Abstract
Adipose tissue homeostasis depends on interactions between stromal cells, adipocytes, and the cytokines and chemokines they produce. The gp130 cytokine, oncostatin M (OSM), plays a role in adipose tissue homeostasis. Mice, lacking the OSM receptor (OSMR) in adipocytes (OsmrFKO mice), exhibit derangements in adipose tissue, insulin sensitivity, and immune cell balance. Here, we describe a possible role for the chemokine stromal-derived factor 1 (SDF-1) in these alterations. We treated 3T3-L1 adipocytes with OSM and observed a suppression of SDF-1 gene expression and protein secretion, an effect which was partially blunted by OSMR knockdown. However, OsmrFKO mice also exhibited decreased SDF-1 gene and protein expression in adipose tissue. These contrasting results suggest that the loss of adipocyte OSM–OSMR signaling in vivo may be indirectly affecting adipokine production and secretion by altering OSM target genes to ultimately decrease SDF-1 expression in the OsmrFKO mouse. We conclude that adipocyte OSM–OSMR signaling plays a role in adipose tissue SDF-1 production and may mitigate its effects on adipose tissue homeostasis.
Keywords: oncostatin M, adipose tissue, stromal-derived factor 1, adipocyte, inflammation
1. Introduction
Interactions between stromal vascular cells (SVCs) and adipocytes are crucial for the maintenance of white adipose tissue (WAT) homeostasis. The chronic, low-grade inflammatory state associated with obesity is characterized by WAT leukocyte infiltration and alterations in the physiological cytokine milieu that favor pro-inflammatory adipokine production and disruption of the stem cell niche. These effects, in combination, can negatively affect overall WAT function and metabolism [1,2,3,4,5,6,7,8].
We have previously focused on the pro-inflammatory WAT adipokine, oncostatin M (OSM), and how its signaling may regulate tissue homeostasis and immune cell balance [9,10]. OSM belongs to the gp130 family of cytokines and regulates a variety of cellular and biological processes in a cell-type-dependent manner [11,12,13]. Unlike any other gp130 cytokine, OSM has its own specific receptor (oncostatin M receptor; OSMR) that heterodimerizes with gp130 to mediate OSM effects [14], which occur via JAK-STAT or ERK pathway activation [15,16,17,18,19,20]. We have demonstrated that, in WAT, SVCs—not adipocytes—produce OSM in conditions of obesity [9,21]. Another group has published data to indicate OSM can be expressed in adipocytes, but we are consistently unable to detect OSM in either cultured adipocytes or adipocytes isolated from mouse adipose tissue [9,21,22,23]. However, we and others have shown that adipocytes do express OSMR [9,10,21,24], which suggests that adipocyte OSM–OSMR signaling may play a role in WAT homeostasis.
We have generated mice with an adipocyte-specific deletion of OSMR (called OsmrFKO mice) [9]. Chow-fed OsmrFKO mice exhibit epididymal adipose tissue (eWAT) insulin resistance and increased eWAT CD45+ leukocyte infiltration, compared to floxed littermates [10]. When challenged with a high-fat diet (HFD), OsmrFKO mice exhibit systemic insulin resistance and increased eWAT pro-fibrotic and pro-inflammatory gene expression [9]. We have not observed any significant differences in fat mass or food intake in OsmrFKO mice on either diet [9,10]. Although our previous data suggest that adipocyte OSM–OSMR signaling may have an important role in the maintenance of WAT immune balance and homeostasis, the mechanisms responsible for this homeostatic role remain unclear.
The alpha chemokine, stromal-derived factor 1 (SDF-1; also known as CXCL12), plays a critical role in hematopoietic stem cell trafficking, homing, and retention, as well as in the accumulation of immune cells positive for the SDF-1 receptor, CXCR4, in injured or inflamed tissues [25]. Recent reports have highlighted the adipose tissue SDF-1–CXCR4 axis in obesity and diabetes. One report postulated that SDF-1 may serve to desensitize adipocytes to insulin [26] and another suggested that SDF-1 modulates angiogenesis in obesity [27]. In addition, a role has been proposed for SDF-1-mediated endothelial cell–stromal interactions, in the maintenance of the physiological adipose tissue cytokine milieu [28]. Direct effects of OSM on SDF-1 expression have been described [29,30,31,32,33], although these studies were not conducted in adipocytes. Here, we provide evidence that adipose tissue SDF-1 levels are affected by the loss of adipocyte OSMR signaling in vivo, and that SDF-1 may have a role in OSM-regulated adipose tissue homeostasis.
2. Materials and Methods
2.1. Animals and Diets
Male floxed oncostatin M receptor (Osmrfl/fl) and OsmrFKO mice, generated as previously described [9], were obtained from our existing colony. Briefly, Osmrfl/fl mice (stock #011081), from Jackson Laboratories, and adiponectin-cre mice, from an in-house colony, were crossed to create the adipocyte-specific OSMR knockout mouse (OsmrFKO) and littermate floxed controls (Osmrfl/fl). Mice were housed in a temperature-controlled (22 °C ± 2 °C) and humidity-controlled (45–55%) room under a 12-h light–dark cycle and were allowed ad libitum access to food and water. Mice were fed a high-fat diet (D12451; 45% fat; Research Diets) or a breeder chow diet (Purina LabDiet #5015; LabDiet, St. Louis, MO, USA), as indicated, for 20 weeks, at which time eWAT was collected for gene and protein expression analyses. Mice were 24–26 weeks old when tissues were collected. As previously reported, no differences in body weight or fat mass were observed between genotypes [9,10]. Where indicated, tissue was dissociated into adipocytes and SVCs, as previously described, [9,10] and gene expression analyses were performed as described below. All studies were approved by the Pennington Biomedical Research Center Institutional Animal Care and Use Committee (protocol #961P).
2.2. 3T3-L1 Adipocyte Culture and Treatment
Murine 3T3-L1 adipocytes were grown to two days post-confluence and differentiated as previously described [9,10]. Cells were treated for 2–48 h with 0.5 nM OSM (R&D Systems, Minneapolis, MN, USA) or vehicle (0.1% bovine serum albumin in phosphate-buffered saline), as indicated. Media was removed after treatments were completed and saved for analysis. Cell monolayers were washed with PBS and harvested in a buffer containing 150 mM NaCl, 10 mM Tris (pH 7.4) 1 mM EDTA, 1 mM EGTA, 0.5% IGEPAL CA-630, 1% Triton X-100, 1 mM PMSF, 1 µM pepstatin, 50 trypsin inhibitory milliunits of aprotinin, 10 µM leupeptin, 1 mM 1, 10-phenanthroline, 0.2 mM sodium orthovanadate, and 100 µM sodium fluoride or RLT buffer (Qiagen, Germantown, MD, USA), for protein and RNA extraction, respectively. The cell suspensions were subjected to a freeze–thaw cycle at −80 °C, passed through a 20-gauge needle five times, and clarified via centrifugation at 13,000× g for 10 min at 4 °C.
2.3. Transient Transfection of 3T3-L1 Adipocytes
Fully differentiated murine adipocytes were transiently transfected with siOsmr or a non-targeting vector, as previously reported [9]. Briefly, cells were trypsinized, resuspended in DMEM+10% fetal bovine serum without antibiotics, counted, and re-plated into 6-well (protein) or 12-well (RNA) plates at a final concentration of 1.16 × 105 cells per cm2. A transfection cocktail containing Dharmafect DUO Transfection Reagent (Dharmacon/Horizon Discovery Group, Lafayette, CO, USA), OptiMEM culture medium (Thermo Fisher, Waltham, MA, USA), and siOsmr or non-targeting siRNA (On-Target Plus Smartpool, Dharmacon; final concentration = 100 nM), was then added to the cells. Cells were incubated for 24 h and media were changed to DMEM + 10% FBS without antibiotics. After an additional 24 h, media were changed to DMEM + 1% fetal calf serum without antibiotics, and OSM or vehicle treatments were initiated. Media was removed after treatments were completed and stored for later analyses. Cells were washed with PBS and harvested for protein or RNA extraction.
2.4. Gene Expression Analyses
Total RNA was isolated from cell monolayers using the RNeasy Mini Kit (Qiagen) and from eWAT homogenates using the RNeasy Lipid Tissue Mini Kit (Qiagen), as previously described [9,10]. RNA concentrations were quantified using a NanoDrop ND-1000 UV–Vis Spectrophotometer. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) with 2 µg of RNA. Quantitative PCR was then performed using the SYBR Premix with ROX Plus (Takara Bio, Mountain City, CA, USA) with 4 ng of cDNA and run on the Applied Biosystems 7900HT system with SDS 2.4 software (Applied Biosystems). Thermal cycling conditions were as follows: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C; dissociation stage: 15 s at 95 °C, 15 s at 60 °C, and 15 s at 95 °C. All target genes were normalized to Ppia (peptidyl prolyl isomerase A), Nono (non-POU domain containing octamer-binding protein), and Ubb (ubiquitin B). The primers used appear in Table 1 below.
Table 1.
Primers used for qPCR.
| Gene Name | Forward | Reverse |
|---|---|---|
| Sdf1/Cxcl12 | GAGCCAACGTCAAGCATCT | CCACTTTAATTTCGGGTCAATGC |
| Timp1 | ACCTGATCCGTCCACAAACA | GGGGTGTGCACAGTGTTTCC |
| Ppia | CCACTGTCGCTTTTCGCCGC | TGCAAACAGCTCGAAGGAGACGC |
| Ubb | CCAGTGGGCAGTGATGG | GCTTACCATGCAACAAAACCT |
| Nono | CATCATCAGCATCACCACCA | TCTTCAGGTCAATAGTCAAGCC |
| Osmr | CGTTCCCCTGTGAGGCCGAG | TCCTCCAAGACTTCGCTTCGGG |
2.5. Immunoblotting
Protein concentrations of cell lysates, tissue homogenates, and media samples were quantified using the BCA Kit for Protein Determination (Sigma-Aldrich, St. Louis, MO, USA). A total of 50–100 µg protein per well were loaded on 5% or 10% polyacrylamide gels and transferred to 0.45 µm nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). Membranes were probed and imaged using standard immunoblotting techniques, as previously described [9,10,21]. Proteins were detected using a horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA) and SuperSignal West Pico PLUS reagents (Thermo Fisher). Anti-STAT5A (E289; ab32043; rabbit monoclonal) and OSMRβ (AF662; goat polyclonal) antibodies were purchased from Abcam and R&D Systems, respectively.
2.6. ELISA and Cytokine Arrays
SDF-1 levels in cell culture media were assessed with a Mouse CXCL12/SDF-1 alpha Quantikine ELISA Kit (catalog number MCX120; R&D Systems), according to manufacturer instructions. Cytokine protein expression levels in eWAT (n = 6 per genotype) were assessed using a Proteome Profiler Mouse Cytokine Array Kit, Panel A (catalog number ARY006; R&D Systems), according to manufacturer instructions.
2.7. Statistical Analyses
Data were analyzed using GraphPad Prism software (Version 8). Differences between groups were calculated using Student’s t-tests. Results were considered significant when p < 0.05.
3. Results
We have previously reported insulin resistance and adipose tissue inflammation in OsmrFKO mice on high-fat or chow diets [9,10]. Direct effects of OSM signaling on SDF-1 expression have been reported [29,30,31,32,33], and recent publications have highlighted a role for adipose tissue SDF-1 in obesity and metabolic disease [26,27]. Given these previous findings, we sought to examine the effects of a loss of adipocyte OSM signaling on adipose tissue SDF-1 expression, using in vitro and in vivo models.
3.1. Effects of In Vivo OSMR Deletion on Adipose Tissue SDF-1 Expression
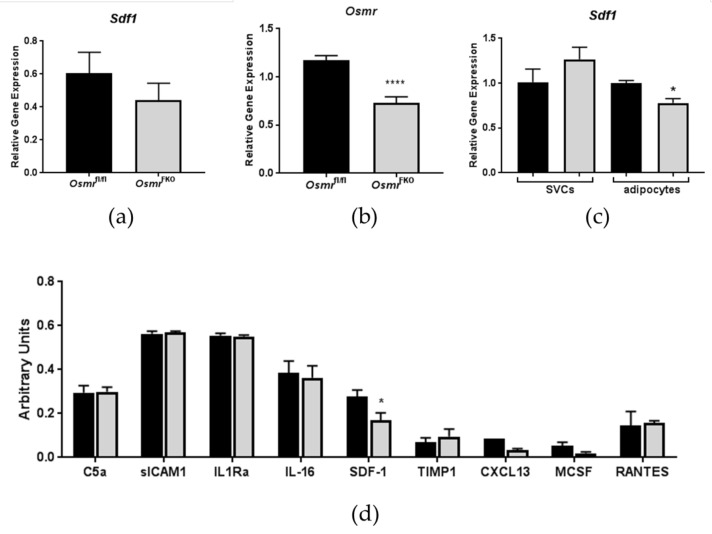
Interestingly, SDF-1 gene expression was decreased in eWAT from chow-fed OsmrFKO mice when compared to floxed controls, although this decrease was not significant (Figure 1a). Osmr gene expression was significantly decreased in the eWAT of HFD-fed OsmrFKO mice (Figure 1b), thereby confirming the knockout in these mice. In the eWAT of HFD-fed OsmrFKO mice that was dissociated into adipocytes and SVCs, Sdf1 gene expression was significantly lower in the adipocytes, but not the SVCs, of OsmrFKO mice (Figure 1c). SDF-1 protein expression levels were also significantly lower in the eWAT of HFD-fed OsmrFKO mice when compared to controls (Figure 1d). These results suggest that the regulation of adipose tissue SDF-1 in the OsmrFKO mouse occurs, in part, at the level of the adipocyte, and that adipocyte OSMR signaling plays an indirect role in the regulation of SDF-1 expression.
Figure 1.
Stromal-derived factor 1 (SDF-1) expression is decreased in epididymal adipose tissue (eWAT) from OsmrFKO mice. Gene expression was analyzed in (a) eWAT from chow-fed OsmrFKO or floxed Osmr (control) mice (n = 6–8/genotype). (b) Osmr knockdown confirmation in eWAT from high fat diet-fed mice (n = 6–8/genotype). (c) Fractionated eWAT from high-fat-diet-fed OsmrFKO mice (n = 2–3 pooled samples/genotype containing tissue from 2–3 mice per sample) and expression levels were normalized to Ppia. (d) Protein levels of various cytokines and chemokines were assessed in eWAT from OsmrFKO and control mice fed a high-fat diet (n = 6/genotype). Black bars = control (floxed) animals; gray bars = OsmrFKO. * p < 0.05, **** p < 0.001 vs. floxed controls.
3.2. Effects of OSM Administration on SDF-1 Expression in 3T3-L1 Adipocytes
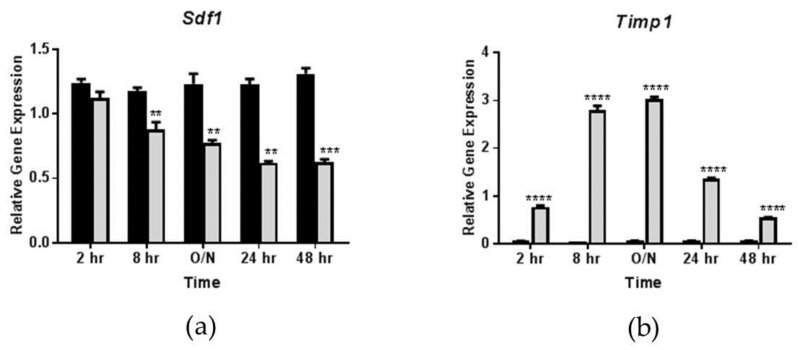
Previous reports indicate that OSM induces SDF-1 gene and protein expression in various cell types and tissues including mouse heart, mouse mesenchymal stem cells, human cardiomyocytes, and human fibroblasts, but not in adipocytes [29,30,31,32,33]. Since we observed decreased SDF-1 expression in the eWAT of OsmrFKO mice, we assessed whether OSM could modulate adipocyte SDF-1 expression. Surprisingly, we found a significant and sustained suppression of Sdf1 expression by OSM in fully differentiated 3T3-L1 adipocytes. In this study, where cells were treated with OSM for time periods varying from 2–48 h, Sdf1 expression was significantly decreased by OSM in a time-dependent manner, with a maximal reduction in expression observed at the 24 h time point (Figure 2a). The effects of OSM on the expression of the tissue inhibitor of metalloproteinases 1 (Timp1), a known OSM target, were examined as a positive control (Figure 2b).
Figure 2.
Oncostatin M (OSM) suppresses adipocyte Sdf1 expression in a time-dependent manner. Fully differentiated 3T3-L1 adipocytes were exposed to 0.5 nM OSM for 2 h, 8 h, ~12 h (overnight; O/N), 24 h, or 48 h. Gene expression was then analyzed and expression levels were normalized to a numerical average of three housekeeping genes (Ppia, Ubb, and Nono). (a) Effects of 0.5 nM OSM on Sdf1 expression in 3T3-L1 adipocytes; (b) Effects of OSM on Timp1 expression in 3T3-L1 adipocytes (shown as a positive control). Black bars = vehicle-treated cells; gray bars = OSM-treated cells. ** p < 0.01, *** p < 0.001, and **** p < 0.0001 vs. vehicle control. This experiment was performed twice.
3.3. Effects of OSM Administration on SDF-1 Expression in OSMR-Deficient 3T3-L1 Adipocytes
To assess the direct involvement of OSM signaling in decreasing Sdf1 expression, we knocked down OSMR in 3T3-L1 adipocytes and, subsequently, treated cells with OSM for 24 h. Indeed, OSM-treated cells, transfected with a scrambled siRNA sequence, exhibited the expected diminution in Sdf1 expression, while cells transfected with siOsmr actually exhibited a slight increase in Sdf1 expression (Figure 3a). Since SDF-1 is a secreted protein, we also assessed its levels in cell culture media from the knockdown experiments. Interestingly, in cells with intact OSMR expression, OSM significantly suppressed SDF-1 secretion. However, in cells lacking OSMR, the suppression of SDF-1 secretion by OSM was blunted (Figure 3b). These results suggest that OSM signaling suppresses adipocyte SDF-1 expression at the gene and protein levels, and that this effect depends on OSMR.
Figure 3.
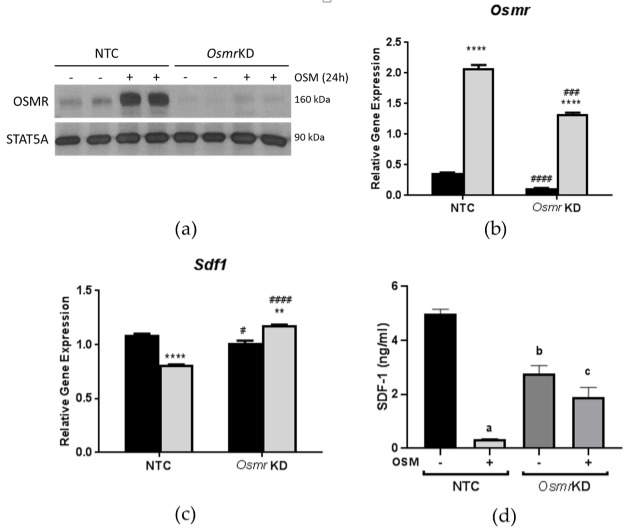
Oncostatin M receptor (OSMR) is required for Sdf1 suppression in adipocytes. Fully-differentiated 3T3-L1 adipocytes were transfected with siOsmr or a non-targeted control siRNA; 48 h after transfection, cells were exposed to 0.5 nM OSM for 24 h. Gene expression was then analyzed and expression levels were normalized to a numerical average of three housekeeping genes (Ppia, Ubb, and Nono). (a,b) Efficiency of OSMR knockdown. (c) The suppressive effect of OSM on Sdf1 expression is lost with OSMR silencing. (d) Effects of OSMR knockdown on SDF-1 secretion into culture media. In panels (a,b), black bars = vehicle-treated cells; gray bars = OSM-treated cells. KD = knockdown. ** p < 0.01 and **** p < 0.0001 vs. vehicle controls. # p < 0.05, ## p < 0.01, and #### p < 0.0001 vs. NTC. In panel (d), a = p < 0.001 vs. NTC; b = p < 0.01 vs. NTC; c = p < 0.05 vs. NTC + OSM. This experiment was performed twice.
4. Discussion
Interactions between adipocytes and SVCs play a role in regulating adipose tissue homeostasis, partly by producing cytokines and chemokines to regulate immune cell balance. We have previously reported that OsmrFKO mice, with no differences in body weight or fat mass, exhibit insulin resistance and eWAT inflammation [9,10]. When OsmrFKO mice are fed a chow diet, insulin resistance is confined to the eWAT level; but when challenged with a high-fat diet, they develop systemic insulin resistance and increased eWAT inflammation [9,10]. Here, we report that some of these phenotypic effects are associated with altered eWAT SDF-1 gene and protein expression.
Previous studies have demonstrated that OSM induces SDF-1 expression, at both the gene and protein levels, in a variety of cell types [29,30,31,32,33]. In our studies, we did not observe increases in SDF-1 with OSM treatment in vitro; instead, we observed suppression of SDF-1 expression. A very recent study of mouse lung reports decreased SDF-1 gene and protein expression upon administration of an OSM-encoding adenoviral vector [34]. Our current results demonstrate that OsmrFKO mice exhibit decreased Sdf1 gene expression in epididymal adipocytes and decreased SDF-1 protein expression in whole eWAT. In an attempt to further assess the cell autonomous effects of OSM–OSMR signaling on adipocyte SDF-1 expression, we treated 3T3-L1 adipocytes with OSM. We observed that OSM decreased Sdf1 expression in a time-dependent manner, and that this effect was partly dependent on adipocyte OSMR expression. In 3T3-L1 adipocytes lacking OSMR, SDF-1 gene expression and protein secretion were increased, compared to OSM-treated control cells. However, in eWAT adipocytes from OsmrFKO mice, SDF-1 gene expression and protein expression levels were significantly decreased. These contrasting in vitro and in vivo results strongly suggest that the decreased SDF-1 expression in our knockout mice was not due to a direct effect of OSM, but may instead be due to an OSM target gene (or genes) in adipocytes whose regulation is altered by the OSMR knockdown. It is likely that this target gene(s) may alter adipokine production, secretion, and signaling to other adipose tissue stromal cells.
A recent report indicates that adipocyte-specific SDF-1 knockout mice are more insulin-sensitive than their floxed littermates, an effect that may result from the loss of the insulin-desensitizing autocrine actions of adipocyte-derived SDF-1 [26]. In this study, it was suggested that stromal cell-derived SDF-1 is responsible for the classic chemotactic effects of the protein [26]. Another study demonstrated a robust increase of SDF-1 in the WAT of obese mice. The systemic administration of an antagonist to CXCR4 (the SDF-1 receptor) in these mice reduced adipose tissue macrophage accumulation and inflammation, and improved whole-body insulin sensitivity [35]. In that report, the authors concluded that SDF-1 was required for the establishment of obesity-induced adipose tissue inflammation and systemic insulin resistance. In contrast to these reports, we observed increased insulin resistance and eWAT inflammation [9,10] associated with decreased SDF-1 expression (Figure 1) in our OsmrFKO mice. These observations support the assertion that the effects on SDF-1 observed in this mouse model are likely an indirect result of alterations in other chemokines and cytokines. Notably, the previous data described here, along with our current data, were obtained from male mice. Given the known in vitro effects of OSM on the estrogen receptor [36], it will be important to conduct similar studies including female mice.
Since we have previously shown that OSM itself is elevated in eWAT in our knockout mice [9,10], the actions of OSM on non-adipocyte cells may be enhancing (or negating) the production of a factor that is altering adipocyte SDF-1 production and secretion. The differences observed in SDF-1 expression between our in vitro and in vivo studies may be attributable to the lack of stromal cells in the in vitro experiments. SDF-1 is produced by several cell types, thus it is possible that adipocyte SDF-1 secretion may act in a paracrine manner to partially regulate adipose stromal cell SDF-1 production. Taken together, our current results, along with previous results from our lab and others, suggest that adipocyte OSM–OSMR signaling may serve as a homeostatic regulator of adipose tissue SDF-1 levels. However, the mechanisms underlying: (1) OSM-induced SDF-1 suppression in adipocytes in vitro; (2) decreased SDF-1 expression in adipose tissue of OsmrFKO mice; and (3) whether similar effects occur in female mice, remain unclear. Future studies will focus on elucidating these mechanisms, as they could further contribute to our understanding of how cytokine signaling between SVCs and adipocytes contribute to the maintenance of adipose tissue homeostasis.
5. Conclusions
In this study, we demonstrated that OSM suppresses SDF-1 gene expression and protein secretion in cultured adipocytes, and that OsmrFKO mice also exhibit decreased adipose tissue SDF-1 gene and protein expression. These results suggest that OSMR mediates decreased adipocyte SDF-1 expression both in vitro and in vivo, but that differing mechanisms govern this regulation. We conclude from these results that adipocyte OSM–OSMR signaling is a regulator of adipose tissue SDF-1 levels.
Acknowledgments
The authors gratefully acknowledge Anik Boudreau for her outstanding technical support.
Author Contributions
Conceptualization, C.M.E.; methodology, H.H., J.L.B., C.M.E.; formal analysis, H.H., J.L.B., C.M.E.; investigation, H.H, J.L.B, C.M.E.; resources, C.M.E.; data curation, H.H., J.L.B., C.M.E.; writing—original draft preparation, H.H., C.M.E.; writing—review and editing, H.H., J.L.B., C.M.E.; supervision, C.M.E.; project administration, C.M.E.; funding acquisition, C.M.E.
Funding
This research was funded by the National Institutes of Health; grant number K01 DK106307 to C.M.E. and the APC was funded by the National Institutes of Health; grant number K01 DK106307 to C.M.E. This work used core facilities that are supported in part by COBRE (NIH P30 GM118430) and NORC (NIH P30 DK072476) center grants from the NIH.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Cipolletta D., Feuerer M., Li A., Kamei N., Lee J., Shoelson S.E., Benoist C., Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Exley M.A., Hand L., O’Shea D., Lynch L. Interplay between the immune system and adipose tissue in obesity. J. Endocrinol. 2014;223:R41. doi: 10.1530/JOE-13-0516. [DOI] [PubMed] [Google Scholar]
- 3.Grant R.W., Dixit V.D. Adipose tissue as an immunological organ. Obesity. 2015;23:512–518. doi: 10.1002/oby.21003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macdougall C.E., Wood E.G., Loschko J., Scagliotti V., Cassidy F.C., Robinson M.E., Feldhahn N., Castellano L., Voisin M.-B., Marelli-Berg F., et al. Visceral Adipose Tissue Immune Homeostasis Is Regulated by the Crosstalk between Adipocytes and Dendritic Cell Subsets. Cell Metab. 2018;27:588–601. doi: 10.1016/j.cmet.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathis D. Immunological Goings-On in Visceral Adipose Tissue. Cell Metab. 2013;17:851–859. doi: 10.1016/j.cmet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurizi G., Della Guardia L., Maurizi A., Poloni A. Adipocytes properties and crosstalk with immune system in obesity-related inflammation. J. Cell. Physiol. 2018;233:88–97. doi: 10.1002/jcp.25855. [DOI] [PubMed] [Google Scholar]
- 7.Schipper H.S., Prakken B., Kalkhoven E., Boes M. Adipose tissue-resident immune cells: Key players in immunometabolism. Trends Endocrinol. Metab. 2012;23:407–415. doi: 10.1016/j.tem.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Seijkens T., Kusters P., Chatzigeorgiou A., Chavakis T., Lutgens E. Immune Cell Crosstalk in Obesity: A Key Role for Costimulation? Diabetes. 2014;63:3982–3991. doi: 10.2337/db14-0272. [DOI] [PubMed] [Google Scholar]
- 9.Elks C.M., Zhao P., Grant R.W., Hang H., Bailey J.L., Burk D.H., McNulty M.A., Mynatt R.L., Stephens J.M. Loss of Oncostatin M Signaling in Adipocytes Induces Insulin Resistance and Adipose Tissue Inflammation in Vivo. J. Biol. Chem. 2016;291:17066–17076. doi: 10.1074/jbc.M116.739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens J.M., Bailey J.L., Hang H., Rittell V., Dietrich M.A., Mynatt R.L., Elks C.M. Adipose Tissue Dysfunction Occurs Independently of Obesity in Adipocyte-Specific Oncostatin Receptor Knockout Mice. Obesity. 2018;26:1439–1447. doi: 10.1002/oby.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fasnacht N., Müller W. Conditional gp130 deficient mouse mutants. Semin. Cell Dev. Biol. 2008;19:379–384. doi: 10.1016/j.semcdb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Muller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/bj20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace P.M., MacMaster J.F., Rouleau K.A., Brown T.J., Loy J.K., Donaldson K.L., Wahl A.F. Regulation of Inflammatory Responses by Oncostatin M. J. Immunol. 1999;162:5547–5555. [PubMed] [Google Scholar]
- 14.Mosley B., De Imus C., Friend D., Boiani N., Thoma B., Park L.S., Cosman D. Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J. Biol. Chem. 1996;271:32635–32643. doi: 10.1074/jbc.271.51.32635. [DOI] [PubMed] [Google Scholar]
- 15.Wong S., Botelho F.M., Rodrigues R.M., Richards C.D. Oncostatin M overexpression induces matrix deposition, STAT3 activation, and SMAD1 Dysregulation in lungs of fibrosis-resistant BALB/c mice. Lab. Investig. 2014;94:1003–1016. doi: 10.1038/labinvest.2014.81. [DOI] [PubMed] [Google Scholar]
- 16.Fritz D.K., Kerr C., Fattouh R., Llop-Guevara A., Khan W.I., Jordana M., Richards C.D. A Mouse Model of Airway Disease: Oncostatin M-Induced Pulmonary Eosinophilia, Goblet Cell Hyperplasia, and Airway Hyperresponsiveness Are STAT6 Dependent, and Interstitial Pulmonary Fibrosis Is STAT6 Independent. J. Immunol. 2011;186:1107–1118. doi: 10.4049/jimmunol.0903476. [DOI] [PubMed] [Google Scholar]
- 17.Mozaffarian A., Brewer A.W., Trueblood E.S., Luzina I.G., Todd N.W., Atamas S.P., Arnett H.A. Mechanisms of Oncostatin M-Induced Pulmonary Inflammation and Fibrosis. J. Immunol. 2008;181:7243–7253. doi: 10.4049/jimmunol.181.10.7243. [DOI] [PubMed] [Google Scholar]
- 18.Levy M.T., Trojanowska M., Reuben A. Oncostatin M: A cytokine upregulated in human cirrhosis, increases collagen production by human hepatic stellate cells. J. Hepatol. 2000;32:218–226. doi: 10.1016/S0168-8278(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 19.Pham Van T., Couchie D., Martin-Garcia N., Laperche Y., Zafrani E.S., Mavier P. Expression of matrix metalloproteinase-2 and -9 and of tissue inhibitor of matrix metalloproteinase-1 in liver regeneration from oval cells in rat. Matrix Biol. 2008;27:674–681. doi: 10.1016/j.matbio.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Canady J., Arndt S., Karrer S., Bosserhoff A.K. Increased KGF Expression Promotes Fibroblast Activation in a Double Paracrine Manner Resulting in Cutaneous Fibrosis. J. Investig. Dermatol. 2013;133:647–657. doi: 10.1038/jid.2012.389. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Infantes D., White U.A., Elks C.M., Morrison R.F., Gimble J.M., Considine R.V., Ferrante A.W., Ravussin E., Stephens J.M. Oncostatin M Is Produced in Adipose Tissue and Is Regulated in Conditions of Obesity and Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2014;99:E217–E225. doi: 10.1210/jc.2013-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komori T., Tanaka M., Senba E., Miyajima A., Morikawa Y. Lack of Oncostatin M Receptor β Leads to Adipose Tissue Inflammation and Insulin Resistance by Switching Macrophage Phenotype. J. Biol. Chem. 2013;288:21861–21875. doi: 10.1074/jbc.M113.461905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komori T., Tanaka M., Senba E., Miyajima A., Morikawa Y. Deficiency of Oncostatin M Receptor β (OSMRβ) Exacerbates High-fat Diet-induced Obesity and Related Metabolic Disorders in Mice. J. Biol. Chem. 2014;289:13821–13837. doi: 10.1074/jbc.M113.542399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moest H., Frei A.P., Bhattacharya I., Geiger M., Wollscheid B., Wolfrum C. Malfunctioning of adipocytes in obesity is linked to quantitative surfaceome changes. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids. 2013;1831:1208–1216. doi: 10.1016/j.bbalip.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Kucia M., Jankowski K., Reca R., Wysoczynski M., Bandura L., Allendorf D.J., Zhang J., Ratajczak J., Ratajczak M.Z. CXCR4–SDF-1 Signalling, Locomotion, Chemotaxis and Adhesion. J. Mol. Histol. 2004;35:233–245. doi: 10.1023/B:HIJO.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 26.Shin J., Fukuhara A., Onodera T., Kita S., Yokoyama C., Otsuki M., Shimomura I. SDF-1 Is an Autocrine Insulin-Desensitizing Factor in Adipocytes. Diabetes. 2018;67:1068–1078. doi: 10.2337/db17-0706. [DOI] [PubMed] [Google Scholar]
- 27.Wada T., Watanabe E.R.I., Kotera Y., Onogi Y., Tsuneki H., Sasaoka T. The SDF1-CXCR4 Signals Regulate Adipose Tissue Expansion by Modulating Angiogenesis in Diet-Induced Obesity in Mice. Diabetes. 2018;67:2005. doi: 10.2337/db18-2005-P. [DOI] [Google Scholar]
- 28.Maan Z.N., Rennert R.C., Duscher D., Januszyk M., Paik K., Chung M.T., Paik K., Fujiwara T., Rodrigues M., Ho N., et al. Abstract 8: SDF-1 Regulates Adipose Niche Homeostasis and Adipose Derived Stromal Cell Function. Plast. Reconstr. Surg. 2014;133:15–16. doi: 10.1097/01.prs.0000444941.23852.ce. [DOI] [PubMed] [Google Scholar]
- 29.Albiero M., Poncina N., Ciciliot S., Cappellari R., Menegazzo L., Ferraro F., Bolego C., Cignarella A., Avogaro A., Fadini G.P. Bone Marrow Macrophages Contribute to Diabetic Stem Cell Mobilopathy by Producing Oncostatin M. Diabetes. 2015;64:2957. doi: 10.2337/db14-1473. [DOI] [PubMed] [Google Scholar]
- 30.Hoermann G., Cerny-Reiterer S., Sadovnik I., Müllauer L., Bilban M., Gröger M., Horny H.-P., Reiter A., Schmitt-Graeff A., Mannhalter C., et al. Oncostatin M is a FIP1L1/PDGFRA-dependent mediator of cytokine production in chronic eosinophilic leukemia. Allergy. 2013;68:713–723. doi: 10.1111/all.12139. [DOI] [PubMed] [Google Scholar]
- 31.Hohensinner P.J., Kaun C., Rychli K., Niessner A., Pfaffenberger S., Rega G., Furnkranz A., Uhrin P., Zaujec J., Afonyushkin T., et al. The inflammatory mediator oncostatin M induces stromal derived factor-1 in human adult cardiac cells. FASEB J. 2009;23:774–782. doi: 10.1096/fj.08-108035. [DOI] [PubMed] [Google Scholar]
- 32.Lee M.J., Song H.Y., Kim M.R., Sung S.-M., Jung J.S., Kim J.H. Oncostatin M stimulates expression of stromal-derived factor-1 in human mesenchymal stem cells. Int. J. Biochem. Cell Biol. 2007;39:650–659. doi: 10.1016/j.biocel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Minehata K.-I., Takeuchi M., Hirabayashi Y., Inoue T., Donovan P.J., Tanaka M., Miyajima A. Oncostatin M Maintains the Hematopoietic Microenvironment and Retains Hematopoietic Progenitors in the Bone Marrow. Int. J. Hematol. 2006;84:319–327. doi: 10.1532/IJH97.06090. [DOI] [PubMed] [Google Scholar]
- 34.Botelho F.M., Rodrigues R., Guerette J., Wong S., Fritz D.K., Richards C.D. Extracellular Matrix and Fibrocyte Accumulation in BALB/c Mouse Lung upon Transient Overexpression of Oncostatin M. Cells. 2019;8:126. doi: 10.3390/cells8020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D., Kim J., Yoon J.H., Ghim J., Yea K., Song P., Park S., Lee A., Hong C.-P., Jang M.S., et al. CXCL12 secreted from adipose tissue recruits macrophages and induces insulin resistance in mice. Diabetologia. 2014;57:1456–1465. doi: 10.1007/s00125-014-3237-5. [DOI] [PubMed] [Google Scholar]
- 36.West N.R., Murphy L.C., Watson P.H. Oncostatin M suppresses oestrogen receptor-alpha expression and is associated with poor outcome in human breast cancer. Endocr. Relat. Cancer. 2012;19:181–195. doi: 10.1530/ERC-11-0326. [DOI] [PubMed] [Google Scholar]