Abstract

A new series of 1,4-naphthoquinones, bearing various cyclic and aliphatic amines on C2, was designed and synthesized to identify antiproliferative agents for triple-negative breast cancer, which represents a clinical challenge without targeted therapies. Among naphthoquinones, 2a and 3a inhibited the proliferation of MDA-MB-231 cells (EC50 = 1.6 and 2.7 μM, respectively), compared to primary human breast cells MCF10A. Furthermore, they did not affect the viability of peripheral blood mononuclear cells (PBMC), suggesting their potential safer use for cancer treatment. Recently, correlations have emerged between the expression of G protein-coupled receptor 55 (GPR55) and both triple-negative breast cancer development and invasion, making it a promising target for the development of targeted therapies. Based on this evidence, molecular docking studies supported the hypothesis of binding to GPR55, and pharmacological tests suggested that compound 3a could exert its antiproliferative activity acting as a GPR55 inverse agonist.
Keywords: Triple-negative breast cancer, naphthoquinone, antiproliferative, GPR55, molecular docking
Breast cancer is the most commonly diagnosed malignancy among the female population, and it is one of the leading causes of cancer-related deaths in women all over the world.1 In addition to surgery and radiotherapy, endocrine therapy usually offers a hope to breast cancer patients. The triple-negative breast cancer (TNBC) encompassed 15–20% of all diagnosed cases and, compared to the other types of breast cancer, it is characterized by a more aggressive nature and poorer prognosis. Furthermore, the risk of relapses is high, and metastases frequently occur with shorter overall survival.2,3 TNBC owes its name to the lacking expression of three markers, estrogen and progesterone receptors (ER and PR) and human epidermal growth factor receptor 2 (HER2), common targets for breast cancer therapies.4 The peculiar heterogeneity of TNBC has always hampered the development of specific targeted therapies.5 Although recently an intense understanding of the molecular mechanisms involved in the physiology of TNBC has opened the perspective for alternative therapeutic approaches, they are still under clinical trials and preclinical studies, and no targeted therapies for TNBC have been approved yet. Currently, systemic chemotherapy is the only available option for patients with TNBC.6,7 Therefore, the identification of therapeutically targeted molecules remains an attracting clinical challenge.
In the last years, the membrane-bound G protein-coupled receptor 55 (GPR55), initially deorphanized as a cannabinoid receptor, has emerged as a potential target for treating cancer, including TNBC.8 In particular, GPR55 is highly expressed in TNBC cell lines, and its activation correlates with cancer proliferation and invasion.9−11 Despite increasing evidence, the paucity of specific ligands of GPR55 has limited its study as anticancer target. Most of them possess electronegative groups and pendant aromatic or heterocyclic rings that make critical hydrogen-bonding and aromatic interactions with specific residues in the receptor binding pocket.12−14 Based on these data, the design and the discovery of new potent ligands of GPR55 may be the key for TNBC targeted therapies.15
The 1,4-naphthoquinone scaffold is often found in many natural and synthetic compounds, and the presence of the two conjugated carbonyl groups on C1 and C4 is responsible for the biological relevance of this system.16 Moreover, the number of specific substituents and their position on the naphthoquinone core are responsible for a wide range of biological activities, including anticancer.17−21 In particular, the antiproliferative activity is favored by the presence of a hydroxyl group on C5.22 Although a number of strides are being made to understand the molecular mechanism of action of 1,4-naphthoquinones, their therapeutic target still remains unknown.
Herein, we report the synthesis of novel 5,8-dihydroxy and 5,8-dimethoxy derivatives of 1,4-naphthoquinone, bearing pendant nonaromatic heterocycles or aliphatic amines on C2, with the main purpose to identify new antiproliferative agents of TNBC. The molecular mechanism of action of the new derivatives was investigated to ascertain their target. Molecular docking studies were also carried out to support the in vitro results. The synthetic steps to obtain compounds 1-6a and 1-3b are outlined in Scheme 1. According to the method of Huot and Brassard,231a was prepared as a tautomeric pair through Friedel–Crafts condensation of 1,4-dimethoxybenzene and 2,3-dichloromaleic anhydride, in a melted mixture of aluminum and sodium chlorides. It was easily purified by crystallization and isolated in high yield. Due to its high reactivity in nucleophilic substitutions, 2-6a were quickly obtained by reaction of 1a with various cyclic and aliphatic amines.
Scheme 1. Synthetic Steps for Compounds 1-6a and 1-4b.
Reagents and conditions: (i) 1. a: AlCl3, NaCl, 140–150 °C; b: 170–175 °C, 5 min; 2. H2O, HCl, rt, overnight, 99%; (ii) amine, Na2CO3, DCM, rt, overnight, 17–28%; (iii) CH3I, Ag2O, CHCl3, rt, 22 h, 56%.
Heterocyclic and diethylamino derivatives were obtained in high yield, while the lengthening of the alkyl amine chain resulted in a lower yield and formation of byproducts. The 5,8-dihydroxyl groups were replaced with methoxy substituents to evaluate their influence on the antiproliferative activity and to rule structure–activity relationship (SAR) considerations. Thus, 1b was obtained in a higher yield compared to its isomer through methylation of 1a with methyl iodide and silver oxide. Following the same reaction described before, 2-3b were obtained by nucleophilic substitutions of 1b with cyclic amines (see Supporting Information for reaction procedures).
To determine whether the new derivatives provide the desired TNBC antiproliferative activity, their EC50 was assessed against the MDA-MB-231 cell line, overexpressing GPR55, using the cell viability assay (Table 1). Although the small number of compounds, the in vitro results indicated the impact of the different substituents on the antiproliferative activity. The dihydroxyl derivative 1a, with two chlorine atoms on C2–3 positions, did not inhibit the cell proliferation. However, the presence of amino substituents on C2 showed some cytotoxic activity. Particularly, the presence of a six-membered ring, as in 2a and 3a, resulted in a potent cytotoxicity (Figure 1A,B). The analysis of dose–response curve profiled compound 3a as ligand for GPR55. In any case, redox activity of compound 2a cannot be ruled out. The morpholino derivative 3a exhibited the best antiproliferative activity, probably due to hydrophobic contacts not found in 1a (see docking results). Substitutions with the 5-membered pyrrolidine ring or dialkylamino groups resulted in a significant loss of potency. Also in this case, reduction of the ring size and its opening into a dialkyl amine led to the lack of hydrophobic contacts with the binding site amino acids. Furthermore, dimethoxy derivatives 1-3b did not affect the proliferation of TNBC cells, even if six-membered rings are attached on C2, as in 2a and 3a. These results suggest that the presence of both six-membered rings on C2 and hydroxyl groups on C5 and C8 are crucial for the antiproliferative activity.
Table 1. Summary of the in Vitro Cytotoxic Activities of the New 1,4-Naphthoquinone Derivatives.
| compd | R | MDA-MB-231a |
|---|---|---|
| 1a | >20 | |
| 2a | piperidin-1-yl | 1.6 |
| 3a | morpholin-1-yl | 2.7 |
| 4a | pirrolidin-1-yl | >10 |
| 5a | –N(Et)2 | >20 |
| 6a | –N(Bu)2 | >20 |
| 1b | >20 | |
| 2b | piperidin-1-yl | >10 |
| 3b | morpholin-1-yl | >10 |
EC50 values were determined by MTT assay after 24 h treatment and are expressed as micromolar concentrations. The values are averages of triplicate (n = 3) data and shown as the mean.
Figure 1.
Dose–response curves of compound 2a (A) and 3a (B).
The most potent compounds 2a and 3a were then evaluated for their safety, testing their cytotoxicity against MCF10A, a primary human breast cell line. The EC50 value of both 2a and 3a against MCF10A cells was higher, suggesting a slightly low antiproliferative activity than that found toward TNBC cell line (Table 2). Since most of chemotherapeutics are administered intravenously, we aimed to investigate the cytotoxicity of the selected compounds also against peripheral blood mononuclear cells (PBMC) to evaluate their safety. Compounds 2a and 3a did not affect the viability of PBMC either at 24 or 48 h of incubation (see Supporting Information), which was over 60% even at the highest test concentration (100 μM, Table 2). These data confirm the selective antiproliferative activity of the new compounds against TNBC and the potential application for the treatment of this deadly type of breast cancer in which combination therapies are emerging as strategy.30
Table 2. Comparison of the in Vitro Cytotoxic Activities of the Best Derivatives against TNBC and PBMC Cell lines.
EC50 values were determined by MTT assay after 24 h treatment and are expressed as micromolar concentrations. The values are averages of triplicate (n = 3) data and shown as the mean.
100 μM test dose. Cell viability of PBMC was evaluated with trypan blue dye exclusion.
Docking simulations were carried out to investigate the binding mode of 3a on GPR55. No three-dimensional structure of GPR55 is currently available from experimental sources. The same docking protocol, already described by us for GPR40 agonists24 and very similar to that applied by others to GPR55 simulations,25,13 has been used to evaluate the putative binding mode of the new 1,4-naphthoquinones on GPR55. The complementarity of the molecular surface of both 3a and GPR55 binding site is illustrated in Figure 2A. Compound 3a, in its best docking pose, showed the oxygen atoms in positions 4 and 5 in hydrogen bond contacts with the terminal ammonium group of Lys80 and the imidazole ring of His170 (Figure 2B). The importance of the 5-OH substituent for compound activity is in agreement with SAR on the naphthoquinones that shows a crucial role of the phenolic group for cytotoxic activity. In fact, docking simulations on methoxylated compounds 1b–3b showed that they lacked the charge-reinforced hydrogen bond constituted by the imidazolium group of His170 and the phenate substituent at position 5 of the ligand.
Figure 2.

(A) Complementarity between the molecular surface of 3a (blue) and that of the GPR55 binding site (gray). (B,C) Graphical representation of the interactions between 3a and LPI and the binding site of GPR55. The GPR55–3a complex (A) is stabilized by two hydrogen bonds (blue dashed lines) involving the ligand oxygens at 4 and 5 positions and by strong hydrophobic contacts (green dashed lines) mainly involving the morpholine ring at C2. Interestingly, LPI conserved the same hydrogen bonds (blue dashed lines, panel B), while its alkyl edge is superposed to the hydrophobic portion of morpholine of 3a. (D) Rough superposition between 3a (pink and atom type) and THM (green and atom type) shows a clear correspondence between the two phenolic hydroxyls of THM and the 4-carbonyl and 5-phenolic group of 3a. Likewise, the overall size and shape of both molecules are comparable.
The two hydrogen bonds of 3a mimic those found between the phosphate group of the lysophosphatidylinositols (LPIs) and the same amino acids (Figure 2C). Moreover, this interaction pattern was also found in the complex between GPR55 and tetrahydromagnolol (THM),13 whose 2,2′-biphenyldiol moiety is reminiscent of the two 1,3-dioxygenated molecular pathways found in 3a (namely, the 1-carbonyl/8-hydroxyl and 4-carbonyl/5-hydroxyl pairs).
In particular, a rough superposition between 3a and THM clearly shows structural similarity (in size and shape) between compounds and match between oxygenated substituents (Figure 2D).
The morpholine ring of 3a was accommodated within an aromatic cage comprising Tyr101, Phe102, Tyr106, Phe190, and Phe246 (Figure 2B), thus mimicking the pendant heteroaromatic ring found in already known GPR55 antagonists.13 This heterocyclic ring was also pointing toward Met105 that is one of the amino acids responsible for GPR55 activation. However, the overall size of 3a, which is significantly smaller than other known GPR55 antagonists since a head region is not present (see below), allows such a compound to have a certain mobility within the binding site, as already hypothesized for other smaller GPR55 antagonists.13 As a consequence, molecular docking simulations cannot predict if 3a is or is not able to prevent the amino acid conformational switch responsible for GPR55 activation. Given the uncertainty about the most appropriate form of GPR55 to be used for molecular modeling, docking simulations have been set starting from either the inactive or active GPR55 and resulted in very similar interaction patterns of 3a. In fact, the key hydrogen bonds involving both Lys80 and His170 were maintained, further supported by hydrophobic interactions with the side chain of the aromatic cage above-described.
The side chains of Val242 and Leu270 provided profitable hydrophobic contacts that contributed to further stabilize the complex (Figure 2B). Docking simulations strongly suggested that such hydrophobic interactions played a fundamental role in defining compound activity. In fact, compounds bearing a small substituent (Cl or a dialkyl amine side chains, such as in 1a, 5a, and 6a, respectively) or a five-membered heterocycle (such as in 4a) instead of the six-membered heterocyclic rings of 2a and 3a, showed a significant reduction or a complete lack of hydrophobic interactions with the two hydrophobic amino acids and with the aromatic cage (not shown), which are probably responsible for their loss of activity. Interestingly, the 8-OH substituent of 3a was superposed to the alcoholic groups of the LPI sugar moiety, although clear electrostatic or hydrogen bond interactions with GPR55 were not evidenced (Figure 2C).
Although 3a was able to maintain the hydrogen bond interaction with Lys80 that is the primary interaction site of the complexes between GPR55 and antagonist compounds,13 the overall structure of 3a was not characterized by the presence of the three structural elements described for GPR55 antagonists (namely, a head edge, a central portion, and a pendant aromatic ring).13 This results in the inability to predict if 3a could act as a GPR55 agonist or antagonist. In this view, we performed a pharmacological test using CID16020046 (thereafter referred to as CID160), a selective antagonist of GPR55 in order to characterize the pharmacodynamics profile of 3a.
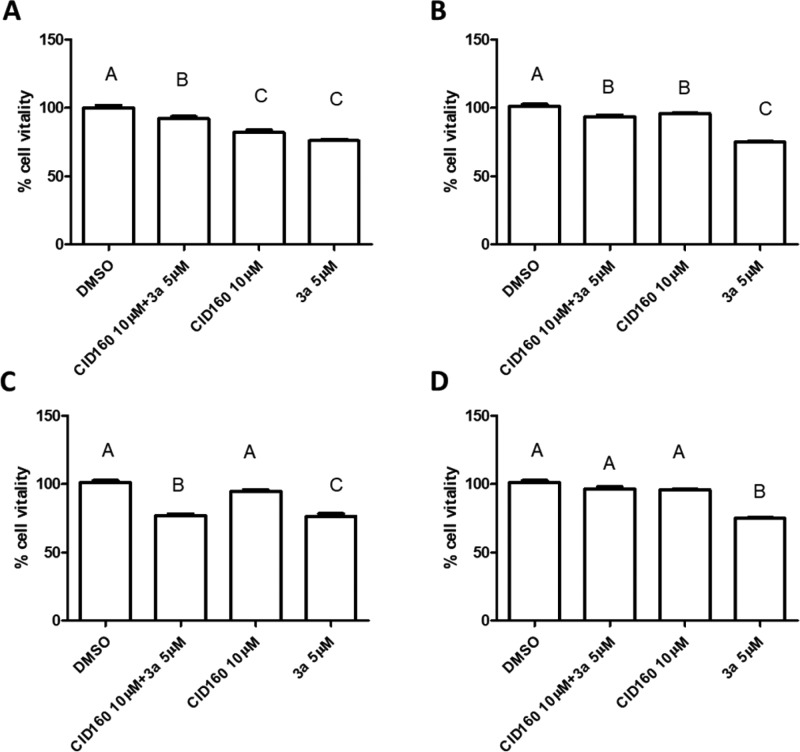
Clinical data identified LPIs as biomarkers of poor prognosis in cancer patients.26 LPIs were found to be endogenous ligands of GPR55.10 Recently, the LPI/GPR55 axis was demonstrated to enhance human breast cancer cell migration independently of the TNBC phenotype.29 Therefore, we tested 3a on both TNBC and hormone-responsive breast cancer cell lines. The exposure of MDA-MB-231 and MDA-MB-468 (Figure 3A,C) as well as MCF-7 and T47D (Figure 3B,D) to 3a led to a significant decrease of the proliferation rate compared to DMSO-treated cell. The evidence that CID160 caused a decrease of cell viability in 3 of the 4 tested cell lines could indicate the presence of endogenously produced ligand.26 In this view, the exposure of MDA-MB-231, MDA-MB-468, and MCF-7 to CID160 for 30 min before treatment with 3a was able to reverse its effect. This suggested that 3a acts as inverse agonist of GPR55. It is of note that the GPCRs exist in an active and an inactive form. In the physiological system, both forms are in equilibrium.27,28 Generally, agonists of GPCRs have higher affinity for the active form. In addition, based on this, 3a is an inverse agonist since it is able to evoke an antiproliferative effect.
Figure 3.
Activity of the GPR55 selective antagonist CID16020046 and 3a, alone and in combination on two TBNC cell line MDA-MB-231 and MDA-MB-468 (A,C) and on two hormone-responsive cell lines MCF-7 and T47D (B,D). Values with the same letter are not significantly different, while values with different letters are significantly different (*p < 0.05 and **p < 0.001 one-way ANOVA followed by Bonferroni’s comparison test, as posthoc; each sample was run in triplicate (n = 3)).
In conclusion, a new class of 1,4-naphthoquinones was designed and synthesized to identify potent antiproliferative agents. On the basis of the data obtained in vitro, we demonstrated that 3a is a GPR55 inverse agonist. Moreover, molecular docking studies showed the interactions between 3a and the binding site residues of GPR55. Further in vivo studies are needed to elucidate the pharmacokinetics profile of 3a as antiproliferative agent in tumors harboring aberrant expression of GPR55, including TNBC.
Acknowledgments
We are indebted to P. H. Reggio and D. Hurst (Department of Chemistry and Biochemistry, University of North Carolina at Greensboro, NC) who kindly provided us with the three-dimensional coordinates of both the active and inactive structures of GPR55 as obtained by homology modeling simulations. Santa Chiara Lab (University of Siena) is also acknowledged for the use of the Schrödinger suite.
Glossary
ABBREVIATIONS
- DCM
dichloromethane
- ER
estrogen receptor
- GPR55
G protein-coupled receptor 55
- HER2
human epidermal growth factor receptor 2
- LPI
lysophosphatidylinositol
- PBMC
peripheral blood mononuclear cells
- PR
progesterone receptor
- THM
tetrahydromagnolol
- TNBC
triple-negative breast cancer
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00333.
General experimental information; full synthetic procedures and characterization data for all new compounds; NMR, GC–MS, and IR spectra; experimental procedure for PBMC isolation and cell viability assays; statistical analysis; computational details (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Siegel R. L.; Miller K. D.; Jemal A. Cancer Statistics, 2018. Ca-Cancer J. Clin. 2018, 68, 7–30. 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- Brouckaert O.; Wildiers H.; Floris G.; Neven P. Update in triple-negative breast cancer: prognosis and management strategies. Int. J. Women's Health 2012, 4, 511–520. 10.2147/IJWH.S18541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitariu A. A.; Cîmpean A. M.; Ribatti D.; Raica M. Triple negative breast cancer: the kiss of death. Oncotarget 2017, 8, 46652–46662. 10.18632/oncotarget.16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho J. S.; Tutt A. N. J. Triple negative tumours: a critical review. Histopathology 2008, 52, 108–118. 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Lehmann B. D.; Bauer J. A.; Chen X.; Sanders M. E.; Chakravarthy A. B.; Shyr Y.; Pietenpol J. A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011, 121, 2750–2767. 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomao F.; Papa A.; Zaccarelli E.; Rossi L.; Caruso D.; Minozzi M.; Vici P.; Frati L.; Tomao S. Triple-negative breast cancer: new perspectives for targeted therapies. OncoTargets Ther. 2015, 8, 177–193. 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon J.; Lousberg L.; Schroeder H.; Jerusalem G. Triple-negative breast cancer: treatment challenges and solutions. Breast Cancer: Targets Ther. 2016, 8, 93–107. 10.2147/BCTT.S69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Illades D.; DeMorrow S. Orphan G protein receptor GPR55 as an emerging target in cancer therapy and management. Cancer Manage. Res. 2013, 5, 147–155. 10.2147/CMAR.S35175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andradas C.; Caffarel M. M.; Pérez-Gómez E.; Salazar M.; Velasco G.; Guzmán M.; Sánchez C. The orphan G protein-coupled receptor GPR55 promotes cancer cell proliferation via ERK. Oncogene 2011, 30, 245–252. 10.1038/onc.2010.402. [DOI] [PubMed] [Google Scholar]
- Andradas C.; Blasco-Benitp S.; Castillo-Lluva S.; Dillenburg-Pilla P.; Diez-Alarcia R.; Juanes-García A.; García-Taboada E.; Hernando-Llorente R.; Soriano J.; Hamann S.; Wenners A.; Alkatout I.; Klapper W.; Rocken C.; Bauer M.; Arnold N.; Quintanilla M.; Megías D.; Vicente-Manzanares M.; Urigüen L.; Gutkind J. S.; Guzmán M.; Pérez-Gómez E.; Sánchez C. Activation of the orphan receptor GPR55 by lysophosphatidylinositol promotes metastasis in triple-negative breast cancer. Oncotarget 2016, 7, 47565–47575. 10.18632/oncotarget.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Guo X.; Song Y.; Zhu C.; Zou W. The LPI/GPR55 axis enhances human breast cancer cell migration via HBXIP and p-MLC signaling. Acta Pharmacol. Sin. 2018, 39, 459. 10.1038/aps.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhouayek M.; Masquelier J.; Muccioli G. G. Lysophosphatidylinositols, from Cell Membrane Constituents to GRP55 Ligands. Trends Pharmacol. Sci. 2018, 39, 586–604. 10.1016/j.tips.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Kotsikorou E.; Sharir H.; Shore D. M.; Hurst D. P.; Lynch D. L.; Madrigal K. E.; Heynen-Genel S.; Milan L. B.; Chung T. D. Y.; Seltzman H. H.; Bai Y.; Caron M. G.; Barak L. S.; Croatt M. P.; Abood M. E.; Reggio P. H. Identification of the GPR55 Antagonist Binding Site Using a Novel Set of High-Potency GPR55 Selective Ligands. Biochemistry 2013, 52, 9456–9469. 10.1021/bi4008885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Aviña M. E.; Lingerfelt M. A.; Console-Bram L. M.; Gamage T. F.; Sharir H.; Gettys K. E.; Hurst D. P.; Kotsikorou E.; Shore D. M.; Caron M. G.; Rao N.; Barak L. S.; Abood M. E.; Reggio P. H.; Croatt M. P. Design, synthesis, and analysis of antagonists of GPR55: Piperidine-substituted 1,3,4-oxadiazol-2-ones. Bioorg. Med. Chem. Lett. 2016, 26, 1827–1830. 10.1016/j.bmcl.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangini M.; Iaccino E.; Mosca M. G.; Mimmi S.; D’Angelo R.; Quinto I.; Scala G.; Mariggió S. Peptide-guided targeting of GRP55 for anti-cancer therapy. Oncotarget 2017, 8, 5179–5195. 10.18632/oncotarget.14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonholo J.; Freitas L. R.; de Abreu F. C.; Azevedo D. C.; Zani C. L.; de Oliveira A. B.; Goulart M. O. F. Electrochemical Properties of Biologically Active Heterocyclic Naphthoquinones. J. Braz. Chem. Soc. 1998, 2, 163–169. 10.1590/S0103-50531998000200008. [DOI] [Google Scholar]
- Janeczko M.; Demchuk O. M.; Strzelecka D.; Kubiński K.; Masłyk M. New family of antimicrobial agents derived from 1,4-naphthoquinone. Eur. J. Med. Chem. 2016, 124, 1019–1025. 10.1016/j.ejmech.2016.10.034. [DOI] [PubMed] [Google Scholar]
- Kehelpannala C.; Kumar N. S.; Jayasinghe L.; Araya H.; Fujimoto Y. Naphthoquinone Metabolites Produced by Monacrosporium ambrosium, the Ectosymbiotic Fungus of tea Shot-Hole Borer, Euwallacea fornicates, in Stems of Tea, Camellia sinensis. J. Chem. Ecol. 2018, 44, 95–101. 10.1007/s10886-017-0913-1. [DOI] [PubMed] [Google Scholar]
- Deniz N. G.; Ibis C.; Gokmen Z.; Stasevych M.; Novikov V.; Komurovska-Porokhnyavets O.; Ozyurek M.; Guclu K.; Karakas D.; Ulukaya E. Design, Synthesis, Biological Evaluation, and Antioxidant and Cytotoxic Activity of Heteroatom-substituted 1,4-Naphtho- and Benzoquinones. Chem. Pharm. Bull. 2015, 63, 1029–1039. 10.1248/cpb.c15-00607. [DOI] [PubMed] [Google Scholar]
- Kretschmer N.; Rinner B.; Deutsch A. J. A.; Lohberger B.; Knausz H.; Kunert O.; Blunder M.; Boechzelt H.; Schaider H.; Bauer R. Naphthoquinones from Onosma paniculata Induce Cell-Cycle Arrest and Apoptosis in Melanoma Cells. J. Nat. Prod. 2012, 75, 865–869. 10.1021/np2006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachayasittikul V.; Pingaew R.; Worachartcheewan A.; Sitthimonchai S.; Nantasenamat C.; Prachayasittikul S.; Ruchirawat S.; Prachayasittikul V. Aromatase inhibitory activity of 1,2-naphthoquinine derivatives and QSAR study. EXCLI J. 2017, 14, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifazi E. L.; Ríos-Luci C.; León L. G.; Burton G.; Padrón J. M.; Misico R. I. Antiproliferative activity of synthetic naphthoquinones related to lapachol. First synthesis of 5-hydroxylapachol. Bioorg. Med. Chem. 2010, 18, 2621–2630. 10.1016/j.bmc.2010.02.032. [DOI] [PubMed] [Google Scholar]
- Huot R.; Brassard P. Friedel-Crafts Condensations with Maleic Anhydrides. III. The Synthesis of Polyhydroxylated Naphthoquinones. Can. J. Chem. 1974, 52, 838–842. 10.1139/v74-132. [DOI] [Google Scholar]
- Kotsikorou E.; Madrigal K. E.; Hurst D. P.; Sharir H.; Lynch D. L.; Heynen-Genel S.; Milan L. B.; Chung T. D. Y.; Seltzman H. H.; Bai Y.; Caron M. G.; Barak L.; Abood M. E.; Reggio P. H. Identification of the GPR55 Agonist Binding Site Using a Novel Set of High-Potency GPR55 Selective Ligands. Biochemistry 2011, 50, 5633–5647. 10.1021/bi200010k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badolato M.; Carullo G.; Perri M.; Cione E.; Manetti F.; Di Gioia M. L.; Brizzi A.; Caroleo M. C.; Aiello F. Quercetin/oleic acid-based G-protein-coupled receptor 40 ligands as new insulin secretion modulators. Future Med. Chem. 2017, 9, 1873–1885. 10.4155/fmc-2017-0113. [DOI] [PubMed] [Google Scholar]
- Ross R. A. L-α-lysophosphatidylinositol meets GPR55: a deadly relationship. Trends Pharmacol. Sci. 2011, 32, 265–269. 10.1016/j.tips.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Bond R. A.; Ijzerman A. P. Recent developments in constitutive receptor activity and inverse agonism, and their potential for GPCR drug discovery. Trends Pharmacol. Sci. 2006, 27, 92–96. 10.1016/j.tips.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Smit M. J.; Vischer H. F.; Bakker R. A.; Jongejan A.; Timmerman H.; Pardo L.; Leurs R. Pharmacogenomic and structural analysis of constitutive g protein-coupled receptor activity. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 53–87. 10.1146/annurev.pharmtox.47.120505.105126. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Guo X.; Song Y.; Zhu C.; Zou W. The LPI/GPR55 axis enhances human breast cancer cell migration via HBXIP and p-MLC signaling. Acta Pharmacol. Sin. 2018, 39, 459–471. 10.1038/aps.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.; Djamgoz M. B. A. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat. Rev. 2018, 62, 110–122. 10.1016/j.ctrv.2017.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.