Abstract
The application of next-generation sequencing in clinical practice is increasing as accuracy and interpretation have improved and the cost continues to decline rapidly. Cell-free DNA is a unique source for next-generation sequencing that could change routine clinical practice in gastroenterology and hepatology. Testing of cell-free DNA in blood and fecal samples is an easy, rapid, and noninvasive method to assess for premalignant, malignant, metabolic, infectious, inflammatory, and autoimmune gastrointestinal and liver diseases. In this review, we describe cell-free DNA technologies, current applications of cell-free DNA testing, and proposed cell-free DNA targets for gastrointestinal and hepatic diseases, with a specific focus on malignancy. In addition, we provide commentary on how cell-free DNA can be integrated into clinical practice and help guide diagnosis, prognosis, disease management, and therapeutic response.
Keywords: cell-free DNA, gastroenterology and hepatology, individualized medicine
Introduction
The existence of extracellular DNA circulating within the blood, termed cell-free DNA (cfDNA), is a promising source of diagnostic material for the management of disease including digestive disorders. Despite the discovery of cfDNA over 60 years ago, it has only recently become feasible to use this type of DNA in the clinical setting. Currently, the main clinical utility of cfDNA testing is for prenatal testing of fetal aneuploidy and sex determination, to gage organ transplant rejection, as well as in oncology for diagnostic and therapeutic purposes as a sensitive and specific surrogate to tissue biopsy. Although this nascent technology has great promise to act as a ‘liquid biopsy’ and noninvasively diagnose disease, there remains many challenges in finding and validating clinically meaningful cfDNA biomarkers.
In this article, we review current advances in and applications of cfDNA technologies, describe how these methodologies can aid in the diagnosis and prognosis of digestive diseases, focusing on malignancy, and discuss the limitations and challenges of these applications.
Background
By definition, cfDNA is extracellular and is found in the blood as well as in other bodily fluids such as urine, bile, pancreatic secretions, or peritoneal fluid (Figure 1). However, the term ‘cell-free’ should not be interpreted that the DNA is circulating completely freely as cfDNA can be found adherent to antibodies, red blood cell membranes, proteins, particularly nucleosomes, in single-stranded, double-stranded, and triple-stranded DNA forms, in complex with RNA, as well as in adducts to proteins. cfDNA should also be differentiated from circulating cells such as circulating tumor cells, which are a source of diagnostic DNA but are not cell-free and therefore must be handled differently for laboratory purposes. In addition, cfDNA should be distinguished from circulating RNA such as messenger or microRNA can also be found in the blood and be used to diagnose diseases and this topic is reviewed elsewhere.1
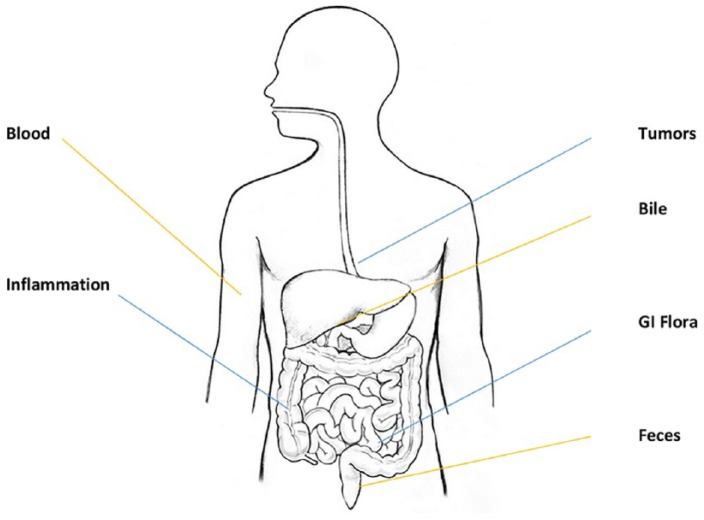
Figure 1.
Sources of cell-free DNA (cfDNA).
cfDNA is released by all tissues of the body including during normal function, but also by sources with differential or abnormal function such as gut flora or cancer (blue lines). cfDNA is usually understood to be in the plasma, but can also be found in other bodily fluids such as the bile, feces, and urine.
There are several sources of cfDNA, and it can be found in the plasma of healthy persons, but an increased concentration of cfDNA can be found in several disease states, especially in inflammatory conditions such as systemic lupus erythematosus as well as with many cancers. Moreover, viral and bacterial DNA can be found circulating in the blood during both healthy and pathologic conditions. In fact, gastrointestinal-derived bacterial DNA can be found circulating in the blood of healthy subjects as well as in increased concentrations in the blood of subjects with inflammatory bowel disease or liver cirrhosis.2,3 The majority of cfDNA research has been completed on blood cfDNA, most commonly in plasma, rather than serum. Far fewer studies have been performed on other bodily fluids such as bile, pancreatic juices, urine, or stool, and therefore this review focuses on circulating cfDNA in the blood.
Despite the discovery of cfDNA in the many varied forms described above, there is still controversy regarding some of its basic properties such as the size ranges found circulating in the blood as well as the mechanism of release of DNA into the extracellular space. Undoubtedly, apoptosis and necrosis of cells are a source, however, there is speculation that even healthy cells may release genomic DNA. Size determination has been hindered by the fragile nature of free, unprotected, DNA and by artifacts that can be introduced by differences in handling and processing of samples and by laboratory equipment. Methods as varied as simple gel electrophoresis, Sanger sequencing, and electron and atomic force microscopy have produced wildly variable estimates in the average cfDNA size. The lower and upper limits of size ranges from a dozen base pairs to tens of thousands of kilobases.
Regardless of size, it cannot be disputed that the total levels cfDNA can be accurately and reliably measured and are found to vary between healthy and certain disease states. Moreover, with recent technological advances the full breadth of the variations and aberrations seen within nuclear genomic DNA can also be reliably measured in cfDNA, including single or multiple base pair substitutions, insertions, deletions, copy number variations and methylation.4
Current advances and applications in cfDNA technologies
In the past, there were two main challenges preventing robust analysis of cfDNA, namely, low concentration and low allele frequencies. Indeed, the overall concentration of total cfDNA is low within the blood and the aberrant allele being tested for might only constitute a small fraction of the total cfDNA. In these scenarios, the mutant allele is obscured during traditional Sanger sequencing by the overwhelming copies of the normal allele. For instance, plasma concentrations of total cfDNA in a cancer patient may be on the order of 2 µg/ml and the frequency of a mutant allele such as KRAS 35G>A (KRasG12D) may only constitute 0.01% of all the circulating DNA (the range of plasma cfDNA in cancer patients in many studies falls ~20 ng/ml–2 µg/ml, as compared with ~0.1–20 ng/ml of cfDNA in the plasma of healthy controls in most studies).2,5
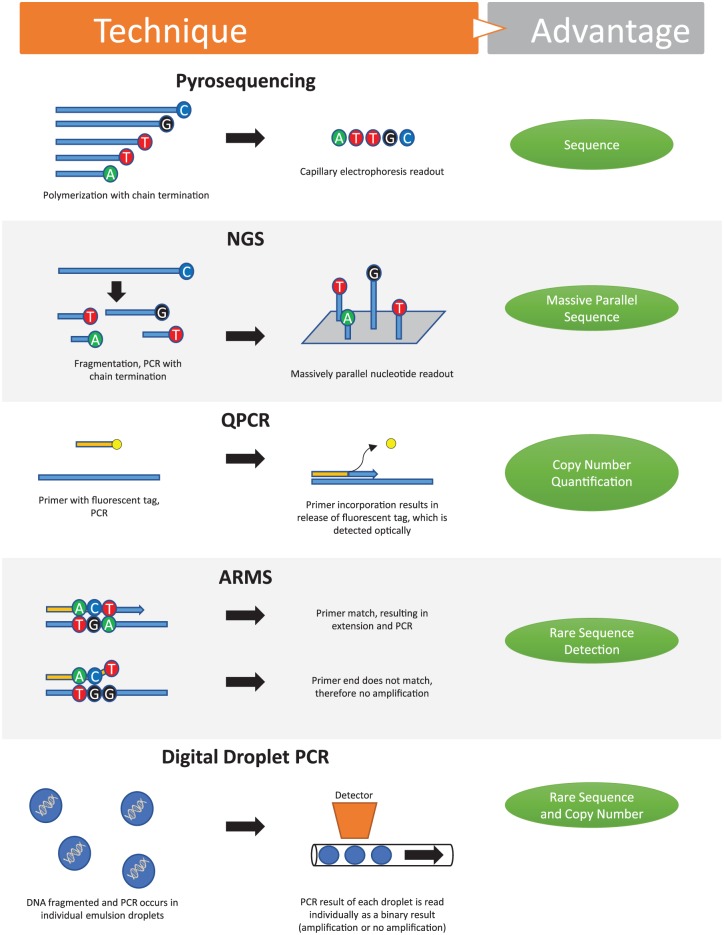
Currently, the challenges of low concentrations and low allele frequencies of cfDNA have been solved by new technological advances. These methods are outlined in Figure 2. Beyond traditional polymerase chain reaction (PCR), the first group of methods involve a priori knowledge of the specific mutant allele being tested. One such method is amplification-refractory mutation system (ARMS) PCR, also known as allele-specific PCR (ASPCR), which has been shown to reliably detect fewer than 100 copies of per milliliter of plasma (subnanogram range).5,6 Assays based on the same principle as ARMS have also been developed using fluorescent probes and quenchers to allow for real-time quantitation.
Figure 2.
Methods of detecting cell-free DNA. ARMS, amplification-refractory mutation system; NGS, next generation sequencing; PCR, polymerase chain reaction; QPCR, quantitative real-time PCR.
Another method for assaying known mutations is to block the amplification of wildtype allele with ‘clamps’ that hybridize to the wildtype allele and block DNA polymerase, or by destruction of the wildtype allele with restriction enzymes. Many ingenious clamps have been designed to efficiently and stably hybridize to target sequences without themselves acting as primers, including protein nucleic acids, 3’-end locked nucleic acids, and xenonucleic acids. These methods allow for the detection of mutant alleles starting with just 15 femtogram (millionth of a nanogram) of genomic DNA dissolved 1:1000 with wildtype DNA.7
Finally, a method that both detects low-frequency alleles and is also able to quantify the exact frequency of the mutant/variant allele is digital droplet PCR. This method relies on separating each strand of cfDNA into its own reaction vesicle or droplet. This is usually accomplished through emulsification of the sample DNA and PCR reagents within a single PCR tube. With only one strand of DNA per reaction there is no chance of competition between wildtype and mutant sequences and, thus, each droplet results in a binary reading of 1 for a positive result or a 0 for a negative result and hence the ‘digital’ reading. All of the droplets are read, usually through a fluorescent PCR reaction, and the results tallied so that the exact frequency of a mutant allele can be calculated.8 One such method is BEAMing, which stems from an abbreviation of the components of the process (beads, emulsion, amplification, and magnetics).9
The main advantage of all the aforementioned assays is that they are highly sensitive, however, they rely on knowledge of the specific mutation and, thus, must be multiplexed to assay for other mutations. The second group of cfDNA assays do not involve a priori knowledge of the specific mutation or variant. These assays rely on massively parallel sequencing or ‘next-generation sequencing’ (NGS) in which all the collected cfDNA is sequenced. These assays all rely on fragmentation of DNA into smaller pieces, separation of individual strands into single reaction vessels, and some form of readout of the sequence, typically optical readout of fluorescently labeled nucleotides added during a polymerase reaction. However, currently, most NGS systems have multiple limitations with regards to analyzing cfDNA. First, because these systems rely on multiple short reads to assemble larger sequences, they do not have the sensitivity to detect small (<1%) copy numbers of aberrant DNA. Second, massive amounts of data are generated and may result in the detection of multiple mutations of unclear significance for clinicians, and distress for patients. Third, these platforms tend to be much more costly than targeted tests. In addition to sequence variation, copy number can also be assayed for in cfDNA. Methods for this detection generally rely on quantitative real-time PCRs (QPCRs) or NGS. Clinically, copy number variants are used to assay for oncogene amplifications such as NMYC or HER2 amplifications, or to assess for duplications and deletions of whole portions of chromosomes.
Similarly, methylation status can be assayed for with any of the previously described methods, the main difference being that samples are bisulfite treated beforehand, which distinguishes methylated cytosine from nonmethylated cytosine. A hidden power of this method is that different tissues have characteristic methylation signatures and, therefore, cfDNA from different tissues may be distinguished in the peripheral blood, allowing for specific analysis of cfDNA originating from the gastrointestinal tissue of interest. More commonly, methylation status of given genes can be used to differentiate healthy versus disease states, which we describe in the next section. Though this technique can be powerful, one major limitation of bisulfite treatment is the loss of significant fragments of cfDNA, particularly smaller (<150 bp) portions that make up a large portion of the cfDNA. Current bisulfite techniques result in recovery of anywhere between 13% and 69% of the input DNA depending on the commercial kit used.10
Specific uses of cfDNA testing in gastrointestinal and liver diseases
A laboratory technique is only useful if there are specific clinical applications; and in this section we will review possible cfDNA targets based on research and current clinical practice, with a summary of putative targets in gastroenterology and hepatology listed in Table 1.
Table 1.
Summary of putative targets in gastroenterology and hepatology.
| Gastrointestinal or liver disease | Specific marker | |
|---|---|---|
| Cancer | ||
| Esophageal Esophageal adenocarcinoma Barrett’s esophagus Esophageal squamous cell carcinoma |
Methylation of APC,11 CDKN2A,12 SFRP1, WIF1, DKK3, and RUNX-313,14
Microsatellite instability15 Chromosomal aberration and microsatellite instability: 9p (CDKN2A), 17p (TP53) and 5q (APC)13,16 |
|
| Gastric Gastric adenocarcinoma |
Mutation of TP5317
Amplification of MYC,18 HER2,19 Alu20,21 |
|
| Intestinal Gastrointestinal stromal tumors |
Microsatellite instability22 |
|
| Hepatic Hepatic adenomas Hepatocellular carcinoma |
Mutations in HFNA123
Total cfDNA levels,24 cfDNA size,25 or chromosomal abberation,26,27 and association with metastasis28 Methylation of GSTP1, p15, p16, RASSF1A associated with HCC29–32 INK4,33 RGS10, ST8SIA6, RUNX2 and VIM34 Microsatellite alterations associated with HCC metastasis29 LINE-1 hypomethylation associated with HCC and poor prognosis35,36 |
|
| Gall bladder and bile duct carcinomas | Mutation of KRAS, BRAF, TP53, ARID1A, IDH1/2, PBRM1, BAP1, PIK3CA, MCL1, PBRM1, ERBB2, SMAD4, FBXW7, GNAS, CDKN2A/2B,STAT3, VHL, APC, PTEN, DCC, TFF1 and NF24,37–41 | |
| Pancreatic Pancreatic adenocarcinoma |
Mutation of KRAS4,42–45 and GNAS46
Methylation of BNC1, ADAMTS1, NPTX2, UCHL1, SARP2, ppENK, p16, RASSF1A, APC, DCC, p14,47 HMLH1, MYOD1, CCND2 and CDKN1C48 |
|
| Colorectal Colon adenocarcinoma |
Mutation KRAS,7,49 BRAF,49 APC and TP53 Amplification of LINE, or Alu50 Methylation of APC,31 RASSF1A,31 HLTF, HPP1/TPEF, hMLH1,51 SEPT9,52 AGBL4, FLI1, and TWIST1 promoter hypermethylation53 |
|
| Autoimmune and inflammatory | ||
| Inflammatory bowel disease | Mutation of XIAP, IL10RA, IL23R, CARD9,54 AICDA, BTK, CD40LG, CYBA, CYBB, DCLRE1C, FOXP3, HPS1, HPS4, HPS6, ICOS, IL2RA, LRBA, MEFV, MVK, NCF2, NCF4, PTEN, RET, SH2D1A, SLC37A4, STXBP2, TTC37 and WAS55
Methylation of TEPP,56 BCL3, PPARG, STAT3, OSM, STAT5, IL12RB, SOX1, COL18A1,57 THRAP2, FANCC, TNFSF4, TNFSF12, FUT7, CARD9, ICAM3, and IL8RB58 |
|
| Ulcerative colitis specific | Microsatellite instability,59 CFI, SPINK4, THY160
Rare copy number variants ABCC4, CLDN10, RNF216, ZNF815, OCM, CCZ1 and KCNK961 |
|
| Crohn’s disease specific | Mutation of NOD254
Methylation of MAPK13, FASLG, PRF1, S100A13, RIPK3, IL-21R, IL-27, IL-19, TNF, MST1, and NOD262 |
|
| PBC PSC |
Methylation of SMARCA1, CD40L, FUNDC2, CXCR3, IL-17, IFN-γ63
None |
|
| Infectious Diseases | ||
| Hepatitis B | Total cfDNA level and size25
Methylation of ZNF300, SLC22A20 and SHISA7 in relation to development of HBV-related HCC64 Methylation of HOXA2, HDAC4 and PPP1R18 |
|
| Hepatitis C | Total cfDNA levels and HCV-related HCC,65,66 and transformation to HCC in HCV28 | |
| Other | ||
| NAFLD and NASH | Total cfDNA levels67
Methylation of PPARγ68 |
|
HCV, Hepatitis C; HCC, hepatocellular carcinoma; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis.
By far, the area with the most potential targets is cancer, and the rest of this review will focus on malignancy, though included in Table 1 are putative cfDNA markers in other digestive diseases. Not only is there a plethora of research into cancer genetics, but also the mutational process that drives cancer makes cfDNA approaches a low-hanging fruit. As mentioned previously, many studies have indicated that the total amount of cfDNA circulating in the blood is increased in many cancers and inflammatory conditions.6,24 Thus, testing for the concentration of cfDNA could be thought of as a general marker akin to the erythrocyte sedimentation rate as a surrogate for inflammation.
Upper gastrointestinal cancers
A use of cfDNA could be to differentiate malignant from premalignant conditions. For instance, patients with Barrett’s esophagus and esophageal adenocarcinoma could be differentiated by a 911 gene signature from a plasma cfDNA methylome proof of concept study.69 Theoretically, this type of signature could be valuable to risk stratify patients with severe gastroesophageal reflux disease (GERD) or Barrett’s esophagus into high and low risk groups for endoscopic screening. Similar targets exist in gastric adenocarcinomas and gastrointestinal stromal tumor (GIST), which could be used to screen for cancer in at risk individuals with concerning symptoms, or to differentiate benign from malignant gastric ulcers (Table 1).
Hepatobiliary and pancreatic cancers
A clinical application of cfDNA could be to diagnose and risk-stratify patients with hepatic masses. For instance, it can be challenging to differentiate atypical focal nodular hyperplasia, hepatocellular adenoma, and hepatocellular carcinoma (HCC) when radiographically ambiguous. Moreover, patients with hepatocellular adenoma need to be screened for malignant transformation to HCC. There are a number of putative markers that can diagnose and differentiate these tumors, some of which have already been found in cfDNA (Table 1). These markers can be used to risk-stratify patients to identify those who require biopsy, ablation, or surgical resection. HFNA1 mutations are found in 30–40% of hepatocellular adenomas and can be used to diagnose these lesions from other tumors when imaging techniques are equivocal.23 In addition, mutation of β-catenin in hepatocellular adenoma is strongly associated with malignant transformation into HCC, therefore identification of the minority of hepatocellular adenomas with β-catenin mutation via cfDNA could help risk-stratify patients and prioritize surgical resection or ablation of those patients with the relevant mutation.23,70 This also applies to HCC screening in patients with hepatitis C (HCV) or cirrhosis, and has even been used to diagnose HCC in patients without elevated levels of traditional markers such as alpha fetoprotein.34
Similarly, radiographic identification of gallbladder cancer can be challenging and may mimic cholecystitis, adenomyomatosis, or polyps of the gallbladder. Pilot studies have demonstrated that the quantity of cfDNA is significantly different between inflammatory conditions and gallbladder carcinoma, suggesting use as a biomarker.71 Biliary strictures or pancreatic cysts that are equivocal for malignancy can be evaluated with endoscopic retrograde cholangiopancreatography (ERCP) brush samples or aspirated pancreatic cyst fluid during endoscopic ultrasound. In either case, cytology is valuable and specific for detecting malignancy but not sensitive. cfDNA from biliary brush samples or cyst aspirates has been shown to increase the sensitivity of detecting malignancy without decreasing specificity when detecting oncogenes such as mutant KRAS or loss of various tumor-suppressors.37,38,72
Lower gastrointestinal cancers
In colorectal cancer, cfDNA has been used to distinguish patients with benign polyps from those with colorectal cancer, as well as to predict progression and treatment resistance.50 For instance, the total level of cfDNA correlates with overall and progression-free survival as closely as the patient’s performance status, and surprisingly even more so than the number of metastatic sites.6,49,73 Moreover, cfDNA can be used to identify resistance to targeted therapies prior radiographic findings of progression. For instance, patients that were previously wildtype for KRAS mutation and treated with the epidermal growth factor receptor (EGFR) inhibitors, were identified to have KRAS mutation in cfDNA and, therefore, resistance to EGFR inhibition, at times months prior to radiographic progression.74–76
Integrating cfDNA testing into current gastrointestinal and liver diseases diagnostics
In the United States, current preventative screening tests for gastrointestinal and liver diseases are limited to interval colonoscopy for colon cancers and one-time HCV screening for individuals born between 1945 and 1965. However, less than two-thirds of eligible Americans are screened for colon cancer per guidelines, with likely one of the most common reasons being unwillingness to undergo colonoscopy. Less-invasive screening techniques such as multitarget stool DNA testing (i.e. Cologuard®) are effective and work by testing for aberrantly methylated BMP3 and NDR54 promoter regions, mutant KRAS, and quantitative immunochemical assay for human hemoglobin.77 Other similar colorectal cancer screening panels have also been devised with targets such as SFRP2 methylation, mutation in APC, TP53, KRAS and microsatellite instability markers with relatively high sensitivity and specificity.78–80 The DNA used in these tests likely represents a combination of cellular DNA from sloughed cells as well as cfDNA, thus assaying the entire gastrointestinal tract at once.
The approach used for noninvasive colon cancer screening, with risk-stratification of patients into those who require colonoscopy versus those who can continue with interval noninvasive screening, could be extended to other gastrointestinal and liver diseases, and specifically applied to diseases diagnosed or treated with esophagogastroduodenoscopy (EGD) or ERCP. For instance, this approach can be used to diagnose celiac disease, for which ~3–5% of patients do not have anti-transglutaminase or anti-endomysial IgA antibodies. Though there is no published cfDNA signature for celiac disease, it is conceivable that a plasma cfDNA signature exists to reflect the T-cell and B-cell activity, which drives small bowel inflammation in celiac disease. A positive cfDNA test would then be followed by a confirmatory EGD.
Another example is pancreatic cancer for which there is currently no preventative screen and is the fourth leading cause of cancer death in the United States. The high mortality is no doubt related to the lack of screening tools and therefore late diagnosis, by which time pancreatic adenocarcinoma is typically quite advanced with a paucity of effective treatments. With early diagnosis, surgical resection is a viable option. Therefore, plasma or stool testing are viable options, and a specific area in which cfDNA testing may be beneficial. Previous investigations have shown that mutant KRAS and methylated BMP3 can be detected in the stool with 90% specificity, but only ~50% sensitivity.81 Several other genetic markers, specifically methylated EYA4, MDFI, and UCHL1, were tested for and found to be significantly increased in stool from patients with pancreatic adenocarcinoma but did not improve the area under the receiver operating characteristic curve. Undoubtedly, with further research a combination of cfDNA markers could be used to develop a routine screen for at risk individuals.
An advantage of stool or plasma cfDNA testing is that it assays the entire gastrointestinal tract and portions of the hepatobiliary system, whereas endoscopies and biopsies evaluate only the particular regions viewed/biopsied. Other strengths of cfDNA testing over current diagnostics are the ease of multiplexing multiple tests for multiple alleles, which can increase both sensitivity and specificity. Though there are current limits to how many cfDNA targets may be multiplexed in a single reaction vessel, there are no limits in the number of separate reactions that be performed in separate vessels for a gene or mutation panel approach to assaying. In addition, as the underlying technology is essentially the same regardless of the allele tested, this allows for rapid translation from the research benchtop to the clinical lab, and reduced expense as all DNA diagnostic technologies continue to fall in price. A reduction in price and ease of running tests could theoretically drive serial testing to monitor disease progression or treatment response,82–84 improve prognostic ability, and allow for rapid detection of treatment resistance so that modification to therapy can be done early.
Overall, we conclude that cfDNA tests can complement traditional gastroenterology and hepatology tools such as endoscopy, colonoscopy, ERCP, and traditional markers (such as plasma inflammatory markers, antibodies, and stool tests). The advantage of cfDNA testing relative to traditional diagnostics is sensitivity, versatility, ease of multiplexing, rapidly reducing costs, and noninvasive nature, which will allow for more frequent and quantitative testing.
Current problems
There are several technical challenges that must be overcome before cfDNA testing becomes widespread. First, there is no standard method of collecting circulating cfDNA, and the method of collection can have a major impact on whether there is cellular lysis resulting in dilution of cfDNA. Several studies have shown that lysis of immature red blood cells (reticulocytes) is a large source of dilution of cfDNA. Though most studies of cfDNA use plasma, others use serum. In addition, there are no standard conditions for the collection and storage of cfDNA samples, although commercial tubes exist that are geared towards reducing cell lysis for the purpose of cfDNA collection (StreckTM tubes). In addition, if not stored appropriately, for instance, without EDTA, endonucleases will rapidly degrade cfDNA. If cfDNA is purified, care must be taken that purification is not biased to certain types of fragments (i.e. larger fragments) when quantitative analysis follows.
In addition to these preanalytical issues with collection, processing, and storage, there are no standard protocols with regards to the analysis of cfDNA. As this technology matures, third-party validation of testing parameters must be taken into account such as the sensitivity, reliability, use of internal and external controls, and regulatory guidance from the FDA as well as laboratory regulatory bodies will be required. These challenges will also have to be balanced with the cost and clinical utility of performing cfDNA testing.
Other inherent problems with cfDNA include the small and fragmented nature, which makes analysis more difficult, and nearly impossible for the detection of mutations such as balanced chromosomal rearrangements.
Future directions
Regular use of cfDNA technologies in the clinical setting will not happen routinely until at least one application gains widespread acceptance as a useful, convenient and affordable tool by clinicians and patients. Noninvasive stool DNA testing for colon cancer may be this first common application that will trigger acceptance and routine use of these technologies, though no plasma testing is commonly used. Plasma testing is likely to begin in oncology before gaining traction in other medical specialties.
Once one plasma test becomes routine, the barrier to other cfDNA tests will fall as clinical laboratories obtain the equipment and expertise required for cfDNA testing. Once cfDNA sequencing, copy number, and methylation analysis of one allele can be performed routinely, the ability to perform the same test on other alleles will be much simpler, spurring further use. As an example, precursors to enzyme-linked immunosorbent assay (ELISA) were invented in the 1960s, and did not come into routine clinical use for some limited applications until the late 1970s, and it took until the 1980s for automated benchtop systems to become available.85 It was after automated systems became available that testing for antibodies and antigens became routine enough that community practitioners started using this technology regularly.
The success of prenatal genetic testing may be a glimpse into the future potential of cfDNA testing in gastroenterology and hepatology. Noninvasive prenatal genetic testing is a cfDNA technology now widely used by pregnant women. These tests are extremely reliable with sensitivities and specificities for most abnormalities being on the order of 95.8–99.7% and ~99.86–99.96%, respectively.86 This reliability is achieved even though it is estimated that fetal DNA concentrations in the maternal plasma are on the order of 1–6 ng/ml.87 In fact, women undergoing in vitro fertilization are now able to have sequencing performed on a single cell harvested from blastomeres to determine which blastomeres to use prior to implantation.88 In addition, proof-of-concept studies have shown that entire fetal genomes can be sequenced from fetal cfDNA in the maternal plasma.89,90 Taken together, the commercial and clinical success of noninvasive prenatal genetic testing demonstrates that it is clinically feasible with the ability to detect disease in small fractions of cfDNA and do so in a highly reliable fashion at acceptable cost. All of these excellent qualities could easily be translated into the diagnosis and management of digestive and liver diseases. As the technology itself matures, the main barrier to routine use will be research to develop markers with robust clinical applicability. cfDNA markers must add meaningfully to diagnosis, prognosis, and disease management. For instance, clinicians must be able to develop reliable algorithms about what to do with findings of high levels of mutant KRAS in the plasma of a patient with weight loss, nausea, and epigastric abdominal pain. For example, should such a patient undergo computed tomography (CT) imaging of the abdomen and pelvis, EGD, or positron emission tomography (PET)-CT? If these studies are negative, what are the next steps? Without preclinical and clinical research to guide clinicians, these types of dilemmas may add more to inability to obtain a diagnosis, cost, and anxiety rather than alleviate these problems. Therefore, to be beneficial results of cfDNA studies must be actionable.
Although these same challenges apply to the use of personalized medicine with genomics and germline DNA, one advantage of cfDNA over germline sequencing is the focus on tissue where the disease occurs. Whereas genomics helps to answer questions more geared towards disease risk over a lifetime and treatment options such as pharmacogenomics, cfDNA analysis is more focused towards changes to the genetics and epigenetics at a tissue level, similar to biopsy. This distinction has led to cfDNA analysis being termed a ‘liquid biopsy’ and similar to traditional biopsy is suited to diagnose disease or predisease states now, aid in treatment decisions (i.e. EGFR inhibitor versus BRAF inhibitor; glucocorticoids versus TNF inhibitor), and to monitor disease progression and treatment effectiveness.
In conclusion, we believe that as cfDNA analysis improves in sensitivity, specificity, cost, and ease of use, this technology will gain more traction in the preclinical realm as it evolves towards clinical use. The theoretical possibilities are enormous and this technology will undoubtedly become part of routine clinical use in gastroenterology and hepatology.
Footnotes
Funding: Chris M. Carlos and Catharine Nicole Jockisch Carlos Endowment Fund in Primary Sclerosing Cholangitis (PSC).
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Konstantinos N. Lazaridis  https://orcid.org/0000-0002-0437-681X
https://orcid.org/0000-0002-0437-681X
Contributor Information
Inuk Zandvakili, Division of Internal Medicine, Mayo Clinic, Rochester, Minnesota, USA.
Konstantinos N. Lazaridis, Division of Gastroenterology and Hepatology, Mayo Clinic, 200 First Street SW, Rochester, MN 55905, USA.
References
- 1. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011; 11: 426–437. [DOI] [PubMed] [Google Scholar]
- 2. Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer–a survey. Biochim Biophys Acta 2007; 1775: 181–232. [DOI] [PubMed] [Google Scholar]
- 3. Barták B, Spisák S, Solymosi N, et al. Metagenome analysis of plasma derived CFDNA in healthy patients and colon diseases. Zeitschrift für Gastroenterologie 2013; 51: A9. [Google Scholar]
- 4. Zill OA, Greene C, Sebisanovic D, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov 2015; 5: 1040–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014; 32: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spindler KL, Pallisgaard N, Andersen RF, et al. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One 2015; 10: e0108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamada T, Iwai T, Takahashi G, et al. Utility of KRAS mutation detection using circulating cell-free DNA from patients with colorectal cancer. Cancer Sci 2016; 107: 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015; 10: e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benesova L, Belsanova B, Suchanek S, et al. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal Biochem 2013; 433: 227–234. [DOI] [PubMed] [Google Scholar]
- 10. Jensen SØ, Ørntoft M-BW, Bramsen JB, et al. Abstract 3804: comprehensive comparison of bisulfite conversion kits: a guide for optimal sensitivity and specificity of circulating cell-free DNA methylation-based biomarkers. Cancer Res 2017; 77: abstract 3804. [Google Scholar]
- 11. Kawakami K, Brabender J, Lord RV, et al. Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst 2000; 92: 1805–1811. [DOI] [PubMed] [Google Scholar]
- 12. Hibi K, Taguchi M, Nakayama H, et al. Molecular detection of p16 promoter methylation in the serum of patients with esophageal squamous cell carcinoma. Clin Cancer Res 2001; 7: 3135–3138. [PubMed] [Google Scholar]
- 13. Ghorbian S, Ardekani AM. Non-invasive detection of esophageal cancer using genetic changes in circulating cell-free DNA. Avicenna J Med Biotechnol 2012; 4: 3–13. [PMC free article] [PubMed] [Google Scholar]
- 14. Liu JB, Qiang FL, Dong J, et al. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol 2011; 17: 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eisenberger CF, Stoecklein NH, Jazra S, et al. The detection of oesophageal adenocarcinoma by serum microsatellite analysis. Eur J Surg Oncol 2006; 32: 954–960. [DOI] [PubMed] [Google Scholar]
- 16. Eisenberger CF, Knoefel WT, Peiper M, et al. Squamous cell carcinoma of the esophagus can be detected by microsatellite analysis in tumor and serum. Clin Cancer Res 2003; 9: 4178–4183. [PubMed] [Google Scholar]
- 17. Hamakawa T, Kukita Y, Kurokawa Y, et al. Monitoring gastric cancer progression with circulating tumour DNA. Br J Cancer 2015; 112: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park KU, Lee HE, Park DJ, et al. MYC quantitation in cell-free plasma DNA by real-time PCR for gastric cancer diagnosis. Clin Chem Lab Med 2009; 47: 530–536. [DOI] [PubMed] [Google Scholar]
- 19. Shoda K, Masuda K, Ichikawa D, et al. HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study. Gastric Cancer 2015; 18: 698–710. [DOI] [PubMed] [Google Scholar]
- 20. Park JL, Kim HJ, Choi BY, et al. Quantitative analysis of cell-free DNA in the plasma of gastric cancer patients. Oncol Lett 2012; 3: 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beeharry MK, Liu WT, Yan M, et al. New blood markers detection technology: A leap in the diagnosis of gastric cancer. World J Gastroenterol 2016; 22: 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rawnaq T, Schwarzenbach H, Schurr PG, et al. Monitoring of loss of heterozygosity in serum microsatellite DNA among patients with gastrointestinal stromal tumors indicates tumor recurrence. J Surg Res 2011; 169: 31–35. [DOI] [PubMed] [Google Scholar]
- 23. Nault JC, Bioulac-Sage P, Zucman-Rossi J. Hepatocellular benign tumors-from molecular classification to personalized clinical care. Gastroenterology 2013; 144: 888–902. [DOI] [PubMed] [Google Scholar]
- 24. Piciocchi M, Cardin R, Vitale A, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int 2013; 7: 1050–1057. [DOI] [PubMed] [Google Scholar]
- 25. Chen H, Sun LY, Zheng HQ, et al. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology 2012; 44: 318–324. [DOI] [PubMed] [Google Scholar]
- 26. Xu H, Zhu X, Xu Z, et al. Non-invasive analysis of genomic copy number variation in patients with hepatocellular carcinoma by next generation DNA sequencing. J Cancer 2015; 6: 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015; 112: E1317–E1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tokuhisa Y, Iizuka N, Sakaida I, et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Cancer 2007; 97: 1399–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang JC, Feng YL, Guo T, et al. Circulating tumor DNA in hepatocellular carcinoma: trends and challenges. Cell Biosci 2016; 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kloten V, Becker B, Winner K, et al. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res 2013; 15: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matthaios D, Balgkouranidou I, Karayiannakis A, et al. Methylation status of the APC and RASSF1A promoter in cell-free circulating DNA and its prognostic role in patients with colorectal cancer. Oncol Lett 2016; 12: 748–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeo W, Wong N, Wong WL, et al. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int 2017; 25: 266–272. [DOI] [PubMed] [Google Scholar]
- 33. Huang G, Krocker JD, Kirk JL, et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med 2014; 52: 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wen L, Li J, Guo H, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015; 25: 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramzy II, Omran DA, Hamad O, et al. Evaluation of serum LINE-1 hypomethylation as a prognostic marker for hepatocellular carcinoma. Arab J Gastroenterol 2011; 12: 139–142. [DOI] [PubMed] [Google Scholar]
- 36. Tangkijvanich P, Hourpai N, Rattanatanyong P, et al. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta 2007; 379: 127–133. [DOI] [PubMed] [Google Scholar]
- 37. Gonda TA, Viterbo D, Gausman V, et al. Mutation profile and fluorescence in situ hybridization analyses increase detection of malignancies in biliary strictures. Clin Gastroenterol Hepatol 2017; 15: 913–919.e911. [DOI] [PubMed] [Google Scholar]
- 38. Kushnir VM, Mullady DK, Das K, et al. The diagnostic yield of malignancy comparing cytology, FISH, and molecular analysis of cell free cytology brush supernatant in patients with biliary strictures undergoing endoscopic retrograde cholangiography (ERC): a prospective study. J Clin Gastroenterol. Epub ahead of print 13 August 2018. DOI: 10.1097/MCG.0000000000001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014; 9: e115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simbolo M, Fassan M, Ruzzenente A, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014; 5: 2839–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rizvi S, Borad MJ, Patel T, et al. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin Liver Dis 2014; 34: 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tjensvoll K, Lapin M, Buhl T, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol 2016; 10: 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mora J, Urgell E, Farre A, et al. Agreement between K-ras sequence variations detected in plasma and tissue DNA in pancreatic and colorectal cancer. Clin Chem 2006; 52: 1448–1449. [DOI] [PubMed] [Google Scholar]
- 44. Kinugasa H, Nouso K, Miyahara K, et al. 761 detection of K-RAS gene mutation by liquid biopsy in patients with pancreatic cancer. Gastroenterology 2015; 148; S145–S146. [DOI] [PubMed] [Google Scholar]
- 45. Maire F, Micard S, Hammel P, et al. Differential diagnosis between chronic pancreatitis and pancreatic cancer: value of the detection of KRAS2 mutations in circulating DNA. Br J Cancer 2002; 87: 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berger AW, Schwerdel D, Costa IG, et al. Detection of hot-spot mutations in circulating cell-free DNA from patients with intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology 2016; 151: 267–270. [DOI] [PubMed] [Google Scholar]
- 47. Henriksen SD, Madsen PH, Krarup H, et al. DNA hypermethylation as a blood-based marker for pancreatic cancer: a literature review. Pancreas 2015; 44: 1036–1045. [DOI] [PubMed] [Google Scholar]
- 48. Liggett T, Melnikov A, Yi QL, et al. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer 2010; 116: 1674–1680. [DOI] [PubMed] [Google Scholar]
- 49. Spindler KL, Pallisgaard N, Vogelius I, et al. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012; 18: 1177–1185. [DOI] [PubMed] [Google Scholar]
- 50. Mead R, Duku M, Bhandari P, et al. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer 2011; 105: 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wallner M, Herbst A, Behrens A, et al. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res 2006; 12: 7347–7352. [DOI] [PubMed] [Google Scholar]
- 52. Toth K, Wasserkort R, Sipos F, et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 2014; 9: e115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin PC, Lin JK, Lin CH, et al. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann Surg Oncol 2015; 22(Suppl. 3): S1419–S1427. [DOI] [PubMed] [Google Scholar]
- 54. Mirkov MU, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol 2017; 2: 224–234. [DOI] [PubMed] [Google Scholar]
- 55. Early onset inflammatory bowel disease: sequencing panel. Emory University, Emory Genetics Laboratory, https://www.ncbi.nlm.nih.gov/gtr/tests/512417/. [Google Scholar]
- 56. Harris RA, Nagy-Szakal D, Pedersen N, et al. Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis 2012; 18: 2334–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin Z, Hegarty JP, Yu W, et al. Identification of disease-associated DNA methylation in B cells from Crohn’s disease and ulcerative colitis patients. Dig Dis Sci 2012; 57: 3145–3153. [DOI] [PubMed] [Google Scholar]
- 58. Cooke J, Zhang H, Greger L, et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis 2012; 18: 2128–2137. [DOI] [PubMed] [Google Scholar]
- 59. Rauh P, Rickes S, Fleischhacker M. Microsatellite alterations in free-circulating serum DNA in patients with ulcerative colitis. Dig Dis 2003; 21: 363–366. [DOI] [PubMed] [Google Scholar]
- 60. Hasler R, Feng Z, Backdahl L, et al. A functional methylome map of ulcerative colitis. Genome Res 2012; 22: 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saadati HR, Wittig M, Helbig I, et al. Genome-wide rare copy number variation screening in ulcerative colitis identifies potential susceptibility loci. BMC Med Genet 2016; 17: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nimmo ER, Prendergast JG, Aldhous MC, et al. Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis 2012; 18: 889–899. [DOI] [PubMed] [Google Scholar]
- 63. Cheung AC, LaRusso NF, Gores GJ, et al. Epigenetics in the primary biliary cholangitis and primary sclerosing cholangitis. Semin Liver Dis 2017; 37: 159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao Y, Xue F, Sun J, et al. Genome-wide methylation profiling of the different stages of hepatitis B virus-related hepatocellular carcinoma development in plasma cell-free DNA reveals potential biomarkers for early detection and high-risk monitoring of hepatocellular carcinoma. Clin Epigenetics 2014; 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Iida M, Iizuka N, Sakaida I, et al. Relation between serum levels of cell-free DNA and inflammation status in hepatitis C virus-related hepatocellular carcinoma. Oncol Rep 2008; 20: 761–765. [PubMed] [Google Scholar]
- 66. Iizuka N, Sakaida I, Moribe T, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res 2006; 26: 4713–4719. [PubMed] [Google Scholar]
- 67. Karlas T, Weise L, Kuhn S, et al. Correlation of cell-free DNA plasma concentration with severity of non-alcoholic fatty liver disease. J Transl Med 2017; 15: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. Epub ahead of print 21 March 2016. DOI: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhai R, Zhao Y, Su L, et al. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia 2012; 14: 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jeannot E, Mellottee L, Bioulac-Sage P, et al. Spectrum of HNF1A somatic mutations in hepatocellular adenoma differs from that in patients with MODY3 and suggests genotoxic damage. Diabetes 2010; 59: 1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumari S, Tewari S, Husain N, et al. Quantification of circulating free DNA as a diagnostic marker in gall bladder cancer. Pathol Oncol Res. Epub ahead of print 30 July 2016. DOI: 10.1007/s12253-016-0087-0. [DOI] [PubMed] [Google Scholar]
- 72. Khalid A, Zahid M, Finkelstein SD, et al. Pancreatic cyst fluid DNA analysis in evaluating pancreatic cysts: a report of the PANDA study. Gastrointest Endosc 2009; 69: 1095–1102. [DOI] [PubMed] [Google Scholar]
- 73. Basnet S, Zhang ZY, Liao WQ, et al. The prognostic value of circulating cell-free DNA in colorectal cancer: a meta-analysis. J Cancer 2016; 7: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Spindler KL, Pallisgaard N, Andersen RF, et al. Changes in mutational status during third-line treatment for metastatic colorectal cancer–results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. Int J Cancer 2014; 135: 2215–2222. [DOI] [PubMed] [Google Scholar]
- 75. Diaz LA, Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486: 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Azuara D, Rodriguez-Moranta F, de Oca J, et al. Novel methylation panel for the early detection of neoplasia in high-risk ulcerative colitis and Crohn’s colitis patients. Inflamm Bowel Dis 2013; 19: 165–173. [DOI] [PubMed] [Google Scholar]
- 77. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014; 370: 1287–1297. [DOI] [PubMed] [Google Scholar]
- 78. Muller HM, Oberwalder M, Fiegl H, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet 2004; 363: 1283–1285. [DOI] [PubMed] [Google Scholar]
- 79. Tagore KS, Lawson MJ, Yucaitis JA, et al. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer 2003; 3: 47–53. [DOI] [PubMed] [Google Scholar]
- 80. Zhai RL, Xu F, Zhang P, et al. The diagnostic performance of stool DNA testing for colorectal cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2016; 95: e2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kisiel JB, Yab TC, Taylor WR, et al. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer 2012; 118: 2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014; 20: 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Frenel JS, Carreira S, Goodall J, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 2015; 21: 4586–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schwarzenbach H, Pantel K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res 2015; 17: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin Chem 2005; 51: 2415–2418. [DOI] [PubMed] [Google Scholar]
- 86. Gil MM, Accurti V, Santacruz B, et al. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 2017; 50: 302–314. [DOI] [PubMed] [Google Scholar]
- 87. Fan HC, Blumenfeld YJ, Chitkara U, et al. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem 2010; 56: 1279–1286. [DOI] [PubMed] [Google Scholar]
- 88. Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell 2015; 58: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med 2012; 4: 137ra176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010; 2: 61ra91. [DOI] [PubMed] [Google Scholar]