Abstract
Alcohol is a well-established risk factor for hepatocellular carcinoma, but the mechanisms are not well understood. Several studies suggested that alcohol promotes tumor growth by altering immune cell phenotypes in the liver. Arginine methylation is a common posttranslational modification generated mostly by a single protein, PRMT1. In myeloid cells PRMT1 is a key regulator of immune response. Myeloid-specific PRMT1 knockout mice are hyperresponsive to LPS and deficient in PPARγ-dependent macrophage M2 polarization. We aimed to define the role of myeloid PRMT1 in alcohol-associated liver tumor progression using a mouse model of DEN injection followed by Lieber–DeCarli alcohol liquid diet feeding. We found that PRMT1 knockout mice showed significantly lower expression of IL-10 and IL-6 cytokines in the liver and downstream STAT3 activation, which correlated with reduced number of surface tumors, reduced proliferation, and reduced number of M2 macrophages in the liver as well as within proliferating nodules. We found that blocking IL-6 signaling in alcohol-fed mice reduced the number of tumors and liver proliferation in wild-type mice but not in knockout mice suggesting that reduced IL-6 in PRMT1 knockout mice contributes to the protection from alcohol. Additionally, PRMT1 knockout did not show any protection in tumor formation in the absence of alcohol. Finally, we confirmed that this mechanism is relevant in humans. We found that PRMT1 expression in tumor-associated macrophages correlated with STAT3 activation in human HCC specimens. Taken together, these data suggest that the PRMT1–IL-6–STAT3 axis is an important mechanism of alcohol-associated tumor progression.
Key words: Liver tumor, Asymmetric dimethyl arginine, Protein arginine methylation, STAT3, Macrophage polarization
INTRODUCTION
Protein arginine methylation is a common posttranslational modification that plays a role in multiple pathways, including cell cycle control, RNA processing, and DNA replication. The protein arginine methyltransferase 1 (PRMT1) is responsible for about 85% of total cellular arginine methylation1 and catalyzes arginine mono- and dimethylation using S-adenosyl methionine (SAM) as a methyl donor. PRMT1 methylates histone H4 at arginine 3, generating H4R3me2a, a transcriptional activation mark1–5, thus contributing to the histone code. As a transcriptional coactivator, PRMT1 is recruited to promoters by a number of different transcription factors3,6,7. Abnormal function of PRMT1 is closely associated with several types of cancer and cardiovascular disease. Arginine methylation impacts gene transcription and splicing as well as upstream signal transduction4. Recently we created a myeloid-specific PRMT1 knockout mouse (MKO) model to investigate the cell type-specific role of PRMT1 in these animals. We found that PRMT1 controls polarization of macrophages through histone arginine methylation at the peroxisome proliferator-activated receptor γ (PPARγ) promoter8,9. Myeloid-specific PRMT1 MKO are hyperresponsive to LPS challenge due to lack of anti-inflammatory (or M2) macrophages.
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related death10,11. Unlike other cancers with known risk factors, causes of HCC are not completely understood. Commonly, HCC is associated with hepatitis B and C and chronic alcohol drinking12,13. About 40% of individuals who chronically consume alcohol develop fatty liver, which can advance to alcoholic hepatitis and cirrhosis. Although there have been conflicting findings, currently it is accepted that alcohol-associated cirrhosis is a medium-high risk factor for HCC. HCC development is a multistep process that involves genetic and epigenetic modifications in the liver that lead to malignant transformation of hepatocytes14. Mouse models of HCC suggest that chronic EtOH consumption increases HCC risk by several mechanisms. Alcohol stimulates hepatocyte proliferation through activation of the Wnt/β-catenin signaling pathway, thus promoting tumorigenesis following an initiating insult in the liver15,16. On the other hand, increased neutrophil and macrophage activation induced by chronic alcohol exposure contributes to chronic tissue inflammation and promotes tumor development16,17. Moreover, chronic alcohol consumption exacerbates DEN-induced hepatocarcinogenesis by enhancing protumor immunity by increasing the number of tumor-associated macrophages (TAMs) and their M2 polarization, impairing antitumor CD8+ T cells and aggravating hepatic pathological injury18.
Here we found that myeloid-specific PRMT1 MKO are protected from alcohol-associated HCC development. MKO showed significantly lower expression of IL-10 and IL-6 cytokines in the liver and downstream STAT3 activation compared to wild-type (WT) mice, which correlated with reduced number of lesions and surface tumors, reduced proliferation, and reduced number of Mrc1+ (M2) macrophages in the liver as well as within proliferating nodules. We found that blocking IL-6 signaling in alcohol-fed mice can reduce the number of lesions and liver proliferation in WT mice but not in MKO, suggesting that reduced IL-6 in PRMT1 MKO contributes to the protection from alcohol-induced HCC development.
MATERIALS AND METHODS
Mice
C57BL/6NTac-Prmt1tm1a(EUCOMM)Wtsi/WtsiCnbc mice were obtained from EUCOMM (EUCOMM project: 40181) and bred with Flp recombinase mice to get homozygous Prmt1 floxed breeders as described previously19. These mice were next crossed with C57B6/LysM-Cre mice (Stock No: 018956; Jackson Labs) to generate mice lacking Prmt1 in myeloid cells. For experiments, PRMT1fl/fl Cre/wt mice were used together with PRMTfl/fl wt/wt littermates as a control.
Mice were injected with 10 mg/kg of DEN intraperitoneally at 2 weeks of age. Mice were fed Lieber–DeCarli diet for 3–7 weeks as indicated, containing 4.8% alcohol (26% of calories from alcohol) or control liquid diet.
All mice were housed in a temperature-controlled, specific pathogen-free environment with 12-h light–dark cycles. All animal handling procedures were approved by the Institutional Animal Care and Use Committees at the University of Kansas Medical Center (Kansas City, KS, USA).
Primary Antibodies
Anti-β-catenin, anti-PCNA, and anti-phospho-STAT3 antibodies were from Cell Signaling. Anti-Mrc-1 antibodies were from Santa Cruz Biotechnology. Rabbit anti-PRMT1 antibody (against aa 300–361) and anti-F4/80 antibody were from Abcam. Mouse anti-β-actin antibodies were from Sigma-Aldrich (St. Louis, MO, USA). Anti-GAPDH was from Ambion.
Secondary Antibodies
IRDye 800CW goat anti-mouse IgG and IRDye 680RD goat anti-rabbit IgG were from Li-COR. General HRP-conjugated secondary antibodies were from Southern Biotechnology Associates (Birmingham, AL, USA).
Cell Culture
Huh7.5 cells20 (obtained from Dr. Charles Rice) were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS, 50 U ml−1 penicillin, and 50 mg ml−1 streptomycin. THP-1 cells were obtained from InvivoGen and maintained according to the manufacturer’s instructions. THP-1 cells were differentiated in the presence of 25 nM PMA. To induce efferocytosis, apoptotic cells were added for 1 h.
AMI-1 was obtained from EMD4 Biosciences and used at 10 μM for 16–24 h. Cytochalasin D was obtained from Sigma-Aldrich and used at 1 μM.
Liver Macrophage Isolation
Liver nonparenchymal cells were freshly isolated from mouse livers by the Cell Isolation Core of the Department of Pharmacology at the University of Kansas Medical Center as described previously21. Macrophages were purified using anti-F4/80 specific magnetic beads (Miltenyi Biotec) according to the manufacturer’s instructions.
Efferocytosis Assay
Apoptotic cells (ACs) were collected by centrifugation, stained with DAPI, and washed with PBS. Equal amounts of ACs were added to macrophages and incubated for 1 h at 37°C. Cells were washed, fixed with 4% paraformaldehyde for 15 min at room temperature, and stained with phalloidin. Coverslips were washed and mounted with FluorSave Reagent (Calbiochem, La Jolla, CA, USA). Slides were observed in a Nikon Eclipse 800 upright epifluorescence microscope (Nikon Instruments, Melville, NY, USA). Images were acquired using a Nikon CoolSNAP camera.
Human Specimens
Deidentified human specimens were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human tissue samples were approved by the Human Subjects Committee of the University of Kansas Medical Center.
Real-Time PCR
RNA was extracted from cultured cells using the RNeasy Mini Kit (Qiagen). cDNA was generated using the RNA reverse transcription kit (Cat. No. 4368814; Applied Biosystems). Quantitative real-time RT-PCR was performed in a CFX96 Real time system (Bio-Rad) using specific sense and antisense primers combined with iQ SYBR Green Supermix (Bio-Rad) for 40 amplification cycles: 5 s at 95°C, 10 s at 57°C, 30 s at 72°C. Human primers were as follows: actin B, gtgctcgatggggtacttcag, tgatggtgggcatgggtcag; PRMT1, caggcggaaagcagtgagaa, gatgccaaagtgtgcgtagg; IL-6, tgaggagacttgcctggtga, cacagctctggcttgttcct; mouse primers, IL-6, ttccatccagttgccttctt, cagaattgccattgcacaac; IL-10, ggttgccaagccttatcgga, acctgctccactgccttgct; TNFa, aggctctggagaacagcacat, tggcttctcttcctgcaccaaa; Mrc1, tggaatcaagggcacagaagt, ctccatttgcattgcccagt; F4/80, ctttggctatgggcttccagtc, gcaaggaggacagagtttatcgtg; Nlrp1b, gccatgggcaattgaactcg, acagtcaggagccaatccca; Nlrp3 gacccacagtgtaacttgcag, cacagaggtcagagctgaacaa; Defb14, aataagaggtggccggtgtg; tttcggcagcattttcgacc; IL-12a, gtgtcttagccagtcccgaa, tcagtttttctctggccgtctt; cyclinB1, cagagttctgaacttcagcctg, ttgtgaggccacagttcaccat; FOXM1, acttggattgaggaccacttcc, gacgaatagagttcttccagcct; GAPDH, cgtcccgtagacaaaatggt, ttgaggtcaatgaaggggtc.
ELISA
ELISA for mouse cytokines was performed using IL-6, TNFα, IL-1β, and AFP ELISA Ready-SET-Go!® kits from Affymetrix/eBioscience according to the manufacturer’s instructions.
Western Blots
Protein extracts (15 μg) were subjected to 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), electrophoretically transferred to nitrocellulose membranes (Amersham Hybond ECL, GE Healthcare), and blocked in 3% BSA/PBS at RT for 1 h. Primary antibodies were incubated overnight at manufacturer-recommended concentrations. Immunoblots were detected with the ECL Plus Western Blotting Detection System (Amersham Biosciences, Piscataway, NJ, USA) or using near-infrared fluorescence with the ODYSSEY Fc, Dual-Mode Imaging system (Li-COR). Expression levels were evaluated by quantification of relative density of each band normalized to that of the corresponding β-actin or GAPDH band density.
Immunohistochemistry
Immunostaining on formalin-fixed sections was performed by deparaffinization and rehydration, followed by antigen retrieval by heating in a pressure cooker (121°C) for 5 min in 10 mM sodium citrate, pH 6.0. Peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 10 min Sections were rinsed three times in PBS/PBS-T (0.1% Tween-20) and incubated in Dako Protein Block (Dako) at room temperature for 1 h. After removal of blocking solution, slides were placed into a humidified chamber and incubated overnight with an antibody, diluted 1:300 in Dako Protein Block at 4°C. Antigen was detected using the SignalStain Boost IHC detection reagent (Cat. No. 8114; Cell Signaling Technology, Beverly, MA, USA), developed with diaminobenzidene (Dako, Carpinteria, CA, USA), counterstained with hematoxylin (Sigma-Aldrich), and mounted. Signal intensity was analyzed by Aperio ImageScope 12.1.
Statistics
Results are expressed as mean ± SD. The Student t-test, paired t-test, Pearson’s correlation, or one-way ANOVA with Bonferroni post hoc test was used for statistical analyses. A value of p < 0.05 was considered significant.
RESULTS
Myeloid PRMT1 Knockout Mice Have Reduced Levels of M2 Macrophages and Anti-Inflammatory Cytokines After 3 Weeks of Alcohol Feeding
Our previous studies showed that loss of PRMT1 in myeloid cells results in a more proinflammatory phenotype in response to infection19. As alcohol is known to induce both proinflammatory and anti-inflammatory genes in liver macrophages, we aimed to test whether macrophage PRMT1 plays any role in either of these effects. To do so we used myeloid-specific PRMT1 MKO together with their WT PRMT1 floxed littermates described previously19. We put mice on a Lieber–DeCarli liquid diet containing 6.4% alcohol for 3 weeks. We found no significant differences between WT and MKO histologically (Fig. 1A), and no difference in serum ALT (Fig. 1B).
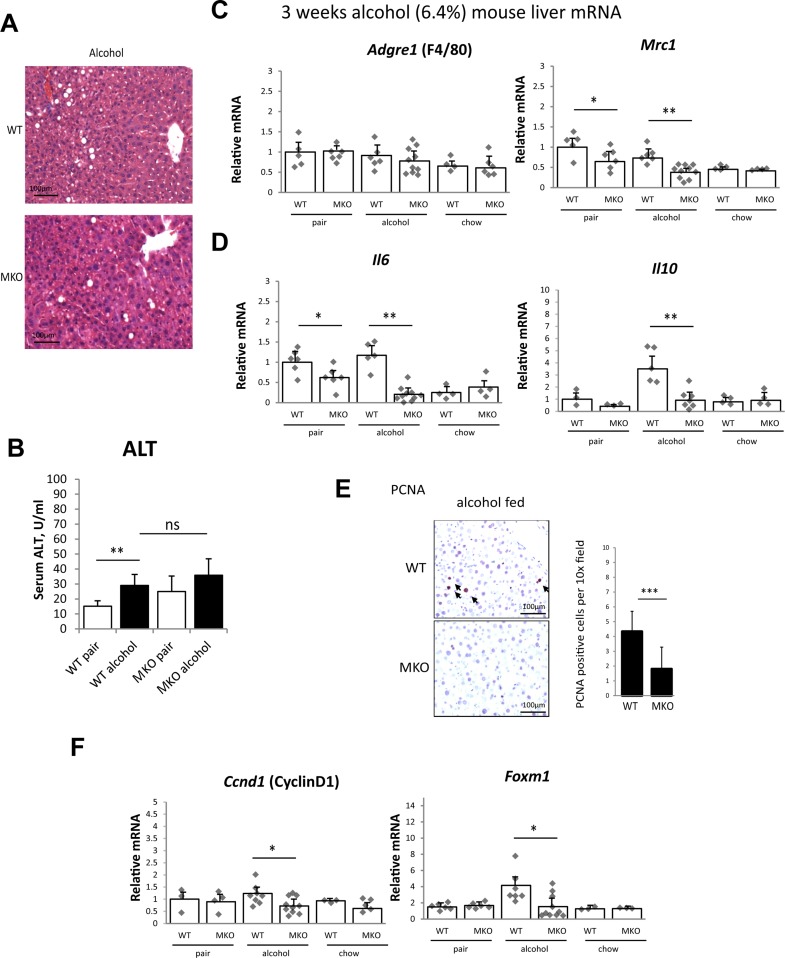
Figure 1.
Myeloid protein arginine methyltransferase 1 (PRMT1) knockout mice (MKO) have reduced levels of M2 macrophages and anti-inflammatory cytokines after 3 weeks of alcohol feeding. PRMT1 flox/flox (WT) and PRMT1 flox/flox LysM Cre (MKO) littermates (8–10 weeks old ) were fed Lieber–DeCarli alcohol liquid diet (alcohol), control liquid diet (pair) for 3 weeks, or left on chow diet (chow). (A) Representative images of H&E staining. Scale bars: 100 μm. (B) Serum ALT levels. Data presented as average ± SD, n = 6–10 animals per group. **p < 0.01. (C, D) Liver mRNA levels. n = 4–6 animals per group. *p < 0.05; **p < 0.01. (E) Representative images of PCNA staining. Right: average number of PCNA per 10× field. Data presented as average ± SD, n = 4 animals per group. ***p < 0.001. (F) Liver mRNA levels. n = 4–6 animals per group. *p < 0.05.
Next, we assessed macrophage phenotype and cytokine expression in the liver. We found that PRMT1 MKO have significantly lower levels of Mrc1 expression either on alcohol or on control liquid diet (pair fed) (Fig. 1C), while chow fed animals showed no difference between genotypes similar to what we described previously19. These data suggest that PRMT1 MKO mice are protected from liquid diet-induced increase in M2 macrophage polarization. PRMT1 MKO mice similarly have significantly lower levels of IL-6 expression either on alcohol or on control liquid diet and IL-10 expression in alcohol liquid diet group (Fig. 1D).
Alcohol effects on the liver are complex. Alcohol increases the number of proliferating hepatocytes, which is thought to play a role in alcohol effect on tumor growth15,16. We found that the increase in hepatocyte proliferation induced by alcohol is mediated by PRMT1 in myeloid cells. PRMT1 MKO mice fed alcohol had significantly lower number of PCNA+ hepatocytes in the liver compared to WT mice (Fig. 1E) and were protected from alcohol-induced increases in cyclin D and FoxM1 expression (Fig. 1F).
Myeloid PRMT1 Controls Tumor Formation in DEN–Alcohol Model of Liver Tumorigenesis
We next tested PRMT1 MKO mice in a model of alcohol-associated HCC adapted from Ambade et al.16 and Yan et al.18. Briefly, we injected WT and MKO littermates at 2 weeks of age with 10 mg/kg of DEN. At 3 month of age mice were put either on control or alcohol Lieber–DiCarli liquid diet (4.8% alcohol) for 7 weeks (Fig. 2A). As expected, in WT mice, alcohol promoted tumor development (Fig. 2B). We found that alcohol increased the number of surface tumors in female and male mice; however, the increase was significant only in male mice. In contrast, PRMT1 MKO mice were protected from the alcohol-induced increase in tumor number (Fig. 2B).
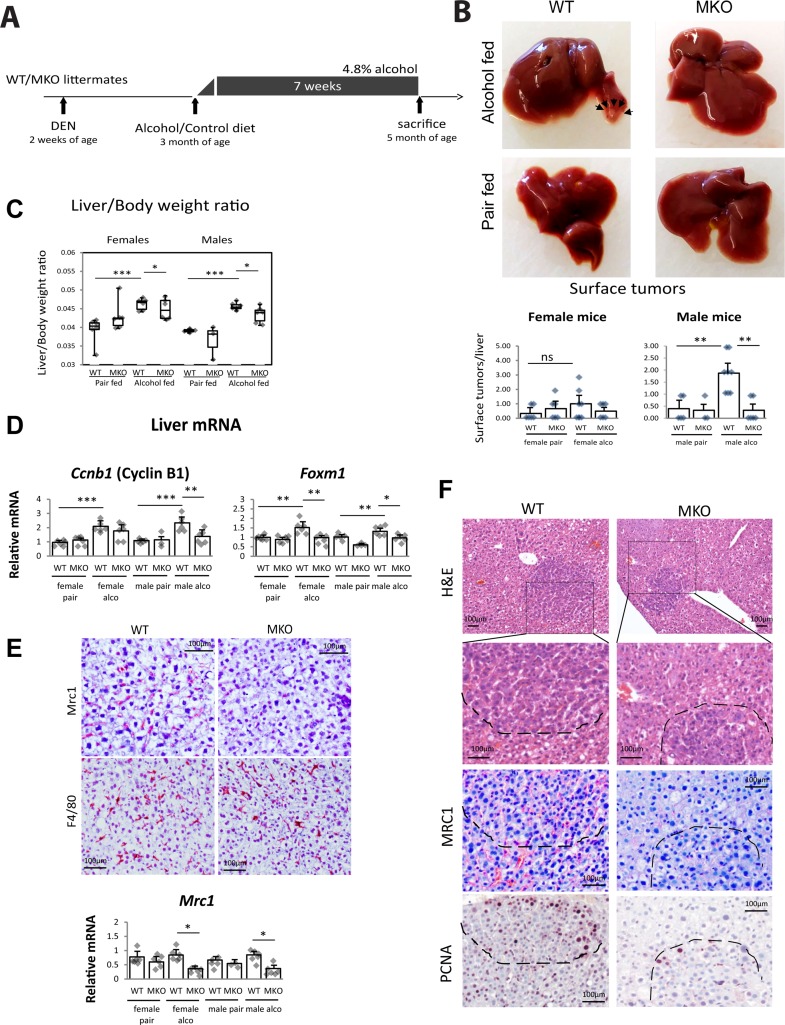
Figure 2.
Myeloid PRMT1 controls tumor formation in DEN–alcohol model of liver tumorigenesis. PRMT1 flox/flox (WT) and PRMT1 flox/flox LysM Cre (MKO) littermates received a single injection of 10 mg/kg of DEN at 2 weeks of age. At 3 months of age mice were randomly separated into two groups and fed Lieber–DeCarli alcohol liquid diet (4.8% alcohol) or control liquid diet (pair) for 7 weeks. Each group had six males and six female mice. (A) Diagram shows the experiment protocol. (B) Surface tumor images and number. **p < 0.01. (C) Liver/body weight ratios. *p < 0.05; ***p < 0.001. (D) Relative liver mRNA. *p < 0.05; **p < 0.01; ***p < 0.001. (E) Immunohistochemistry staining for Mrc1 (CD206) and F4/80. Scale bars: 100 μm. Bottom. Relative liver mRNA of Mrc1. *p < 0.05. (F) Representative images of nodules in WT and myeloid-specific PRMT1 MKO fed alcohol. H&E staining, Mrc1 staining, and PCNA staining in consecutive sections. Scale bars: 100 μm.
PRMT1 MKO mice had significantly lower liver/body weight ratio on alcohol diet compared to WT mice (Fig. 2C) and reduced expression of cyclin B1 and FoxM1 in these livers (Fig. 2D).
We found that alcohol-fed PRMT1 MKO mice had a reduced number of Mrc1+ cells (M2 macrophages) but no difference in total number of macrophages in the liver (Fig. 2E). We confirmed that lesions present in WT mice were infiltrated with Mrc1+ cells, while MKO lesions were mostly Mrc1−. This difference corresponded to reduced number of PCNA+ cells within lesions (Fig. 2F).
PRMT1 Knockout in Myeloid Cells Results in More Proinflammatory Phenotype Compared to Wild-Type Mice
Alcohol increased serum AFP levels in both female and male mice; we found no difference between WT and MKO mice in serum AFP levels after alcohol (Fig. 3A).
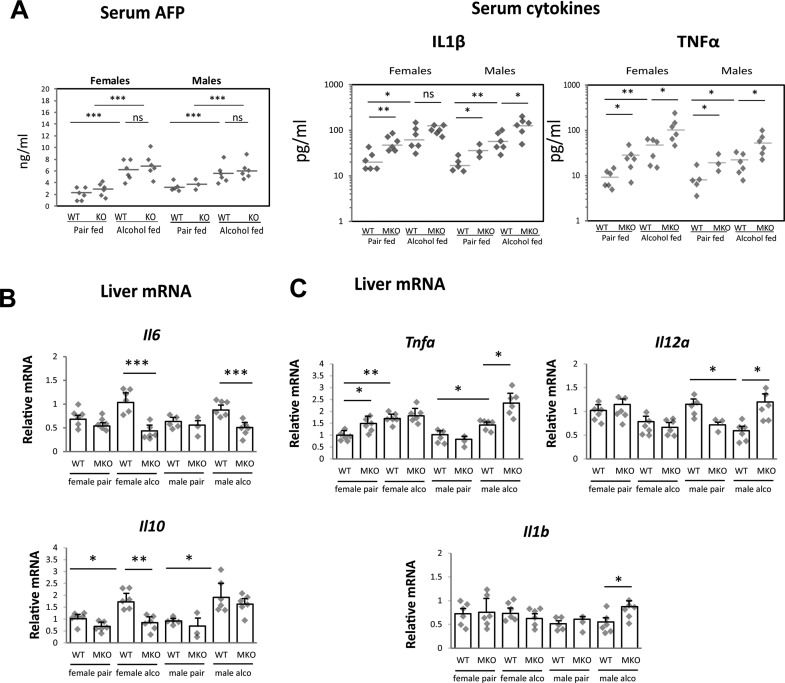
Figure 3.
PRMT1 knockout in myeloid cells results in more proinflammatory phenotype compared to wild-type (WT) mice. PRMT1 flox/flox (WT) and PRMT1 flox/flox LysM Cre (MKO) littermates received a single injection of 10 mg/kg of DEN at 2 weeks of age. At 3 month of age, mice were randomly separated into two groups and fed Lieber–DeCarli alcohol liquid diet (4.8% alcohol) or control liquid diet (pair) for 7 weeks. Each group had six males and six female mice. Serum cytokine and AFP levels (A) and relative liver mRNA levels (B, C) in these mice.
Next, we studied serum cytokine levels in WT and MKO in more detail. We found that serum levels of TNF-α and IL-1β were significantly higher in MKO mice compared to WT littermates (Fig. 3A). In the liver we found a similar situation. Although there were gender differences, MKO mice had less anti-inflammatory (Fig. 3B) and more proinflammatory cytokines (Fig. 3C). These data suggest that PRMT1 MKO mice have enhanced proinflammatory signaling similar to what were described before19.
Macrophage PRMT1 Controls Expression of IL-6 and IL-10 and Downstream STAT3 Activation in Hepatocytes
To elucidate which of these cytokine changes are due to PRMT1 knockout in liver macrophages, we isolated total liver macrophages using anti-F4/80 magnetic beads from alcohol-fed mice and studied gene expression changes using a PCR array for immune response genes (Bio-Rad). We found that PRMT1 KO macrophages had significantly less IL-6, IL-10, and NLRP1b as well as other inflammasome pathway genes IL-1B and NLRP3 (Fig. 4A). In contrast PRMT1 KO macrophages had significantly increased expression of IL-12 and antimicrobial peptide gene Defb14 (Fig. 4A). By comparing isolated macrophage gene expression data with total liver mRNA data presented in Figures 1 and 3, we concluded that liver macrophages are a major source of total hepatic IL-6 and IL-10. Additionally, IL-6 and IL-10 production by macrophages is PRMT1 dependent. As the effect of PRMT1 knockout on IL-6 expression was higher in magnitude compared to the effect on IL-10 expression we focused further studies on IL-6. IL-6 is a potent hepatocyte mitogen; activation of the IL-6 signaling pathway is a major contributor in the development of liver tumors22. We hypothesized that reduced IL-6 levels in PRMT1 MKO result in reduced proliferation within the liver and reduced tumor number.
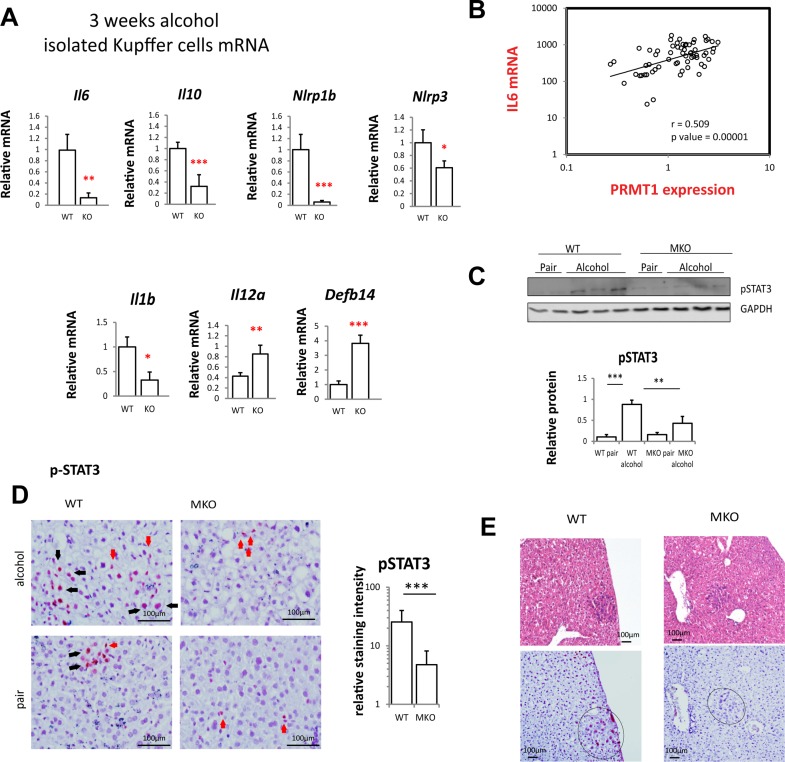
Figure 4.
Kupffer cell PRMT1 controls expression of IL-6 and IL-10 and downstream STAT3 activation in hepatocytes. (A) PRMT1 flox/flox and PRMT1 flox/flox LysM Cre mice were fed alcohol for 3 weeks. Kupffer cells WT or KO were isolated from nonparenchymal cell fraction using anti-F4/80 beads. Relative mRNA levels in freshly isolated cells. Data presented as average ± SD, n = 3 animals per group. *p < 0.05; **p < 0.01; ***p < 0.001. (B) Correlation between PRMT1 mRNA and IL-6 mRNA expression in human blood monocytes (CD14+) of patients. N = 62. r = 0.508; p = 0.00001. (C) Relative pSTAT3 (Y705) levels in whole liver nuclear extracts from mice as in Figure 2. n = 6 mice per group. **p < 0.01; ***p < 0.001. (D) Immunohistochemistry staining of pSTAT3 in liver section from mice as in Figure 2. Black arrows, positive hepatocytes; red arrows, positive nonparenchymal cells. Scale bars: 100 μm. Right: pSTAT3 staining in hepatocytes was quantified using Aperio Image Scope. Data presented as average ± SD, n = 4 animals per group. ***p < 0.001. (E) Representative images of immunohistochemistry staining of pSTAT3 in liver section from mice as in Figure 2 showing a proliferating nodule. Scale bars: 100 μm.
We found a strong correlation between PRMT1 expression and IL-6 expression in human blood monocytes from n = 91 patients with liver disease presenting to the University of Kansas Hospital between January and June 2015 (Fig. 4B), suggesting that PRMT1-dependent IL-6 expression is relevant in humans.
We then tested whether PRMT1-dependent IL-6 expression activates its downstream effector STAT3. We found that alcohol feeding results in about a fivefold increase in pSTAT3 in the whole liver, while MKO mice had reduced levels of alcohol-induced pSTAT3 (Fig. 4C). We analyzed localization of pSTAT3 in the liver by immunohistochemistry. We found that alcohol-induced pSTAT3 nuclear staining both in hepatocytes as well as in nonparenchymal cells in WT mice. However, in PRMT1 MKO mice pSTAT3 was present mostly in nonparenchymal cells (Fig. 4D), while in hepatocytes pSTAT3 staining intensity was fourfold lower than in WT mice (Fig. 4D). Similarly, pSTAT3 staining was higher in the nodules from WT compared to MKO mice (Fig. 4E).
Macrophage Polarization by Apoptotic Cells Requires PRMT1 and PPARγ
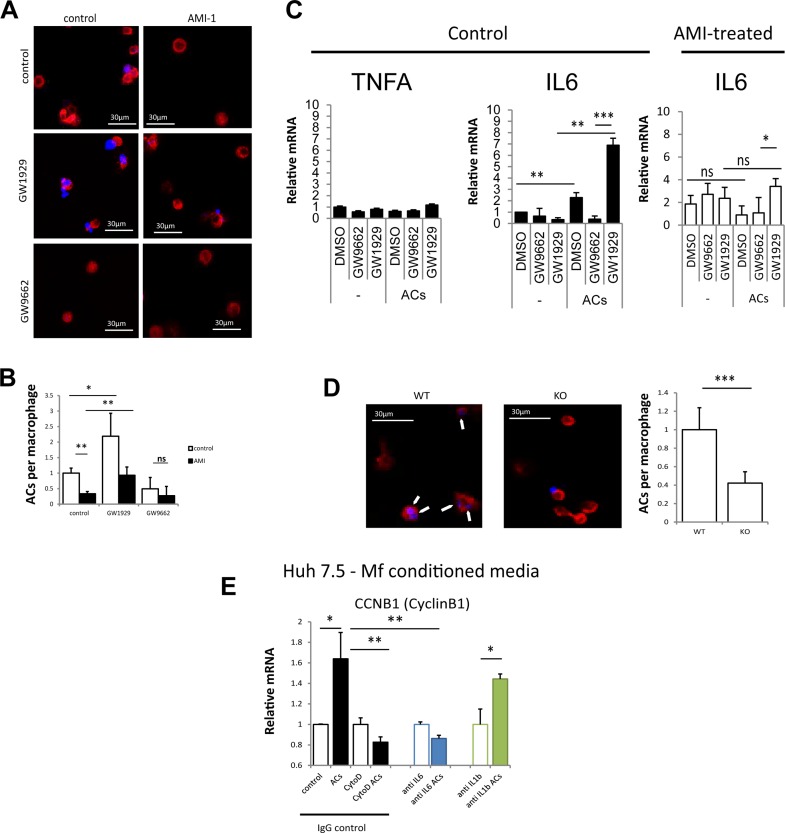
We next investigated the mechanism of PRMT1-dependent IL-6 regulation in alcohol-fed mice. IL-6 can be induced by LPS as well as by M2 signals such as IL-4 or apoptotic cells22–24. It is generally accepted that Kupffer cells get polarized by dead cells (hepatocytes) to produce IL-6 and promote hepatocyte proliferation22. Macrophage efferocytosis, or phagocytosis of apoptotic cells, promotes M2 polarization and tumor progression25. Efferocytosis is a PPARγ-dependent process26. Figure 5A shows that THP-1 macrophages show increased efferocytosis ability in the presence of the PPARγ agonist GW1929. In contrast, the PPARγ antagonist GW9662 blocked the uptake of apoptotic cells. THP-1 macrophages treated with the PRMT1 inhibitor AMI-1 showed reduced ability to take in apoptotic bodies (Fig. 5A and B). In response to apoptotic cells, macrophages polarize toward a more M2-like phenotype with increased expression of IL-6 but not TNFα. AMI-1-treated macrophages show reduced ability to induce IL-6 in response to apoptotic cells (Fig. 5C).
Figure 5.
Macrophage polarization by apoptotic cells requires PRMT1 and PPARγ. (A) THP-1 cells were differentiated into macrophage in the presence or absence of PRMT1 inhibitor AMI-1. Macrophages labeled with phalloidin-568 (red) were treated with apoptotic cells (UV-irradiated hepatocytes) labeled with DAPI (blue) for 1 h in the presence of PPARγ agonist GW1929 or PPARγ antagonist GW9662 where indicated. Scale bars: 30 μm. (B) Average number of phagocytosed apoptotic cells (ACs) per macrophage as in (A) in n = 3 independent experiments. Data presented as average ± SD. *p < 0.05; **p < 0.01. (C) THP-1 monocytes as in (A). Relative mRNA levels after 24 h of ACs in n = 3 independent experiments. Data presented as average ± SD. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Efferocytosis assay as in (A) using isolated macrophages from WT or MKO mice. Arrows indicate internalized apoptotic cells. Scale bars: 30 μm. ***p < 0.001. (E) THP-1 cells were treated with ACs as in (A) in the presence or absence of cytochalasin D. Conditioned media was added to Huh 7.5 cells in the presence of anti-IL-6, anti-IL-1β, or control IgG. Relative mRNA levels of cyclin B1 in n = 3 independent experiments. Data presented as average ± SD *p < 0.05; **p < 0.01.
We confirmed that PRMT1 KO macrophages isolated from myeloid-specific MKO have reduced ability for efferocytosis (Fig. 5D). In contrast to WT macrophages, KO macrophages had dramatically lower ability to uptake apoptotic cell debris (Fig. 5D).
Finally, we tested whether IL-6 from macrophages incubated with apoptotic cells can induce hepatocellular proliferation. We found that conditioned media from apoptotic cell-treated macrophages induced cyclin B1 expression in Huh 7.5 cells. This effect was blocked if apoptotic cells were added in the presence of cytochalasin D, a blocker of efferocytosis, or in the presence of IL-6 neutralizing antibody, but not IgG control (Fig. 5E).
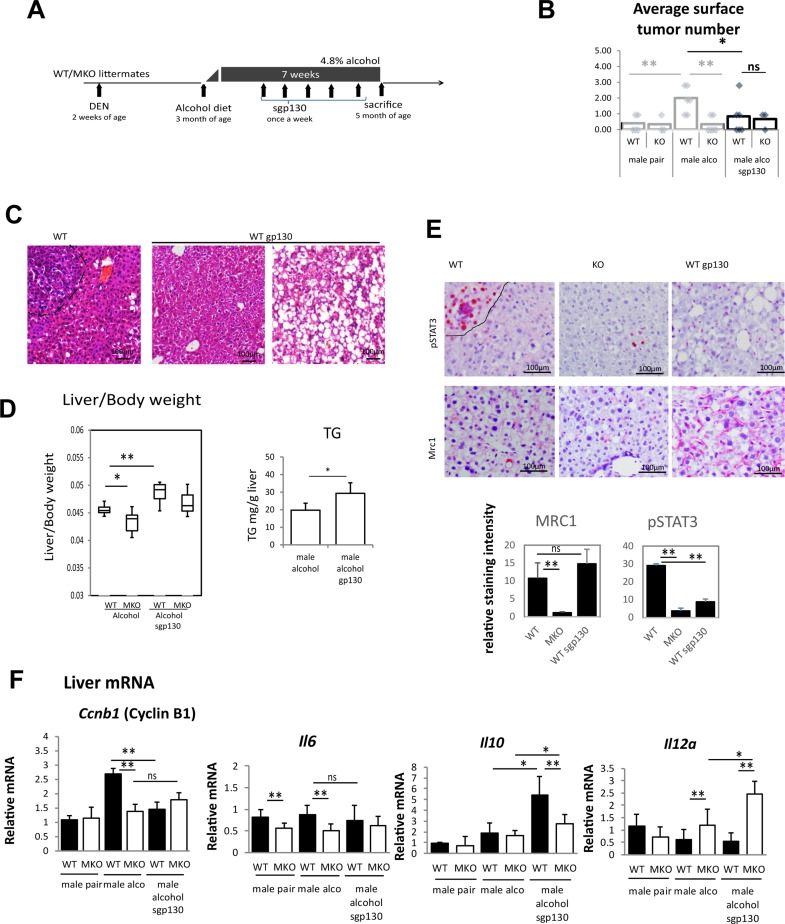
Blocking IL-6 Signaling in Mice Reduces Tumor Formation in the DEN–Alcohol Model of Liver Tumorigenesis
To test the role of PRMT1-dependent IL-6 signaling in alcohol-associated tumor development, we treated mice with the soluble IL-6R blocker gp13024. We gave sgp130 injections to WT and PRMT1 MKO male mice starting 2 weeks after beginning alcohol diet (Fig. 6A). This treatment reduced the number of surface tumors in WT mice (Fig. 6B). Interestingly, sgp130 promoted lipid accumulation in the liver (Fig. 6C), which resulted in higher liver/body weight ratio (Fig. 6D) due to higher TG content. These data are in accordance with previously published results on the role of IL-6 in alcohol-induced steatosis27.
Figure 6.
Blocking IL-6 signaling in mice reduces tumor formation in DEN–alcohol model of liver tumorigenesis. Male PRMT1 flox/flox (WT) and PRMT1 flox/flox LysM Cre (MKO) littermates received a single injection of 10 mg/kg of DEN at 2 weeks of age. At 3 months of age, mice were fed Lieber–DeCarli alcohol liquid diet (4.8% alcohol) for 7 weeks. Starting at week 3, mice received weekly injections of soluble gp130 protein. n = 6 mice per group. (A) Diagram shows the experiment protocol. (B) Surface tumors number. *p < 0.05; **p < 0.01; ns: p > 0.05. (C) H&E staining of liver sections showing normal area and steatosis area. Scale bars: 100 μm. (D) Left: liver/body weight ratios. Right: average TG content in male mice on alcohol untreated or treated with sgp130. *p < 0.05; **p < 0.01. (E) Immunohistochemistry staining for Mrc1 (CD206) and pSTAT3. Scale bars: 100 μm. Bottom: Relative staining intensity. n = 4 mice per group. **p < 0.01; ns: p > 0.05. (F) Relative liver mRNA. *p < 0.05; **p < 0.01; ns: p > 0.05.
We confirmed that sgp130 treatment reduced hepatocyte pSTAT3 activation without affecting the number of Mrc1+ cells in the liver (Fig. 6E). Treatment with sgp130 prevented the alcohol-induced increase in proliferation (Fig. 6F). Interestingly sgp130 treatment did not affect IL-6 expression in the liver, but significantly increased levels of IL-10 in WT and MKO mice, as well as IL-12 levels in MKO mice.
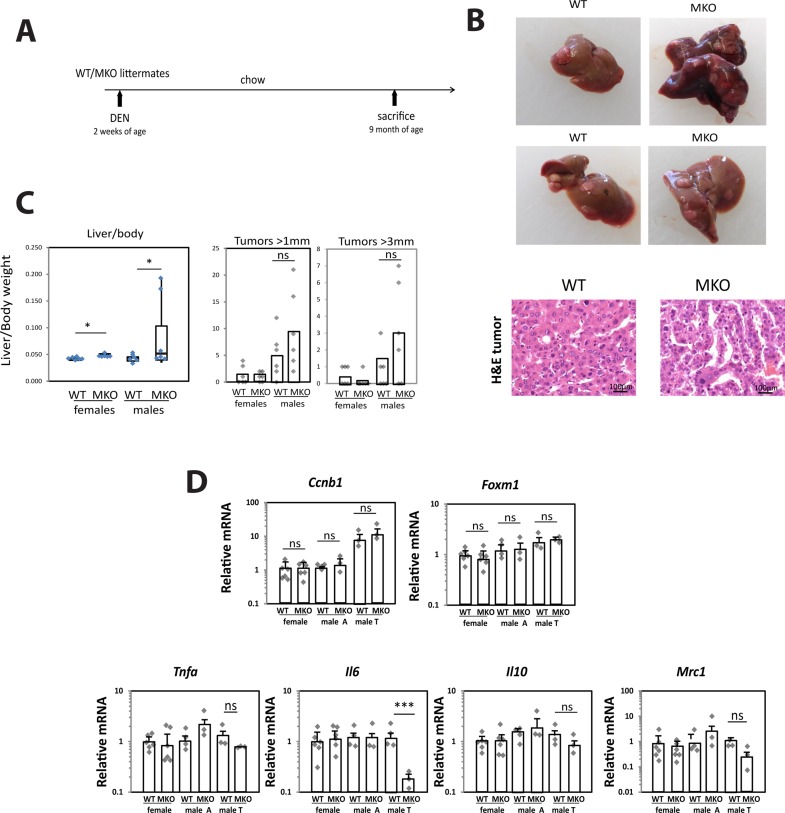
DEN Treatment Alone Promotes Tumor Formation in PRMT1-Independent Manner
To test whether the effect of PRMT1 KO is alcohol specific, we compared PRMT1 WT and MKO mice that had been injected with DEN at 2 weeks of age and left on chow diet for 9 months (Fig. 7A). In contrast to the situation in which tumors were promoted by alcohol, we found that in the absence of alcohol tumors developed at a later time, but PRMT1 MKO mice had similar numbers of tumors as WT mice (Fig. 7B). In contrast to the alcohol-fed group, MKO mice on chow had higher liver/body weight ratios compared to WT mice and larger (>3 mm) surface tumors in male mice, although the difference was not significant (Fig. 7C). We isolated mRNA from livers of these mice, and in male mice we isolated tumor and adjacent tissue mRNA. We found that WT and MKO mice had similar expression levels of cyclin B, FoxM1, and Mrc1. Interestingly, we found that although IL-6 expression was not affected by PRMT1 in the nontumor liver, in tumor tissues from MKO mice IL-6 expression was 10-fold lower than in tumors from WT mice (Fig. 7D).
Figure 7.
DEN treatment alone promotes tumor formation in PRMT1-independent manner. PRMT1 flox/flox (WT) and PRMT1 flox/flox LysM Cre (MKO) littermates received a single injection of 10 mg/kg of DEN at 2 weeks of age. Mice were sacrificed at 9 months of age. Each group had six males and six female mice. (A) Diagram shows the experiment protocol. (B) Surface tumors images and H&E staining of tumor tissue. Scale bars: 100 μm. (C) Liver/body weight ratios and tumor number. *p < 0;05; ns: p > 0.05. (D) Relative liver mRNA and adjacent (A) or tumor (T) tissue mRNA where indicated. ***p < 0.001; ns: p > 0.05.
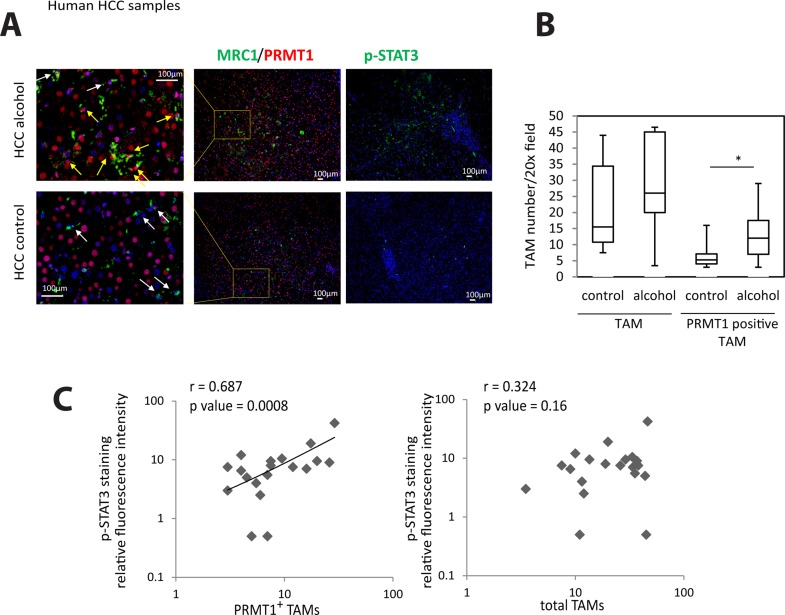
PRMT1 Expression in TAMs Correlates With STAT3 Activation in Human HCC
Finally, we studied whether the PRMT1–IL-6–STAT3 pathway is relevant in humans. We obtained human HCC specimens from patients undergoing liver transplantation for HCC. We hypothesized that PRMT1 expression in TAMs will correlate with STAT3 activation in these tumors. We costained tumor-associated macrophages using Mrc1 and PRMT1 since 98% of TAMs are MRC1+ 28. We found that PRMT1+ TAMs were significantly more numerous in HCCs associated with alcohol compared to HCCs not associated with alcohol (Fig. 8A and B). Notably, the number of PRMT1+ TAMs correlated with pSTAT3 staining intensity in these specimens, while the total number of TAMs did not correlate with STAT3 activation (Fig. 8C).
Figure 8.
PRMT1 expression in TAMs correlates with STAT3 activation in human hepatocellular carcinoma (HCC). (A) Immunofluorescence staining of human HCC sections using anti-Mrc1, anti-PRMT1, and anti-pSTAT3 antibodies. Scale bars: 100: μm. (B) Average number of Mrc1+/PRMT1+ cells per 20× field or number of Mrc1+ cells per 20× field in HCC sections of control HCC (n = 15) or HCC associated with alcohol (n = 5). *p < 0.05. (C) pSTAT3 staining intensity plotted versus average number of Mrc1+/PRMT1+ cells per 20× field or total number of Mrc1+ cells per 20× field. N = 20.
DISCUSSION
Alcohol abuse is a significant risk factor for liver diseases. About 40% of individuals who chronically consume alcohol develop fatty liver, which can advance to alcoholic hepatitis, cirrhosis, and HCC. Epidemiological data have suggested that alcohol abuse contributes to HCC development. Alcohol interacts with other causes of liver disease, including hepatitis B and C, and conditions, such as diabetes and obesity, to increase the risk for developing HCC, either synergistically or additively. HCC risk in patients with ALD-associated cirrhosis increases with age and with quantity and duration of alcohol consumption29.
Recently, several mouse models were developed to study the mechanism of alcohol effects on cancer development in the liver. All models involved a combination of one or multiple injections of DEN with alcohol feeding of mice for 6–16 weeks15,16,18. Results of these studies suggested that alcohol promotes HCC progression in mice via increasing hepatocyte proliferation15,16, increasing inflammation, increasing tumor-associated macrophage infiltration16,18, reducing antitumor CD8+ T cells18, exacerbating fibrosis16, and promoting EMT16,18. However relative contributions of these pathways are unknown.
In our study, we employed a single injection of DEN at 2 weeks of age followed by 7 weeks of Lieber–DeCarli alcohol feeding beginning at 3 months of age. The Lieber–DeCarli feeding model is an accepted model of ALD that results in features of human ALD including steatosis, inflammation, and liver fibrosis30. Seven weeks of feeding was sufficient to induce an increase in serum AFP of threefold over pair-fed control mice and induce visible surface tumors in 50% of female and 100% of male mice.
Interestingly, we did not find a difference in serum AFP levels between male and female mice even though the number of surface tumors was twofold lower in females compared to males. AFP is an oncofetal protein expressed by neoplastic hepatocytes but not in normal adult hepatocytes. An increase in serum AFP level is a convenient indicator of HCC progression. However, it cannot be used to compare HCC development between male and female mice since there are many other factors that control tumor number under these conditions. The FGF19 transgenic mouse is another example when AFP levels are elevated to similar extent in male and female mice, while at the same time HCC incidence is fivefold higher in females31. These findings combined with our results suggest that gender differences cannot be explained by different levels of AFP expression.
HCC is an inflammation-driven cancer, and alcohol exposure exacerbates the inflammation and protumor immune environment18. Many observations indicate that tumor-associated macrophages (TAMs) express several M2-associated protumor functions, including promotion of angiogenesis, matrix remodeling, and suppression of adaptive immunity. To investigate the role of macrophage polarization, specifically M2 polarization, in alcohol-associated HCC development, we used previously the developed myeloid-specific PRMT1 MKO model. PRMT1 MKO mice are lacking PPARγ-dependent M2 or anti-inflammatory macrophage polarization8,9.
In the current study, we demonstrated that myeloid-specific PRMT1 MKO are protected from alcohol-induced increases in tumor development. PRMT1 MKO mice had significantly lower levels of IL-6 and IL-10 in the livers, a reduced number of Mrc1+ cells and were protected from alcohol-induced increases in hepatocellular proliferation.
IL-6 is induced in the livers of alcohol-fed mice most likely through exposure of macrophages to apoptotic cells (hepatocytes) as described previously22. We found that this process, efferocytosis, is lacking in PRMT1 knockout macrophages resulting in sixfold lower expression of IL-6 in liver macrophages isolated from alcohol-fed MKO compared to WT cells. In accordance with these data, PRMT1 MKO had fourfold lower levels of pSTAT3 in hepatocytes after alcohol feeding.
IL-10 is another cytokine involved in STAT3 activation, which can contribute to tumor formation in our model. Previously, we found that IL-10 is regulated by PRMT1 through histone arginine methylation of the IL-10 gene promoter9. However, in the current study, we found that the change in IL-10 expression in the livers of male mice was not significantly different between WT and MKO and could not explain the change in surface tumor number and proliferation.
We tested the role of IL-6 signaling in alcohol-associated cancer progression using an IL-6 receptor blocker, gp13024. WT mice receiving sgp130 were partially protected from alcohol-induced tumor development, suggesting that this can be one of the potential mechanisms of enhanced tumor progression.
Taken together, our data suggest that PRMT1-dependent macrophage polarization might be a promising target to prevent alcohol-associated HCC development.
Our results are in line with previous reports on the role of PRMT1 in cancer32–39. In addition, there is evidence that PRMT1 can contribute to the progression of liver fibrosis, cirrhosis, and HCC by several other mechanisms as well. Based on the previous studies, PRMT1 can contribute to fibrosis and inflammation and can directly and indirectly regulate fibroblast activities37. PRMT1 regulates the TGF-β pathway activation via arginine methylation of Smad7, which is an important contributor to progression of cancer and fibrosis40.
PRMT1 has been further associated with inflammation via epithelial and immune cell lines. Findings indicated both proinflammatory and anti-inflammatory functions of PRMT1. PRMT1 serves as an epigenetic cofactor that induces the expression of Cox27, PRMT1 regulates inflammation in a rat asthma model41–44, PRMT1 can modulate inflammation through CITED regulation45, PRMT1 directly modulates NF-κB activity7, and PRMT1 promotes chronic inflammation through regulation of CIITA46.
Using an in vivo model, we found that in liver myeloid cells PRMT1 promotes anti-inflammatory macrophage differentiation via regulation of PPARγ19, and the present results in the alcohol-HCC mouse model show that myeloid PRMT1 promotes a protumor immune environment. Further studies are necessary to define the cell type-specific roles of PRMT1 in liver disease progression, alcohol-induced liver injury, fibrosis, and cirrhosis.
ACKNOWLEDGMENTS
This study was supported by grants AA012863 from the National Institute on Alcoholism and Alcohol Abuse, COBRE Pilot grant NCRR/NIH P20 RR021940, and 2016 Pinnacle Award from AASLD. This work was supported in part by grants from the National Institute of General Medical Sciences (P20GM103549 and P30GM118247) of the National Institutes of Health.
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275(11):7723–30. [DOI] [PubMed] [Google Scholar]
- 2. Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell 2014;53(3):484–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrero MJ, Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol Cell 2006;24(2):233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell 2009;33(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chittka A. Dynamic distribution of histone H4 arginine 3 methylation marks in the developing murine cortex. PLoS One 2010;5(11):e13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 2004;117(6):735–48. [DOI] [PubMed] [Google Scholar]
- 7. Hassa PO, Covic M, Bedford MT, Hottiger MO. Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1. J Mol Biol. 2008;377(3):668–78. [DOI] [PubMed] [Google Scholar]
- 8. Tikhanovich I, Zhao J, Bridges B, Kumer S, Roberts B, Weinman SA. Arginine methylation regulates c-Myc-dependent transcription by altering promoter recruitment of the acetyltransferase p300. J Biol Chem. 2017;292(32):13333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tikhanovich I, Zhao J, Olson J, Adams A, Taylor R, Bridges B, Marshall L, Roberts B, Weinman SA. Protein arginine methyltransferase 1 modulates innate immune responses through regulation of peroxisome proliferator-activated receptor gamma-dependent macrophage differentiation. J Biol Chem. 2017;292(17):6882–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–27. [DOI] [PubMed] [Google Scholar]
- 11. Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, Forman D, Bray F. Global burden of cancer in 2008: A systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 2012;380(9856):1840–50. [DOI] [PubMed] [Google Scholar]
- 12. Sanyal AJ, Yoon SK, Lencioni R. The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist 2010;15(Suppl 4):14–22. [DOI] [PubMed] [Google Scholar]
- 13. Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109(4):542–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27(1):55–76. [DOI] [PubMed] [Google Scholar]
- 15. Mercer KE, Hennings L, Sharma N, Lai K, Cleves MA, Wynne RA, Badger TM, Ronis MJ. Alcohol consumption promotes diethylnitrosamine-induced hepatocarcinogenesis in male mice through activation of the Wnt/beta-catenin signaling pathway. Cancer Prev Res. (Phila) 2014;7(7):675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ambade A, Satishchandran A, Gyongyosi B, Lowe P, Szabo G. Adult mouse model of early hepatocellular carcinoma promoted by alcoholic liver disease. World J Gastroenterol. 2016;22(16):4091–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Szabo G, Petrasek J. Inflammasome activation and function in liver disease. Nat Rev Gastroenterol Hepatol. 2015;12(7):387–400. [DOI] [PubMed] [Google Scholar]
- 18. Yan G, Wang X, Sun C, Zheng X, Wei H, Tian Z, Sun R. Chronic alcohol consumption promotes diethylnitrosamine-induced hepatocarcinogenesis via immune disturbances. Sci Rep. 2017;7(1):2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tikhanovich I, Zhao J, Olson J, Adams A, Taylor R, Bridges B, Marshall L, Roberts B, Weinman SA. Protein arginine methyltransferase 1 modulates innate immune responses through regulation of peroxisome proliferator-activated receptor gamma-dependent macrophage differentiation. J Biol Chem. 2017;292(17):6882–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28(2):167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z, Zhao J, Tikhanovich I, Kuravi S, Helzberg J, Dorko K, Roberts B, Kumer S, Weinman SA. Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ. 2016;23(4):583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J Hepatol. 2016;64(6):1403–15. [DOI] [PubMed] [Google Scholar]
- 23. Liu Y, Lin J. Blocking the IL-6-STAT3 signaling pathway: Potential liver cancer therapy. Future Oncol. 2011;7(2):161–4. [DOI] [PubMed] [Google Scholar]
- 24. Hong J, Wang H, Shen G, Lin D, Lin Y, Ye N, Guo Y, Li Q, Ye N, Deng C, et al. Recombinant soluble gp130 protein reduces DEN-induced primary hepatocellular carcinoma in mice. Sci Rep. 2016;6:24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang M, Liu J, Piao C, Shao J, Du J. ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis. 2015;6:e1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Croasdell A, Duffney PF, Kim N, Lacy SH, Sime PJ, Phipps RP. PPARgamma and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015;2015:549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Assal O, Hong F, Kim WH, Radaeva S, Gao B. IL-6-deficient mice are susceptible to ethanol-induced hepatic steatosis: IL-6 protects against ethanol-induced oxidative stress and mitochondrial permeability transition in the liver. Cell Mol Immunol. 2004;1(3):205–11. [PubMed] [Google Scholar]
- 28. Shirabe K, Mano Y, Muto J, Matono R, Motomura T, Toshima T, Takeishi K, Uchiyama H, Yoshizumi T, Taketomi A, et al. Role of tumor-associated macrophages in the progression of hepatocellular carcinoma. Surg Today 2012;42(1):1–7. [DOI] [PubMed] [Google Scholar]
- 29. Joshi K, Kohli A, Manch R, Gish R. Alcoholic liver disease: High risk or low risk for developing hepatocellular carcinoma? Clin Liver Dis. 2016;20(3):563–80. [DOI] [PubMed] [Google Scholar]
- 30. Ghosh Dastidar S, Warner JB, Warner DR, McClain CJ, Kirpich IA. Rodent models of alcoholic liver disease: Role of binge ethanol administration. Biomolecules 2018;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nicholes K, Guillet S, Tomlinson E, Hillan K, Wright B, Frantz GD, Pham TA, Dillard-Telm L, Tsai SP, Stephan JP and others. A mouse model of hepatocellular carcinoma: Ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am J Pathol. 2002;160(6):2295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathioudaki K, Papadokostopoulou A, Scorilas A, Xynopoulos D, Agnanti N, Talieri M. The PRMT1 gene expression pattern in colon cancer. Br J Cancer 2008;99(12):2094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell 2008;31(2):212–21. [DOI] [PubMed] [Google Scholar]
- 34. Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, Arold ST, Chou CK, Tsou PH, Yamaguchi H, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125(12):4529–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng X, Von Keudell G, Suzuki T, Dohmae N, Nakakido M, Piao L, Yoshioka Y, Nakamura Y, Hamamoto R. PRMT1 promotes mitosis of cancer cells through arginine methylation of INCENP. Oncotarget 2015;6(34):35173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Avasarala S, Van Scoyk M, Karuppusamy Rathinam MK, Zerayesus S, Zhao X, Zhang W, Pergande MR, Borgia JA, DeGregori J, Port JD, et al. PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J Biol Chem. 2015;290(21):13479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuniati L, van der Meer LT, Tijchon E, van Ingen Schenau D, van Emst L, Levers M, Palit SA, Rodenbach C, Poelmans G, Hoogerbrugge PM, et al. Tumor suppressor BTG1 promotes PRMT1-mediated ATF4 function in response to cellular stress. Oncotarget 2016;7(3):3128–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chuang CY, Chang CP, Lee YJ, Lin WL, Chang WW, Wu JS, Cheng YW, Lee H, Li C. PRMT1 expression is elevated in head and neck cancer and inhibition of protein arginine methylation by adenosine dialdehyde or PRMT1 knockdown downregulates proliferation and migration of oral cancer cells. Oncol Rep. 2017;38(2):1115–23. [DOI] [PubMed] [Google Scholar]
- 39. Hsu JH, Hubbell-Engler B, Adelmant G, Huang J, Joyce CE, Vazquez F, Weir BA, Montgomery P, Tsherniak A, Giacomelli AO, et al. PRMT1-mediated translation regulation is a crucial vulnerability of cancer. Cancer Res. 2017;77(17):4613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katsuno Y, Qin J, Oses-Prieto J, Wang H, Jackson-Weaver O, Zhang T, Lamouille S, Wu J, Burlingame A, Xu J, et al. Arginine methylation of SMAD7 by PRMT1 in TGF-beta-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J Biol Chem. 2018;293(34):13059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Q, Yang X, Zhong B, Jiao F, Li C, Li D, Lan X, Sun J, Lu S. Upregulated protein arginine methyltransferase 1 by IL-4 increases eotaxin-1 expression in airway epithelial cells and participates in antigen-induced pulmonary inflammation in rats. J Immunol. 2012;188(7):3506–12. [DOI] [PubMed] [Google Scholar]
- 42. Sun Q, Liu L, Roth M, Tian J, He Q, Zhong B, Bao R, Lan X, Jiang C, Sun J and others. PRMT1 upregulated by epithelial proinflammatory cytokines participates in COX2 expression in fibroblasts and chronic antigen-induced pulmonary inflammation. J Immunol. 2015;195(1):298–306. [DOI] [PubMed] [Google Scholar]
- 43. Sun Q, Liu L, Mandal J, Molino A, Stolz D, Tamm M, Lu S, Roth M. PDGF-BB induces PRMT1 expression through ERK1/2 dependent STAT1 activation and regulates remodeling in primary human lung fibroblasts. Cell Signal. 2016;28(4):307–15. [DOI] [PubMed] [Google Scholar]
- 44. Sun Q, Liu L, Wang H, Mandal J, Khan P, Hostettler KE, Stolz D, Tamm M, Molino A, Lardinois D and others. Constitutive high expression of protein arginine methyltransferase 1 in asthmatic airway smooth muscle cells is caused by reduced microRNA-19a expression and leads to enhanced remodeling. J Allergy Clin Immunol. 2017;140(2):510–524 e3. [DOI] [PubMed] [Google Scholar]
- 45. Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36(10):3202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fan Z, Li J, Li P, Ye Q, Xu H, Wu X, Xu Y. Protein arginine methyltransferase 1 (PRMT1) represses MHC II transcription in macrophages by methylating CIITA. Sci Rep. 2017;7:40531. [DOI] [PMC free article] [PubMed] [Google Scholar]