes algorithms | London dG, FlexX, DrugScore, Mcdock | Customizable, available source-code, gives binding affinity score, shows interacting amino acids with position, and is user-friendly. | [25] |
| 2. | PyRx | Lamarckian genetic algorithm | Binding energy, Internal energy, Internal energy, Unbound energy | Temperature Resistance. Pyrex’s excellent thermal properties at both high and low temperatures are one of its key features. | [26] |
| 3. | Glide (Grid-based Ligand Docking with Energetics) |
Monte Carlo | Glide score | Lead discovery and lead optimization | [27] |
| 4. | AutoDock | Lamarckian genetic algorithm | Empirical free-energy function | Adaptability to user-defined input | [28] |
| 5. | GOLD (Genetic Optimization for Ligand Docking) | Genetic algorithm | GoldScore, ChemScore, ASP (Astex Statistical Potential), CHEMPLP (Piecewise Linear Potential), User-defined | Allows atomic overlapping between protein and ligand | [29] |
| 6. | Surflex | Surflex-Dock search Algorithm |
Bohm’s scoring function | High accuracy level by extending force fields | [30] |
| 7. | FlexX | Incremental reconstruction | Modified Bohm scoring function | Provides a large number of conformations | [31] |
| 8. | ICM (Internal Coordinate Modeling) |
Monte Carlo minimization | Virtual library screening scoring function | Allows side chain flexibility to find a parallel arrangement of two rigid helixes | [32] |
| 9. | MVD (Molegro Virtual Docker) | Evolutionary algorithm | MolDock score | High accuracy level of predicting binding mode | [33] |
| 10. | Fred (Fast Rigid Exhaustive Docking) |
Exhaustive search algorithm | Gaussian scoring function | Nonstochastic approach to examine all possible poses within a protein active site | [34] |
| 11. | LigandFit | Monte Carlo method | LigScore, Piecewise Linear Potential (PLP), Potential of Mean Force (PMF) | Generates good hit rates based on LigScore | [35] |
| 12. | FITTED (Flexibility Induced Through Targeted Evolutionary Description) | Genetic algorithm | Potential of Mean Force (PMF), Drug Score | Analyzes the effect of water molecules on protein–ligand complexes | [36] |
| 13. | GlamDock | Monte Carlo method | ChillScore | Provides provision of two-dimensional analysis to screen ligands by targeting protein | [37] |
| 14. | iGEMDOCK | Genetic algorithm | Empirical scoring function | Integrates the structure-based virtual screening and post-screening analysis. Provides a graphical integrated environment for virtual screening | [38] |
MOE was selected for docking among various available resources as it has a user-friendly graphical interface. It represents a good graphical view of results by showing ligand and receptor binding residues with their positions and interactions. In MOE, receptor–ligand binding affinities with all possible binding geometries are prioritized on the basis of a numerical value called S-score. MOE has multi-disciplinary applications, such as in structure-based design, fragment-based design, pharmacophore discovery, medicinal chemistry applications, biologics applications, protein and antibody modeling, molecular modeling and simulations, cheminformatics and QSAR, and methods’ development and deployment. MOE identifies salt bridges, hydrogen bonds, hydrophobic interactions, sulfur-LP, cation-π, and solvent exposure, and gives the S score. Interactions of inhibitors with receptor proteins are predicted on the basis of the S score [24].
2.7. Lipinski’s Rule of Five for Drug-Likeness or ADME (Absorption, Distribution, Metabolism, and Excretion) Analysis
Drug-likeness of our inhibitors, including absorption, distribution, metabolism, and excretion of these inhibitors within the body, was predicted using SwissADME (Swiss Institute of Bioinformatics, Switzerland) [39]. The Egan BOILED-Egg method available in SwissADME tool was used for the determination of the absorption of the inhibitors in the gastrointestinal tract and brain. BOILED-Egg (Brain Or IntestinaL EstimateD permeation predictive model), also called Egan egg, provides a threshold (WLOGP ≤ 5.88 and TPSA ≤ 131.6) and a clear graphical representation of how far a molecular structure is from the ideal one for good absorption [40]. In this 2D graphical representation, the yolk area represents the molecules that can passively permeate through the blood–brain barrier (BBB), whereas the molecules located in the white region are predicted to be passively absorbed by the gastrointestinal (GI) tract.
Herein, we particularly focused on ACE, which has been reported as a key enzyme in hypertension [9]. Various substances containing naturally occurring compounds from herbal and animal sources (e.g., Allicin and teprotide) have come forward as potential inhibitors of ACE (Table 2) and may control high blood pressure.
Table 2.
Synthetic, herbal, and animal source inhibitors of angiotensin-converting enzyme (ACE).
| No. | Ligand | Features | Source | Function | Citation |
|---|---|---|---|---|---|
| 1. | Benazepril | 97% protein binding, a half-life of 10–11 h, pregnancy category: D | Synthetic | Cures hypertension | [41] |
| 2. | Captopril | 25–30% protein binding, a half-life of 2 h, pregnancy category: D | Synthetic | Controls blood pressure | [42] |
| 3. | Cilazapril | A half-life of 1 to 4 h | Synthetic | ACE inhibition | [43] |
| 4. | Lisinopril | Pregnancy category: D, does not bind serum proteins other than ACE |
Synthetic | Inhibition of ACE | [44] |
| 5. | Moexipril | Pregnancy category: D, <90% protein binding and 1 h half-life |
Synthetic | Treatment of hypertension and congestive heart failure | [45] |
| 6. | Trandolapril | Half-life 6 to 10 h, pregnancy category: D | Synthetic | Controls high blood pressure | [46] |
| 7. | Enalapril | Pregnancy category: D, half-life of 11 h |
Synthetic | ACE inhibition to control hypertension | [47] |
| 8. | Fosinopril | 12 h half-life, pregnancy category: D, ≥95% protein-binding capacity |
Synthetic | Normalizes blood pressure | [48] |
| 9. | Perindopril | 20% protein binding, pregnancy category: D and 1–2 h half life | Synthetic | Controls blood pressure | [49] |
| 10. | Quinapril | 97% protein binding, 2 h biological half-life, pregnancy category: D | Synthetic | Inhibition of ACE | [50] |
| 11. | Ramipril | Protein binding 73% (ramipril), 56% (ramiprilat), half-life of 2–4 h |
Synthetic | Congestive heart failure control | [51] |
| 12. | Allicin | Has water solubility of 24 mg/mL at 10 °C, solid, melting point >25 °C | Garlic and onion | Inhibition of ACE | [52] |
| 13. | Teprotide | Has 10 hydrogen bond donors, 13 hydrogen bond acceptors, and 79 heavy atoms | Snake venom | Antihypertensive agent | [53] |
3. Results and Discussion
3.1. Physiochemical Properties of ACE
Human ACE was found to exhibit a molecular weight of 67,993.2 Daltons and isoelectric pH 5.82. It is a stable protein with an aliphatic index of 78.86, whereas its instability index was predicted to be 39.46. The prediction of GRAVY value of −0.4441 demonstrates that ACE is a hydrophilic peptide (Table 3).
Table 3.
Physiochemical properties of ACE predicted by ProtParam.
| Serial Number | Property | Value |
|---|---|---|
| 1. | Number of amino acids | 589 |
| 2. | Total number of atoms | 9457 |
| 3. | Molecular weight | 67,993.20 Dalton |
| 4. | Theoretical pI | 5.82 |
| 5. | Extinction coefficient * | 143,240 at Abs 0.1% 2.112, assuming all pairs of Cys residues form cystines |
| 6. | Instability index | 39.46 |
| 7. | Aliphatic index | 78.86 |
| 8. | Grand average of hydropathicity (GRAVY) | −0.441 |
| 9. | Chemical Formula | C3076H4656N818O883S24 |
| 10. | Charge | Negative |
* Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
3.2. Membrane Topology of ACE
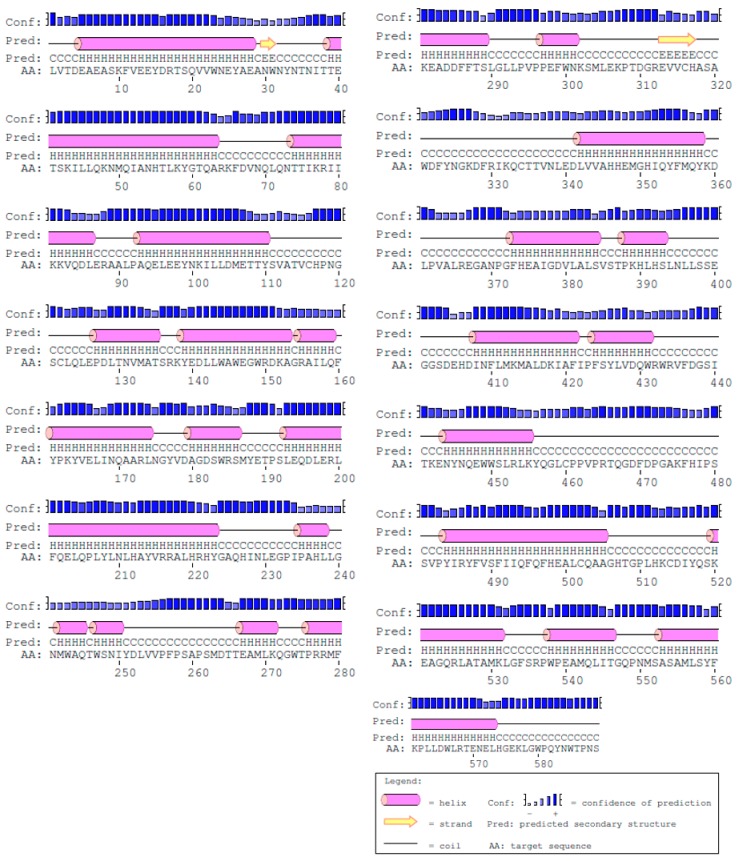
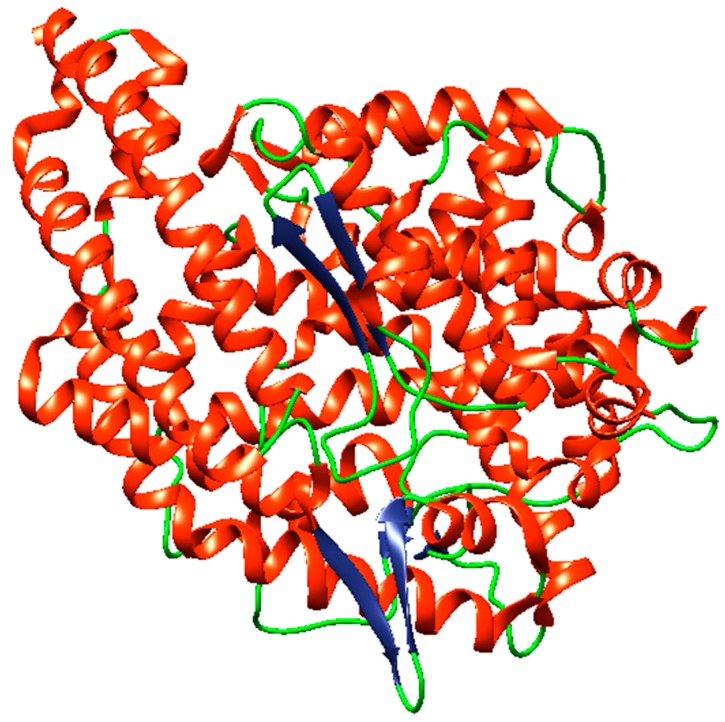
The use of diverse computational tools including TOPCON, Signal p4.1, OCTOPUS, PolyPhobius, SCAMPI, and SPOCTOPUS predicted that ACE is an extracellular enzyme, present outside of the cell membrane (Figure 1). The structure of human ACE was predicted to comprise 333 helices, 9 strands, and 247 coils (Figure 2). NCBI Conserved Domain Database has shown that human ACE has only one domain and belongs to the family Peptidase M2, peptidyl-dipeptidase A, with the interval from 11 to 571 amino acid numbers. However, MEME predicted that ACE has the sequences “VCHPNGSC” in position 115–122, “HHEMGHIQYFMQYK” at 346–359, and “FHEALC” at 373–378. The 3D structure of human ACE was retrieved from the PDB (PDB ID: 1o8A) online protein database and visualized with a desktop tool, i.e., CHIMERA (Figure 3). ACE was predicted to have 14 active sites for interactions using InterPro (EMBL-EBI, Cambridgeshire, UK) (Table 4).
Figure 2.
Secondary structure of human ACE; pink cylinders, yellow arrows, and black lines show helixes, strands, and coils, respectively.
Figure 3.
3D structure of human ACE (PDB ID: 1o8A), visualized through CHIMERA.
Table 4.
Active sites of human ACE.
| Amino Acid | Position | Amino Acid | Position |
|---|---|---|---|
| Histidine | 317 | Alanine | 318 |
| Serine | 319 | Histidine | 347 |
| Glutamic Acid | 348 | Histidine | 351 |
| Glutamic Acid | 375 | Phenylalanine | 421 |
| Lysine | 475 | Phenylalanine | 476 |
| Histidine | 477 | Valine | 482 |
| Tyrosine | 484 | Tyrosine | 487 |
3.3. Reported Inhibitors of ACE
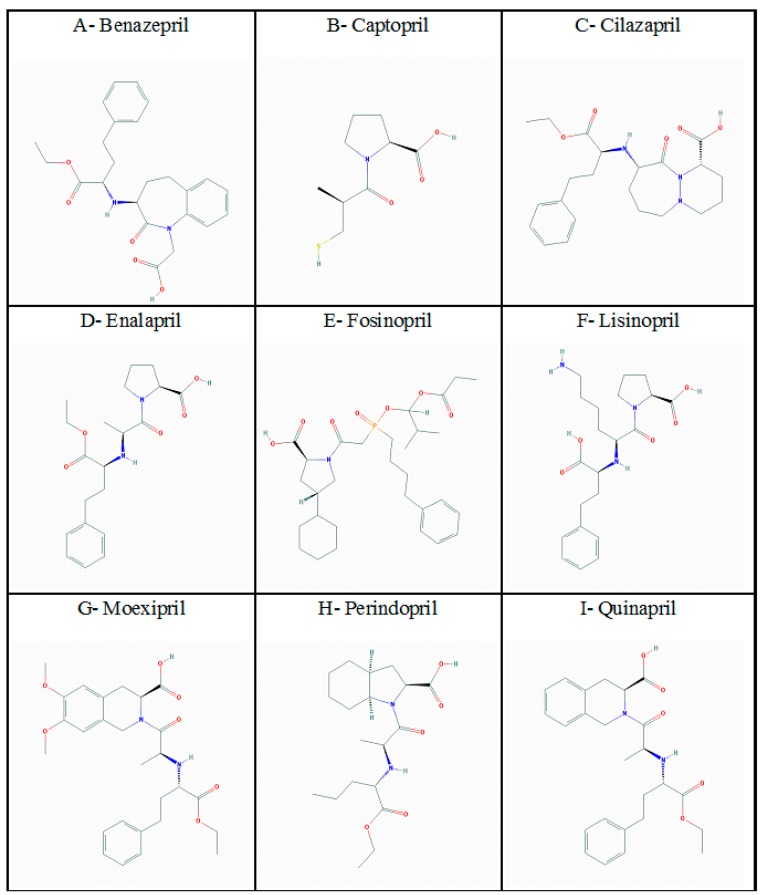
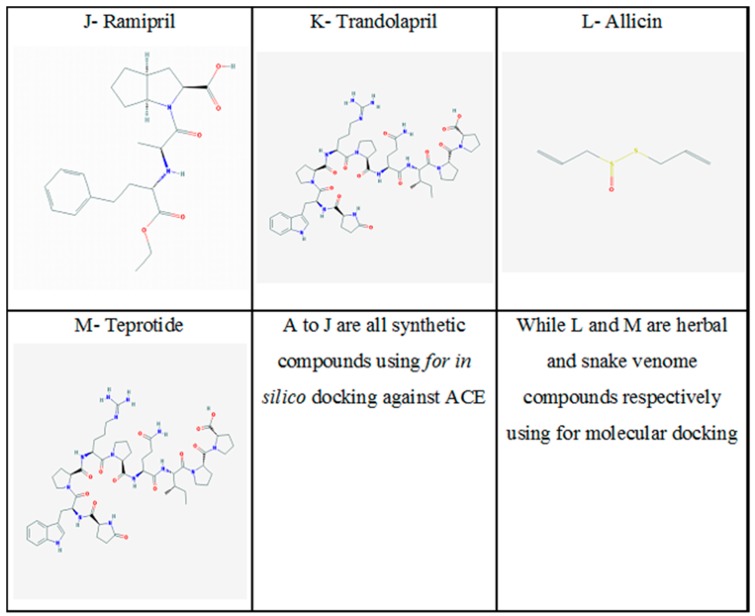
The 2D structures of reported inhibitors of ACE were downloaded from PubChem in SDF format and are portrayed in Figure 4.
Figure 4.
2D structures of various ACE inhibitors including (A) Benazepril, (B) Captopril, (C) Cilazapril, (D) Enalapril, (E) Fosinopril, (F) Lisinopril, (G) Moexipril, (H) Perindopril, (I) Quinapril, (J) Ramipril, (K) Trandolapril, (L) Allicin, and (M) Teprotide.
Herbal and Natural Inhibitors
3.4. Molecular Docking
In silico docking of human ACE against selected inhibitors was performed using MOE against all the predicted active sites. The results showed that all selected inhibitors were in the pocket of the target protein (ACE), exhibiting a possible interaction with ACE. The docking results were manipulated using the GBVI/WSA dG scoring function with the generalized Born solvation model (GBVI). The GBVI/WSA dG is a force field-based scoring function, which estimates the free energy of binding of the ligand from a given orientation. Interaction results were evaluated with the S score. Inhibitors with the lowest S score tend to establish a strong interaction with ACE on specific active sites (Table 5). After in silico docking, we identified a ligand showing the minimum S score among all the inhibitors. Teprotide, which is present in snake venom, showed a minimum S score of −20.1163; therefore, it establishes the strongest interaction with ACE among all the inhibitors discussed in this study. Fosinopril is another widely used and effective drug against hypertension. It was predicted to exhibit a strong binding affinity for ACE, with an S score of −18.9225. Earlier studies demonstrated that fosinopril doses of 10 and 20 mg could inhibit 85% and 93% of ACE activity, respectively, within 24 h of administration [55]. Hayek et al. used ACE as a receptor and fosinopril as an inhibitor to cure hypertension and concluded, after 12 weeks of treatment, that fosinopril remarkably reduces blood pressure in mice [56]. Heart Outcomes Prevention Evaluation Study Investigators evaluated the role of ramipril in reducing the overactivity of ACE and showed that ramipril significantly lessens the rates of myocardial infarction and stroke in a wide range of high-risk patients [57].
Table 5.
Inhibitors ranked on the basis of their S-values.
| No. | Name | S-Values |
|---|---|---|
| 1. | Teprotide | −20.1163 |
| 2. | Fosinopril | −18.9225 |
| 3. | Moexipril | −16.816 |
| 4. | Quinapril | −13.456 |
| 5. | Lisinopril | −12.502 |
| 6. | Cilazapril | −12.493 |
| 7. | Trandolapril | −12.2673 |
| 8. | Enalapril | −11.7516 |
| 9. | Ramipril | −11.3562 |
| 10. | Captopril | −10.8282 |
| 11. | Benazepril | −9.3245 |
| 12. | Perindopril | −8.105 |
| 13. | Allicin | −5.5448 |
3.5. Drug-Likeness and ADME Predictions of Our Inhibitors
The antagonistic interaction of inhibitors with a receptor protein or enzyme cannot guarantee the suitability of an inhibitor as a drug; therefore, ADME analysis of inhibitors is important in the drug development [58]. ADME is based on Lipinski’s rule of five [59] and helps to make decisions on the approval of inhibitors for biological systems. Poor ADME characteristics and unfavorable toxicology for a biological system are the major cause of the failure of most medicines in clinical experiments.
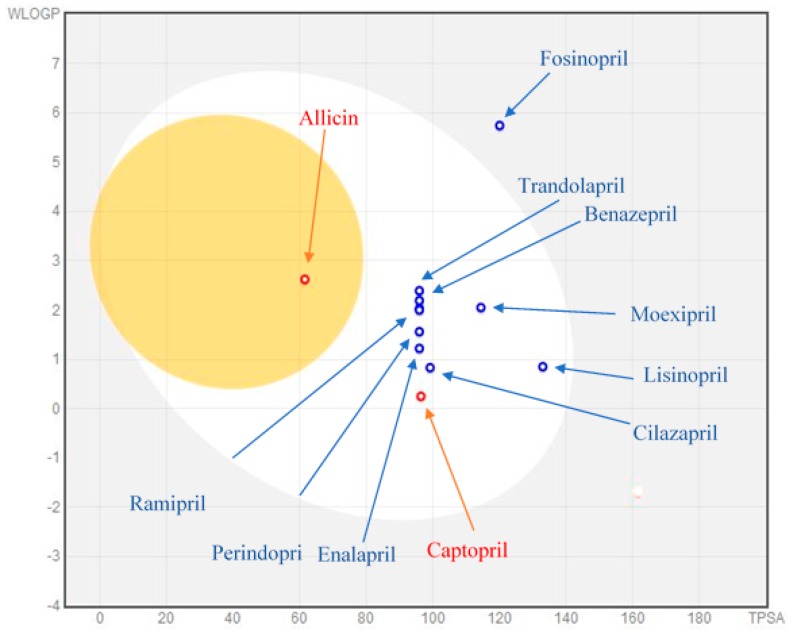
The Lipinski’s rule of five was published in 1997 by Christopher A. Lipinski and is also known as the Pfizer’s rule of five or Rule of five (Ro5). It is a rule of thumb to evaluate the drug-likeness and to determine if a chemical compound with a certain pharmacological or biological activity has properties that would make it a likely orally active drug in humans. Ro5 depends on four simple physiochemical parameter ranges: the molecular weight (MW), which should be less than 500 g/mol, lipophilicity (Log P) less than 5, and number of hydrogen bond donors and acceptors less than 5 and 10, respectively, as seen for 90% of orally functional drugs that have obtained phase II clinical status. These parameters are connected with intestinal permeability and aqueous solubility and determine the first step of oral bioavailability. These rules explain molecular properties valuable for a drug’s pharmacokinetics in the human body, including their absorption, distribution, metabolism, and excretion (ADME). If a ligand fails to fulfill the parameters of Ro5, then it is highly probable that it will cause trouble if ingested [1]. ADME predictions of our inhibitors are shown in Table 6. BOILED-egg results, showing the possibility of absorption and penetration of inhibitors in the GI attract and brain using WLOGP and TPSA parameters are presented in Figure 5.
Table 6.
Lipinski’s rule of five for ADME analysis of our inhibitors (ligands).
| No. | Name | Lipinski’s Rule of Five | Drug-Likeness | ||||
|---|---|---|---|---|---|---|---|
| Molecular Weight (g/mol) | Lipophilicity (MLog P) |
Hydrogen Bond Donors | Hydrogen Bond Acceptors | No. of Rule Violations | |||
| Less than 500 Dalton | Less than 5 | Less than 5 | Less than 10 | Less than 2 Violations | Lipinski’s Rule Follows | ||
| 1. | Teprotide | 1101.26 | −3.11 | 10 | 13 | 3: MW > 500, NH or OH > 5, N or O > 10, 1: MW > 500 |
No |
| 2. | Fosinopril | 563.66 | 3.74 | 1 | 7 | 0 | Yes |
| 3. | Moexipril | 498.57 | 1.54 | 2 | 8 | 0 | Yes |
| 4. | Quinapril | 438.52 | 2.17 | 2 | 6 | 0 | Yes |
| 5. | Lisinopril | 405.49 | −1.46 | 4 | 7 | 0 | Yes |
| 6. | Cilizapril | 417.50 | 1.79 | 2 | 7 | 0 | Yes |
| 7. | Trandolapril | 430.54 | 2.19 | 2 | 6 | 0 | Yes |
| 8. | Enalapril | 376.45 | 1.32 | 2 | 6 | 0 | Yes |
| 9. | Ramipril | 416.51 | 1.98 | 2 | 6 | 0 | Yes |
| 10. | Caprtopril | 217.29 | 0.45 | 1 | 3 | 0 | Yes |
| 11. | Benzapril | 424.49 | 2.23 | 2 | 6 | 0 | Yes |
| 12. | Perindopril | 368.47 | 1.36 | 2 | 6 | 0 | Yes |
| 13. | Allicin | 162.27 | 1.18 | 0 | 1 | 0 | Yes |
Figure 5.
Evaluation of the analyzed ligands by the BOILED-Egg method.
All of the inhibitors or ligands discussed herein satisfy the Lipinski’s rule, except for teprotide, which significantly violates three parameters (MW > 500, number of hydrogen bond donors > 5 and number of hydrogen bond acceptors > 10); furthermore, it also violates the BOILED-egg method. Although teprotide has the highest binding affinity for human ACE among all the inhibitors, it is not proposed as an orally active drug due to violation of the Lipinski’s rule. An Egan’s egg graph for the inhibitors was generated using SwissADME. The graph showed that only allicin, a herbal compound, is absorbed by the brain, though in the acceptable range. The remaining inhibitors showed gastrointestinal absorption within an acceptable range, except for teprotide and lisinopril (WLOGP >5.88 and TPSA >131.6) (Figure 5).
So, on the basis of Egan’s boiled-egg rule threshold values (WLOGP ≤ 5.88 and TPSA ≤ 131.6), only allicin penetrates the blood–brain barrier, though within acceptable limits. The blue dots indicate molecules predicted to be effluated from the CNS by P-glycoprotein, and the red dots indicate molecules predicted not to be effluated from the CNS by P-glycoprotein.
The above-mentioned results of the molecular docking that were obtained using molecular operating environment (MOE) allowed us to observe ligand–receptor. Analysis of the interactions with the Protein–Ligand Interaction Profiler between ACE and the inhibitor teprotide revealed the best binding affinity. By analyzing the drug’s score (S-value), teprotide showed the lowest S-value (−20.1163), resulting as the best ligand among our selected ligands to inhibit the activity of ACE. However, it violates three parameters (MW > 500D, H-bond donors >5 and H-bond acceptors >10) of the Lipinski’s rule, as well as Egan’s parameters (WLOGP > 5.88 and TPSA > 131.6). Although teprotide is proposed to be a potential therapeutic inhibitor of ACE, it may fail as an orally active drug because it deviates from the Lipinski’s rule and from the Egan’s rule. Compared to teprotide, the higher S-value (−18.9225) of fosinopril demonstrates its lower binding affinity for ACE. Notably, fosinopril satisfies all parameters of the Lipinski’s rule, except for MW >500D, and also complies to the BOILED-egg approach, showing no brain and GI tract absorption, which renders it a more suitable potential orally active drug and therapeutic inhibitor of ACE to be tested in clinical trials, compared to teprotide. Allicin is a herbal compound found in garlic that helps to inhibit the activity of ACE and satisfy all Lipinski’s rules with minimal absorption in the brain. Nonetheless, it displayed lower binding affinity and placed last among our selected inhibitors because of its highest S-value (−5.5448). Although allicin has a lowest binding affinity for ACE, it can be used in drug design for the treatment of hypertension because of its herbal nature; also, garlic can be used as food to bring the blood pressure within normal range. Therefore, from various investigations, it is clear that teprotide, which is extracted from a snake (B. jararaca) venom, has the highest binding affinity for ACE compared to other inhibitors but it cannot be used as an orally active drug. In contrast, fosinopril, a synthetic compound, showed the second highest binding affinity for ACE and therefore could be used as a potentially therapeutic compound for the development of orally active drugs inhibiting the activity of ACE and thereby useful to treat hypertension. Fosinopril also does not exhibit any type of absorption in the brain and gastrointestinal tract. Similarly, allicin can also be used to develop orally active drugs for the management of hypertension.
4. Conclusions
In this study, fosinopril was predicted as the best ACE inhibitor (with maximum binding affinity for ACE after teprotide) to be used as a potentially therapeutic orally active drug (on the basis of Lipinski’s rule of five and BOILED-egg approach) for the treatment of hypertension. Among the animal inhibitors, teprotide showed the highest binding affinity compared to all other ligands studied here; however, according to Lipinski’s rule and BOILED-egg method, it is not recommended as a suitable therapeutic agent. Furthermore, allicin, a herbal ligand, exhibited reasonable binding affinity for ACE and follows Lipinski’s rule of five but can only be used as food because of its slight absorption in the brain. In conclusion, our study provides a clearer insight into the interaction properties of known putative synthetic inhibitors of ACE and bioactive inhibitors, including interactions with the blood–brain barrier. In recent years, consumers have paid attention to natural bioactive compounds as potential medicines because of their effectiveness in promoting health, associated with less adverse effects. In future, we will be able to use the knowledge of inhibitors’ pharmacological properties, including those of bioactive compounds such as allicin, to make effective therapeutic drugs based on ACE inhibition to cure hypertension.
Acknowledgments
We are thankful to the Center for Advanced Studies in Agriculture and Food Security (CAS-AFS), University of Agriculture, Faisalabad, Pakistan, for providing the infrastructural support. The authors also acknowledge the support from the Department of Computer Science, UAF, for providing literature, computational, and all other facilities. The authors (S.M. and K.A.A.-G.) would like to express their sincere appreciation to the Deanship of Scientific Research at King Saud University, KSA, for its funding through the Prolific Research Group Project No. PRG-1436-011, and to the Higher Education Commission (HEC), Pakistan, for funding through the Project # 2049/SRGP.
Author Contributions
M.Z.N., M.A.A., M.B., R.M.A., and S.M., designed the study. S.A.A., M.H., M.U., and Q.A. performed the computational analysis. M.Z.N., M.A.A., and K.A.A.-G. wrote the manuscript, in consultation with all other authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mendis S., Puska P., Norrving B., World Health Organization . Global Atlas on Cardiovascular Disease Prevention and Control. World Health Organization; Geneva, Switzerland: 2011. [Google Scholar]
- 2.Catalá López F., Tabarés Seisdedos R. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;8:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santulli G. Epidemiology of cardiovascular disease in the 21st century: Updated numbers and updated facts. J. Cardiovasc. Dis. 2013;1:1–2. [Google Scholar]
- 4.Bundy J.D., He J. Hypertension and Related Cardiovascular Disease Burden in China. Ann. Glob. Health. 2016;82:227–233. doi: 10.1016/j.aogh.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Campbell N.R., Lackland D.T., Lisheng L., Niebylski M.L., Nilsson P.M., Zhang X.H. Using the Global Burden of Disease study to assist development of nation-specific fact sheets to promote prevention and control of hypertension and reduction in dietary salt: A resource from the World Hypertension League. J. Clin. Hypertens. 2015;17:165–167. doi: 10.1111/jch.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruppert V., Maisch B. Genetics of human hypertension. Herz. 2003;28:655–662. doi: 10.1007/s00059-003-2516-6. [DOI] [PubMed] [Google Scholar]
- 7.Newhouse S.J., Wallace C., Dobson R., Mein C., Pembroke J., Farrall M., Clayton D., Brown M., Samani N., Dominiczak A. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Hum. Mol. Genet. 2005;14:1805–1814. doi: 10.1093/hmg/ddi187. [DOI] [PubMed] [Google Scholar]
- 8.Brand E., Chatelain N., Paillard F., Tiret L., Visvikis S., Lathrop M., Soubrier F., Demenais F. Detection of putative functional angiotensinogen (AGT) gene variants controlling plasma AGT levels by combined segregation-linkage analysis. Eur. J. Hum. Genet. 2002;10:715. doi: 10.1038/sj.ejhg.5200874. [DOI] [PubMed] [Google Scholar]
- 9.Jimsheena V., Gowda L.R. Angiotensin I-converting enzyme (ACE) inhibitory peptides derived from arachin by simulated gastric digestion. Food Chem. 2011;125:561–569. doi: 10.1016/j.foodchem.2010.09.048. [DOI] [Google Scholar]
- 10.Riordan J.F. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003;4:225. doi: 10.1186/gb-2003-4-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passos-Silva D.G., Brandan E., Santos R.A.S. Angiotensins as therapeutic targets beyond heart disease. Trends Pharmacol. Sci. 2015;36:310–320. doi: 10.1016/j.tips.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Kouranov A., Xie L., de la Cruz J., Chen L., Westbrook J., Bourne P.E., Berman H.M. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelhoch H.J.B. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 14.McGuffin L.J., Bryson K., Jones D.T.J.B. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 15.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. PubChem substance and compound databases. Nucleic Acids Res. 2015;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scholz C., Knorr S., Hamacher K., Schmidt B. DOCKTITE—A Highly Versatile Step-by-Step Workflow for Covalent Docking and Virtual Screening in the Molecular Operating Environment. J. Chem. Inf. Model. 2015;55:398–406. doi: 10.1021/ci500681r. [DOI] [PubMed] [Google Scholar]
- 19.Bajorath J. Integration of virtual and high-throughput screening. Nat. Rev. Drug Discov. 2002;1:882. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- 20.Jorgensen W.L. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 21.Kuntz I.D., Blaney J.M., Oatley S.J., Langridge R., Ferrin T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-X. [DOI] [PubMed] [Google Scholar]
- 22.Kalyaanamoorthy S., Chen Y.-P.P. Structure-based drug design to augment hit discovery. Drug Discov. Today. 2011;16:831–839. doi: 10.1016/j.drudis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Chemical Computing Group Inc. Molecular Operating Environment (MOE) Chemical Computing Group Inc.; Montreal, QC, Canada: 2016. H3A 2R7. [Google Scholar]
- 24.Clark A.M., Labute P. 2D depiction of protein–ligand complexes. J. Chem. Inf. Model. 2007;47:1933–1944. doi: 10.1021/ci7001473. [DOI] [PubMed] [Google Scholar]
- 25.Vilar S., Cozza G., Moro S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 26.Yuliana D., Bahtiar F.I., Najib A. In silico screening of chemical compounds from roselle (Hibiscus Sabdariffa) as angiotensin-I converting enzyme inhibitor used PyRx program. ARPN J. Sci. Technol. 2013;3:1158–1160. [Google Scholar]
- 27.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 28.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdonk M.L., Cole J.C., Hartshorn M.J., Murray C.W., Taylor R.D. Improved protein–ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 30.Jain A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 31.Schellhammer I., Rarey M. FlexX-Scan: Fast, structure-based virtual screening. Proteins Struct. Funct. Bioinform. 2004;57:504–517. doi: 10.1002/prot.20217. [DOI] [PubMed] [Google Scholar]
- 32.Abagyan R., Totrov M., Kuznetsov D. ICM—A new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994;15:488–506. doi: 10.1002/jcc.540150503. [DOI] [Google Scholar]
- 33.Molegro A. MVD 5.0 Molegro Virtual Docker. CLC bio; Aarhus, Denmark: 2011. [Google Scholar]
- 34.Kellenberger E., Rodrigo J., Muller P., Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins Struct. Funct. Bioinform. 2004;57:225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 35.Venkatachalam C.M., Jiang X., Oldfield T., Waldman M. LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 2003;21:289–307. doi: 10.1016/S1093-3263(02)00164-X. [DOI] [PubMed] [Google Scholar]
- 36.Corbeil C.R., Englebienne P., Moitessier N. Docking ligands into flexible and solvated macromolecules. 1. Development and validation of FITTED 1.0. J. Chem. Inf. Model. 2007;47:435–449. doi: 10.1021/ci6002637. [DOI] [PubMed] [Google Scholar]
- 37.Tietze S., Apostolakis J. GlamDock: Development and validation of a new docking tool on several thousand protein−ligand complexes. J. Chem. Inf. Model. 2007;47:1657–1672. doi: 10.1021/ci7001236. [DOI] [PubMed] [Google Scholar]
- 38.Hsu K.-C., Chen Y.-F., Lin S.-R., Yang J.-M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011;12:S33. doi: 10.1186/1471-2105-12-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daina A., Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamerson K., Weber M.A., Bakris G.L., Dahlöf B., Pitt B., Shi V., Hester A., Gupte J., Gatlin M., Velazquez E.J. Benazepril plus amlodipine or hydrochlorothiazide for hypertension in high-risk patients. N. Engl. J. Med. 2008;359:2417–2428. doi: 10.1056/NEJMoa0806182. [DOI] [PubMed] [Google Scholar]
- 42.Nagaoka S., Kawasaki S., Kawasaki H., Kamei K. The angiotensin converting enzyme (ACE) inhibitor, captopril disrupts the motility activation of sperm from the silkworm, Bombyx mori. J. Insect Physiol. 2017;103:18–28. doi: 10.1016/j.jinsphys.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Narayanam M., Sahu A., Singh S. Use of LC–MS/TOF, LC–MSn, NMR and LC–NMR in characterization of stress degradation products: Application to cilazapril. J. Pharm. Biomed. Anal. 2015;111:190–203. doi: 10.1016/j.jpba.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 44.Younas F., Aslam B., Muhammad F., Mohsin M., Raza A., Faisal M.N., Hassan S.-U., Majeed W. Haematopoietic effects of Angelica sinensis root cap polysaccharides against lisinopril-induced anaemia in albino rats. Pharm. Biol. 2017;55:108–113. doi: 10.1080/13880209.2016.1230635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy C.P., Ramakrishna K., Narayana Rao K. Structural identification of degradants of moexipril by LC-MS/MS. Biomed. Chromatogr. 2017;31:e4004. doi: 10.1002/bmc.4004. [DOI] [PubMed] [Google Scholar]
- 46.Elgendy I.Y., Bavry A.A., Gong Y., Handberg E.M., Cooper-DeHoff R.M., Pepine C.J. Long-term mortality in hypertensive patients with coronary artery disease: Results from the US cohort of the International Verapamil (SR)/Trandolapril Study. Hypertension. 2016;68:1110–1114. doi: 10.1161/HYPERTENSIONAHA.116.07854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaddy R., Canter C., Halnon N., Kochilas L., Rossano J., Bonnet D., Bush C., Zhao Z., Kantor P., Burch M. Design for the sacubitril/valsartan (LCZ696) compared with enalapril study of pediatric patients with heart failure due to systemic left ventricle systolic dysfunction (PANORAMA-HF study) Am. Heart J. 2017;193:23–34. doi: 10.1016/j.ahj.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Araiza-Saldaña C.I., Pedraza-Priego E.F., Torres-López J.E., Rocha-González H.I., Castañeda-Corral G., Hong-Chong E., Granados-Soto V. Fosinopril Prevents the Development of Tactile Allodynia in a Streptozotocin-Induced Diabetic Rat Model. Drug Dev. Res. 2015;76:442–449. doi: 10.1002/ddr.21280. [DOI] [PubMed] [Google Scholar]
- 49.El-Bagary R.I., Elkady E.F., Mowaka S., Attallah M.A. A Validated HPLC Method for Simultaneous Determination of Perindopril Arginine, Amlodipine, and Indapamide: Application in Bulk and in Different Pharmaceutical Dosage Forms. J. AOAC Int. 2017;100:992–999. doi: 10.5740/jaoacint.16-0279. [DOI] [PubMed] [Google Scholar]
- 50.Wojewodzka-Zelezniakowicz M., Kisiel W., Kramkowski K., Gromotowicz-Poplawska A., Zakrzeska A., Stankiewicz A., Kolodziejczyk P., Szemraj J., Ladny J.R., Chabielska E. Quinapril decreases antifibrinolytic and prooxidative potential of propofol in arterial thrombosis in hypertensive rats. J. Renin-Angiotensin-Aldosterone Syst. 2016;17:1470320316647239. doi: 10.1177/1470320316647239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mancia G., Schumacher H., Böhm M., Redon J., Schmieder R.E., Verdecchia P., Sleight P., Teo K., Yusuf S. Relative and Combined Prognostic Importance of On-Treatment Mean and Visit-to-Visit Blood Pressure Variability in ONTARGET and TRANSCEND PatientsNovelty and Significance. Hypertension. 2017;70:938–948. doi: 10.1161/HYPERTENSIONAHA.117.09714. [DOI] [PubMed] [Google Scholar]
- 52.Reuter H., Koch H., Lawson L. The Science and Therapeutic Application of Allium sativum L. and Related Species. Williams and Wilkins; Baltimore, MD, USA: 1996. [Google Scholar]
- 53.Cragg G.M., Newman D.J., Snader K.M. Natural products in drug discovery and development. J. Nat. Prod. 1997;60:52–60. doi: 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- 54.El-Sheakh A.R., Ghoneim H.A., Suddek G.M., Ammar E.S. Attenuation of oxidative stress, inflammation, and endothelial dysfunction in hypercholesterolemic rabbits by allicin. Can. J. Physiol. Pharmacol. 2015;94:216–224. doi: 10.1139/cjpp-2015-0267. [DOI] [PubMed] [Google Scholar]
- 55.Ford N.F., Fulmor I.E., Nichola P.S., Alpin P.G., Herron J.M. Fosinopril monotherapy: Relationship between blood pressure reduction and time of administration. Clin. Cardiol. 1993;16:324–330. doi: 10.1002/clc.4960160407. [DOI] [PubMed] [Google Scholar]
- 56.Hayek T., Attias J., Coleman R., Brodsky S., Smith J., Breslow J.L., Keidar S.J. The angiotensin-converting enzyme inhibitor, fosinopril, and the angiotensin II receptor antagonist, losartan, inhibit LDL oxidation and attenuate atherosclerosis independent of lowering blood pressure in apolipoprotein E deficient mice. Cardiovasc. Res. 1999;44:579–587. doi: 10.1016/S0008-6363(99)00239-4. [DOI] [PubMed] [Google Scholar]
- 57.Heart Outcomes Prevention Evaluation Study Investigators Effects of an angiotensin-converting–enzyme inhibitor, ramipril, on cardiovascular events in high-risk patients. N. Engl. J. Med. 2000;342:145–153. doi: 10.1056/NEJM200001203420301. [DOI] [PubMed] [Google Scholar]
- 58.Lipinski C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Lipinski C.A. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv. Drug Deliv. Rev. 2016;101:34–41. doi: 10.1016/j.addr.2016.04.029. [DOI] [PubMed] [Google Scholar]