Abstract
It is now clear that heredity is not determined purely by Mendelian genetic inheritance; sometimes, epigenetic signals can be passed from parent to progeny for multiple generations. This phenomenon is termed transgenerational epigenetic inheritance (TEI), and examples have now been observed in multiple organisms including plants, flies, mice, and nematodes. Here we discuss the recent findings that TEI is a multi-step process and that the putative chromatin modifiers SET-25 and SET-32 are important in the establishment but not maintenance of silencing.
Keywords: Caenorhabditis elegans, chromatin, chromatin modifiers, H3K9me3, histone, histone modifications, RNA interference, SET domain, SET-25, SET-32, SET-domain proteins, transgenerational epigenetic inheritance
Comment on: Woodhouse, R.M., Buchmann, G., Hoe, M., Harney, D.J., Low, J.K.K., Larance, M., Boag, P.R., and Ashe, A. Chromatin Modifiers SET-25 and SET-32 Are Required for Establishment but Not Long-Term Maintenance of Transgenerational Epigenetic Inheritance. Cell Rep. 2018 Nov 20;25(8):2259–2272.e5. doi: 10.1016/j.celrep.2018.10.085. PubMed PMID: 30463020. https://www.ncbi.nlm.nih.gov/pubmed/30463020
Transgenerational epigenetic inheritance (TEI) – the transmission of epigenetic signals between generations – is known to occur in a range of organisms (reviewed in Miska and Ferguson-Smith1); however, the mechanism(s) by which it works remain unclear. In this study, Woodhouse et al2 used the nematode Caenorhabditis elegans to study TEI, employing a system that relies on RNA interference (RNAi)-induced silencing. In C elegans, RNAi is usually performed by growing animals on bacteria that express dsRNA fragments complementary to a target gene. Ingested dsRNA is processed by endogenous machinery and ultimately initiates silencing of the target gene. Silencing is caused by post-transcriptional gene silencing of target gene mRNA in the cytoplasm3 and/or transcriptional gene silencing in the nucleus,4 both being epigenetic processes, with no change to DNA sequence.
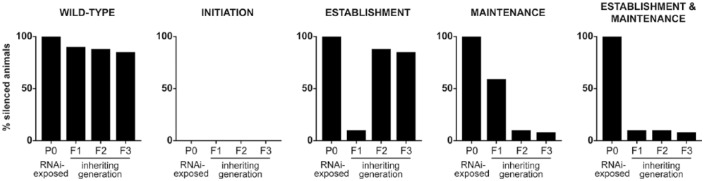
Generally, RNAi-induced silencing affects the exposed animals and occasionally the next generation.5,6 In our recent study, we used a system developed by Ashe et al7 involving RNAi-induced silencing of a single-copy, germline-expressed green fluorescent protein (GFP) transgene. What makes this system suited to the study of TEI is that GFP silencing is observed not only in the RNAi-exposed generation and its unexposed offspring, but also in at least 3 additional unexposed generations. Furthermore, use of the GFP transgene provides a visual signal of silencing, enabling precise quantification of inheritance of silencing over generations, as well as the ability to sort animals into silenced and non-silenced groups: this precision allows investigation into the particular requirements of these different populations. Consequently, this approach uniquely enables an examination of the precise genetic requirements of TEI in exposed individuals, their offspring, and following generations, and the identification of 3 distinct steps of TEI: initiation, establishment, and maintenance (Figure 1 and see below).
Figure 1.
The RNAi-induced heritable silencing assay distinguishes roles in initiation, establishment, and maintenance of TEI. Our study used a system of RNAi-induced silencing of a germline GFP transgene, originally developed by Ashe et al.7 Selection of silenced individuals following the removal of the RNAi trigger enables precise quantification of silencing proportions and reveals requirements of genes in particular generations. Animals mutant in a particular candidate gene are tested in the assay, and the resulting output reveals a role for the gene in initiation, establishment, maintenance, or establishment and maintenance of TEI.
GFP, green fluorescent protein; RNAi, RNA interference; TEI, transgenerational epigenetic inheritance.
Our recent work used this powerful system to characterise the role of the putative histone lysine methyltransferases SET-25 and SET-32 in TEI.
TEI Comprises Three Genetically Distinct Steps
We previously published our GFP transgene as a tool to detect TEI, in which we observed cumulative loss of silencing over generations in mutant strains.7 When testing set-25 and set-32 mutant strains, we observed that mutant animals were efficiently silenced in the generation exposed to RNAi, showing that set-25 and set-32 are dispensable for RNAi-induced silencing in the exposed generation. However, F1 offspring displayed a dramatic failure to inherit silencing, demonstrating a defect in transgenerational inheritance of silencing that was more severe than any we had previously observed. Extremely surprisingly, when the few F1 animals which escaped inheritance failure and remained silenced were propagated, their offspring (F2s) showed silencing proportions comparable with wild-type animals of the same generation.2 This finding revealed 3 distinct steps within TEI; SET-25 and SET-32 are dispensable for the initiation of silencing of the RNAi-exposed generation, required for the inheritance of silencing to the first unexposed generation, and then dispensable for the ongoing maintenance of heritable silencing in the following generations (Figure 2).
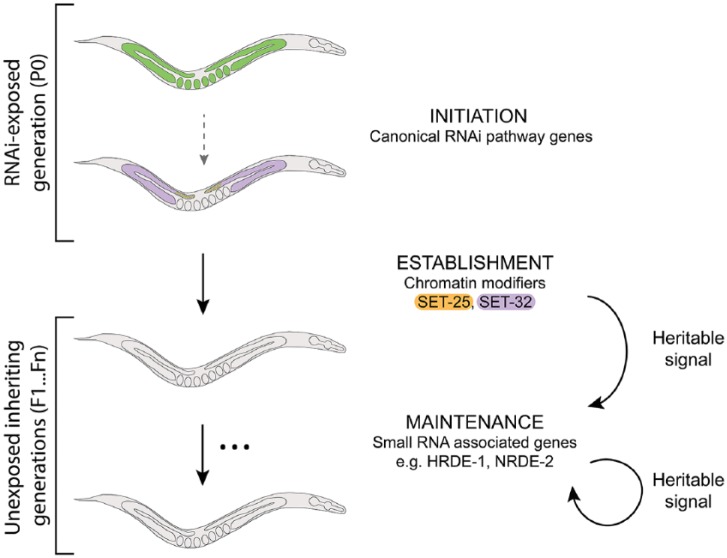
Figure 2.
TEI is a 3-step process. We propose that stable heritable silencing requires 3 phases, each with distinct genetic requirements. In response to RNAi, silencing is initiated in the exposed P0 generation by canonical RNAi pathway genes. In most cases, offspring do not inherit gene silencing, but occasionally an unknown trigger results in the establishment of a heritable silencing signal by the putative histone methyltransferases SET-25 and SET-32 in the germline of the P0 individuals. Silencing is then maintained transgenerationally in subsequent generations by small-RNA-associated genes, including HRDE-1 and NRDE-2. The heritable signal passed between generations remains to be elucidated.
HRDE-1, heritable RNAi defective-1; NRDE-2, nuclear RNAi defective-2; RNAi, RNA interference; TEI, transgenerational epigenetic inheritance.
The silencing defect we observed in set-25 and set-32 mutants in the F1 generation could conceivably have been due to either a failure to establish a heritable silencing signal in the exposed P0 generation, or a failure to receive a silencing signal in the inheriting F1 generation. We therefore investigated the generation in which set-25 and set-32 promote transgenerational silencing, to determine the generation in which the set-25/set-32-dependent heritable signal is established. We found that unexposed F1 homozygous mutant set-25 or set-32 animals, which had segregated from RNAi-exposed heterozygous P0 parents, showed normal silencing inheritance.2 This demonstrated that set-25 and set-32 are not required for silencing in the inheriting generation, and that therefore their role in TEI must be to establish heritable silencing in the RNAi-exposed generation.
The Gu group copublished a study8 alongside ours which provides further evidence of distinct requirements for establishment and maintenance of TEI. Kalinava et al exposed animals to continual RNAi against the endogenous oma-1 gene at every generation and showed that set-32 mutants had defective transcriptional silencing for 4 generations of exposure, but the defect lessened in each generation such that silencing was equivalent to wild-type by the fifth generation. Furthermore, in parallel experiments, set-32 was dispensable for transcriptional silencing in subsequent unexposed ‘maintenance’ generations.8,9 These findings suggest that set-32 mutants have delayed transcriptional silencing, and hence that set-32 is necessary for promoting the establishment of transgenerational silencing but not required for silencing maintenance, supporting our results. Together, these 2 studies2,8 demonstrate a role for set-32 in the establishment of TEI in both transgene and endogenous gene contexts.
Whereas our study indicated a role for set-25 in the establishment of TEI, Kalinava et al8 found no such requirement for heritable silencing of oma-1. However, they found that set-25 is required for silencing a small proportion of endogenous nuclear RNAi targets, suggesting that the requirement of set-25 is locus-specific and may depend on factors such as local chromatin structure, epigenetic state, or gene expression level.
The conclusions from these studies are twofold: TEI is a multi-step process with distinct genetic requirements at each stage, and set-32 and in some cases set-25 are required for the establishment but not ongoing maintenance of heritable silencing.
How Might set-25 and set-32 Establish Heritable Silencing?
Further results from our study provide some clues as to the possible mechanism of set-25- and set-32-dependent heritable silencing establishment. We investigated the expression patterns of set-25 and set-32 throughout development and found that both are expressed in the germline of larval stage 2 animals onwards. set-25 is expressed in nuclei in the proliferative mitotic zone and in all nuclei of embryos from about the 8- to 16-cell stage. set-32 is expressed in both the cytoplasm and nuclei throughout the entire germline, and expression peaks in larval stage 4 just prior to adulthood. Thus, set-25 and set-32 likely function in the germline of RNAi-exposed individuals to establish a silencing signal, with set-32 potentially having a more extensive role.
What could this silencing signal be? SET-25 has been shown previously to deposit H3K9me3 in immunofluorescence and quantitative mass spectrometry studies.10,11 We confirmed this finding by also showing (by quantitative mass spectrometry on whole worm extracts) an almost complete loss of H3K9me3 in set-25 mutants. set-32 mutants, in contrast, showed subtle defects in a range of histone modifications in the same experiment, leaving no conclusive evidence for its substrate specificity. SET-32 may modify a small subset of loci or function at a particular developmental stage, masked in our whole-animal, mixed-age study, or require a trigger such as RNAi for activation. Possibly, both SET-domain proteins may have non-histone targets through which they exert gene regulation and silencing, with several recent papers identifying non-histone targets of previously identified histone methyltransferases.12,13 Hence, the establishment of silencing may involve modifications of histones or non-histone proteins.
Turning to other mechanisms through which silencing may be established, SET-25 is necessary for complete attachment of heterochromatic transgenic arrays to the nuclear periphery,11 a region associated with gene silencing.14 Interestingly, the recruitment of SET-25 to heterochromatin at the nuclear periphery requires the presence of H3K9me3, but is independent of the SET domain.11 Furthermore, MORC family CW-type zinc finger-1 (MORC-1), a member of the conserved GHKL-type ATPase family, has recently been shown to be required for transgenerational inheritance of RNAi-induced silencing of a germline GFP transgene.15,16 morc-1 is also required for positioning of heterochromatin at the nuclear periphery,16 suggesting that the establishment of heritable silencing may involve localisation of the silenced locus to the nuclear periphery. Therefore, establishment of TEI could involve SET-25, SET-32, and other factors, including MORC-1, establishing a heritable silencing signal through the deposition of repressive histone modifications or other protein modifications, localisation of the targeted locus to the nuclear periphery, or a combination of these factors.
This signal may then be ‘read’ by TEI maintenance factors, which propagate the silencing signal to subsequent generations. Directed by RNAi-produced small silencing RNAs, the nuclear Argonaute protein heritable RNAi defective-1 (HRDE-1) and its interaction partners nuclear RNAi defective (NRDE)-1, -2, and -4 mediate gene silencing in the nucleus by inhibiting RNA polymerase II during transcriptional elongation4 and by promoting repressive chromatin modifications.4,17–20 While dispensable for silencing of the RNAi-exposed generation, these 4 core small RNA factors are all required for TEI,7,21 and HRDE-1 and NRDE-2 are required to maintain silencing across every inheriting generation.7,21,22 HRDE-1 and NRDE-1, -2, and -4 therefore likely form part of a maintenance machinery, alongside other TEI-related factors. Whether these factors also promote silencing establishment needs to be clarified.
The question remains: what is the heritable agent that passes between the RNAi-exposed P0 generation and F1 offspring, and between subsequent generations? Past research has highlighted small RNAs as the likely candidate; 22-nucleotide-long secondary RNAs (so called due to their method of synthesis) antisense to the RNAi target sequence can be detected both in RNAi-exposed individuals and in each subsequent heritably silenced generation.7,21,23,24 We separated F1 worms according to their inherited silencing status (GFP-silenced or GFP-expressing) and, in wild-type and SET-25 mutant populations, found minimal differences between these 2 groups in the abundance of GFP-targeting secondary RNAs. Despite this lack of difference in small RNAs, in both these strains GFP-silenced F1s produce F2 offspring with high proportions of inherited silencing, whereas GFP-expressing F1s produce 100% GFP-expressing F2s. In other words, the proportion of secondary RNAs did not correlate well with the heritable silencing phenotype, bringing into question whether secondary RNAs are in fact the main heritable agent. Further research is required to investigate whether secondary RNAs interact with another heritable agent to execute TEI, or whether a novel, unidentified mechanism is at work.
We therefore propose a model whereby, during RNAi exposure, classical RNAi machinery initiates silencing in the exposed generation, SET-25 and SET-32 establish a transgenerational silencing signal in the exposed generation, and then small RNA-related factors, including HRDE-1 and NRDE-2, maintain silencing over subsequent generations (Figure 2). Other factors previously implicated in TEI should be placed in this model and, importantly, future work should be designed in such a way that roles in initiation, establishment, and maintenance can be distinguished.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: RMW was supported by an Australian Government Research Training Program Scholarship and AA by an Australian Research Council Discovery Early Career Researcher Award and Future Fellowship.
Declaration Of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: RMW and AA wrote the manuscript.
ORCID iD: Alyson Ashe  https://orcid.org/0000-0001-7334-0389
https://orcid.org/0000-0001-7334-0389
References
- 1. Miska EA, Ferguson-Smith AC. Transgenerational inheritance: models and mechanisms of non-DNA sequence-based inheritance. Science. 2016;354:59–63. [DOI] [PubMed] [Google Scholar]
- 2. Woodhouse RM, Buchmann G, Hoe M, et al. Chromatin modifiers SET-25 and SET-32 are required for establishment but not long-term maintenance of transgenerational epigenetic inheritance. Cell Rep. 2018;25:2259.e5–2272.e5. [DOI] [PubMed] [Google Scholar]
- 3. Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [DOI] [PubMed] [Google Scholar]
- 6. Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. [DOI] [PubMed] [Google Scholar]
- 7. Ashe A, Sapetschnig A, Weick EM, et al. PiRNAs can trigger a multi-generational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalinava N, Ni JZ, Gajic Z, Kim M, Ushakov H, Gu SG. C. elegans Heterochromatin Factor SET-32 plays an essential role in transgenerational establishment of nuclear RNAi-mediated epigenetic silencing. Cell Rep. 2018;25:2273.e3–2284.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kalinava N, Ni JZ, Peterman K, Chen E, Gu SG. Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenetics Chromatin. 2017;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snyder MJ, Lau AC, Brouhard EA, et al. Anchoring of heterochromatin to the nuclear lamina reinforces dosage compensation-mediated gene repression. PLoS Genet. 2016;12:e1006341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Towbin BD, Gonzalez-Aguilera C, Sack R, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. [DOI] [PubMed] [Google Scholar]
- 12. Tsusaka T, Kikuchi M, Shimazu T, et al. Tri-methylation of ATF7IP by G9a/GLP recruits the chromodomain protein MPP8. Epigenetics Chromatin. 2018;11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilkinson AW, Diep J, Dai S, et al. SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature. 2019;565:372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meister P, Taddei A. Building silent compartments at the nuclear periphery: a recurrent theme. Curr Opin Genet Dev. 2013;23:96–103. [DOI] [PubMed] [Google Scholar]
- 15. Spracklin G, Fields B, Wan G, et al. The RNAi inheritance machinery of Caenorhabditis elegans. Genetics. 2017;206:1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weiser NE, Yang DX, Feng S, et al. MORC-1 integrates nuclear RNAi and transgenerational chromatin architecture to promote germline immortality. Dev Cell. 2017;41:408.e7–423.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011;7:e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:19683–19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mao H, Zhu C, Zong D, et al. The Nrde pathway mediates small-RNA-directed histone H3 lysine 27 trimethylation in Caenorhabditis elegans. Curr Biol. 2015;25:2398–2403. [DOI] [PubMed] [Google Scholar]
- 21. Buckley BA, Burkhart KB, Gu SG, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minkina O, Hunter CP. Stable heritable germline silencing directs somatic silencing at an endogenous locus. Mol Cell. 2017;65:659.e5-670e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houri-Ze’evi L, Korem Y, Sheftel H, et al. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. [DOI] [PubMed] [Google Scholar]
- 24. Shirayama M, Seth M, Lee HC, et al. PiRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]