Abstract
One of the most promising applications of DNA origami is its use as an excellent evolution of nanostructured intelligent systems for drug delivery, but short in vivo lifetime and immune-activation are still major challenges to overcome. On the contrary, stealth liposomes have long-circulation time and are well tolerated by the immune system. To overcome DNA origami limitations, we have designed and synthesized a compact short tube DNA origami (STDO) of approximately 30 nm in length and 10 nm in width. These STDO are highly stable ≥48 h in physiological conditions without any postsynthetic modifications. The compact size of STDO precisely fits inside a stealthy liposome of about 150 nm and could efficiently remotely load doxorubicin in liposomes (LSTDO) without a pH driven gradient. We demonstrated that this innovative drug delivery system (DDS) has an optimal tumoral release and high biocompatible profiles opening up new horizons to encapsulate many other hydrophobic drugs.
Keywords: DNA origami, remote loading, liposome, cancer, drug delivery
The unspecific large biodistribution of chemotherapeutic agents results in the poor accumulation of drugs in the tumor site that ultimately leads to high cytotoxic and other side effects. The side effects are known to make chemotherapy painful and, in some cases, can lead to therapy failure.1 In order to limit side effects and increase the therapeutic index of anticancer drugs, nanoparticles could improve physicochemical aspects of the drug such as water solubility, pharmacokinetics and biodistribution profiles, and drug half-life.2 Taking advantages from biocompatibility, biodegradability, low toxicity,3 and desirable accumulation on tumoral tissues,4 liposomal technology is under very active investigation as a DDS. Unfortunately, liposomes are efficiently loaded only with weak base or acid molecules limiting its application to a few drugs.5 Many researchers tried to solve this issue, but still there are no general methods to load different drugs in liposome efficiently. In the last decades, the attention of many research groups has been focused on DNA nanotechnology. DNA nanostructures have many advantages such as easy and custom synthesis, robust and precise nanopatterning, and mechanical rigidity, features that are ideal for the applications in the biomedical field.6 Successful applications include artificial ion channels,7 logical templates for proteins,8,9 vesicles,10−13 and cellular devices.14,15 The efficacy of complex DNA origami as an in vivo drug delivery vehicle for cancer therapy16−22 has been recently highlighted by Zhang et al., demonstrating that DNA origami with triangular shape are able to deliver doxorubicin (doxo) and passively accumulate in the tumor.19 Although attractive from a biotechnological perspective, one of the main shortcomings of DNA origami is that DNA is a biodegradable material in biological environments.23−26 DNA origami activate the innate immune system, thus severely limiting possible in vivo applications. Encapsulation of nanostructured DNA in double-layer membranes, polymer, or protein coating could avoid DNase degradation and immune system activation.26−28
Thus, there was a need of a versatile system to load different drugs, having long circulation lifetime and without the activation of immune system. Herein, we try to solve this issue by presenting an innovative remote loading system based on a compact short tube DNA origami, which is highly stable in biological environment, easily customizable, and accurately designed to fit inside a liposome. The remote loading process does not depend on the acid dissociation constant of the drug but the interaction with DNA origami.
To design a biocompatible molecular-scale device that is able to freely circulate in the blood, accumulate in the tumor, and slowly release the drug, a short tube DNA origami (STDO) structure was optimized to fit in pegylated liposomes of about 150 nm (Figure 1A,B). STDO was designed using the software CADnano (Version 2) with a six-helix bundle as template (Figure S1A,B in the Supporting Information). An easy one-step reaction was used to assemble the STDO with a very compact structure (theoretical dimension: about 30 nm in length and 10 nm in width) (Table S1 in the Supporting Information). After the synthesis, the STDO was purified by PEG precipitation29 and analyzed by agarose gel electrophoresis (AGE) (Figure S1B in the Supporting Information) and transmission electron microscopy (TEM) (Figure S1C in the Supporting Information). The dimension of the STDO was 30.1 ± 0.67 nm in length and 10.2 ± 0.09 nm in width as predicted (Figure S1D–F in the Supporting Information). PEG purification eliminates more than 95% of staple strands in excess.
Figure 1.
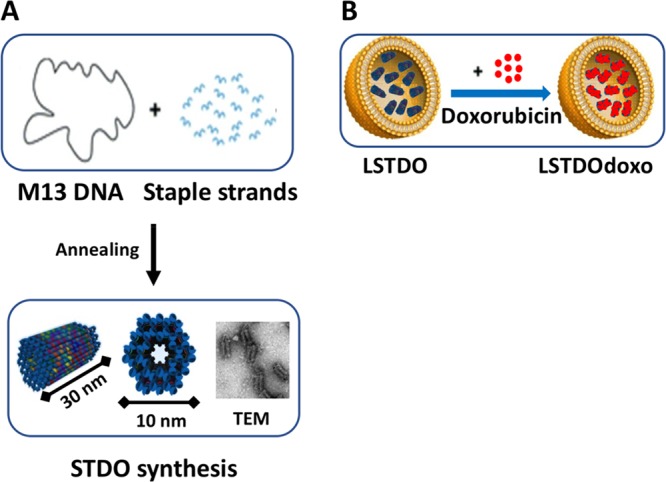
Schematic representation of (A) STDO. The single strand circular genome of M13mp18 is folded to form a DNA origami tube of 30 nm in length, through the hybridization of 113 rationally designed staple strands. (B) LSTDO. Pegylated liposomes were extruded in the presence of STDO to create a hybrid drug delivery system. Following, the preassembled LSTDO was actively loaded with doxo taking advantage by the intrinsic property of DNA to interact with anthracyclines.
We speculate that the compact structure of STDO could limit the number of accessible sites for DNases and thus prevent in vivo rapid degradation. To support our hypothesis, a triangular (TrDO) DNA origami was synthesized accordingly with Rothemund’s work,6,19 and their stability to DNases was compared to STDO. A time-course stability assay was done in the presence of 10% of FBS at 37 °C. Interestingly, the STDO exhibits significant stability up to 48 h compared to TrDO, which is stable only for 8 h (Figure 2A,B).
Figure 2.
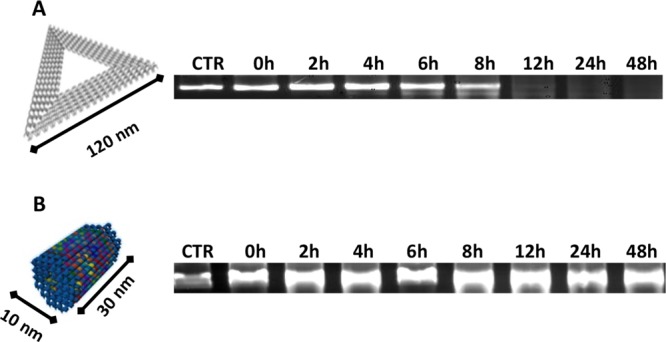
AGE analysis of DNA origami’s stability: (A) TrDO. (B) STDO. The stability test was performed in DMEM 10% FBS at 37 °C. Samples were collected at indicated time points and run on 1% agarose gel. STDO has a long stability up to 48 h.
The structures of DNA origami were verified by TEM and dynamic light scattering (DLS) analyses. The calculated hydrodynamic radii were 136.8 ± 93.4 nm for the TrDO (Figure S2A,B in the Supporting Information) and 49.0 ± 13.4 nm for STDO (Figure 3A), in line with their structural geometry.
Figure 3.
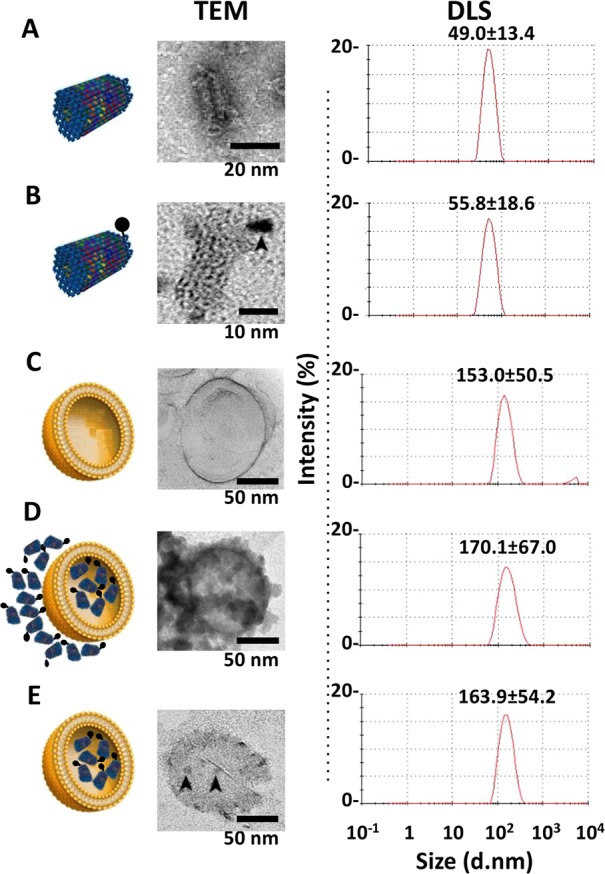
Characterization of liposomes, STDO, and LSTDO. (A) STDO was synthesized and characterized by TEM and DLS. (B) To visualize STDO inside liposome, a Cd–Se quantum dot (QD, arrowhead) was conjugated to STDO. (C) Pegylated liposomes were synthesized and (D) loaded with STDO (LSTDO). DLS was able to detect only liposomes (LSTDO). (E) Excess of STDO was eliminated incubating LSTDO with a cationic resin that binds unloaded STDO but not liposomes. Arrowheads indicate QD-STDO inside liposomes.
These observations suggest that the small and compact DNA origami structures exhibit higher stability due to their high resistance toward nuclease degradation and thus the increased in vitro and in vivo stability compared to larger and flexible structures. This enhanced stability is contributed by two phenomena: (i) the size of smaller origami makes them harder to find by the enzymes and thus slower to degrade; and (ii) a more compact structure exposes fewer reaction sites to the enzymes, further slowing their efficiency and thus the origami degradation. However, more observations are required to fully understand this distinctive behavior of these STDO nanostructures since other factors may be at play. The stability of DNA origami in vivo is an important subject on its own and will be investigated in a further work.
A liposomal/STDO nanostructure (LSTDO) was created utilizing a lipid composition derived from liposomal doxo5 applying the membrane extrusion method. The new DDS was analyzed by TEM and DLS. STDOs were modified with a biotinylated staple and conjugated with an electron-dense streptavidin quantum dot (QD) in order to visualize it inside the liposome by TEM (Figure 3B). The calculated hydrodynamic radii of the STDO-QD and of the LSTDO before and after purification from unencapsulated STDO were 55.8 ± 18.6, 153.0 ± 50.5, 170.1 ± 67.0, and 163.9 ± 54.2 nm, respectively (Figure 3B–E). TEM and DLS analyses showed the presence of STDOs inside the liposomes, confirming that the proposed coating method works as expected.
For the remote loading approach, the LSTDO was incubated with doxo. The unloaded doxo was removed by dialysis, and the loading efficiency was calculated to be about 50% (w/w) (Figure 4A). Following the loading process, the kinetics of release of doxo was tested. The fast and anabolic metabolism of cancer cell stimulates the lowering of pH in the tumor microenvironment.30 At pH 7.4, doxo is a cationic amphipathic weak base with a primary amino group in equilibrium between an ionized form and a nonionized form, while DNA presents a net negative charge. Lowering pH at 5.5, the protonation of DNA can affect the stability of the structure and doxo could be released more quickly than in physiological condition.31 As suggested in the literature,18 the release test was performed at different pH. As shown in Figure 4B, the release of LSTDO but not of liposomes depends on pH. At pH 5.5, the doxo was released faster than at pH 7.4. At pH 7.4, LSTDO released 50% of doxo 2.4-fold slowly than liposomes. Detailed release kinetics of each structure at different pH are shown in Figure S3 in the Supporting Information.
Figure 4.
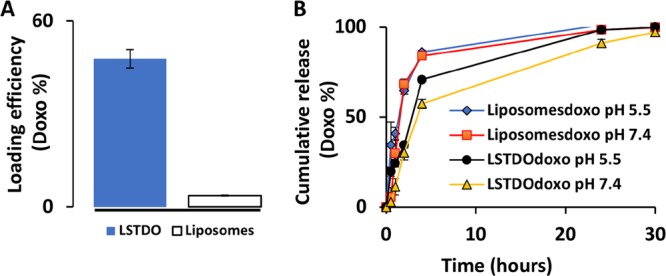
(A) Doxo loading efficiency of LSTDO. LSTDO and liposomes were incubated for 24 h with doxo and dialyzed for 2 h, and the doxo was quantified by absorbance at 450 nm (three independent experiments). (B) Doxo cumulative release of LSTDO.
This result suggests that in the extracellular space of the tumor and in subcellular compartments, the acidic conditions promote a high ratio of tumoral/nontumoral drug concentration when delivered with the LSTDO, thereby elevating the therapeutic index of doxo.
Other than the remote loading concept, LSTDO is intended to avoid the activation of the immune system by naked DNA.32−34 To support this hypothesis, we performed an ELISA test on the plasma of FVB/N mice on different cytokines (Il-1a, Il-1B, Il-2, Il-4, Il-6, Il-12, Il-17A, INFγ, TNFα). Mice were treated i.v. with the same amount of STDO and LSTDO and that should be employed for 3 mg/kg of doxo treatment based on the loading efficiency. The results demonstrated that STDO induces a high release of pro-inflammatory IL-6 compared to LSTDO and liposomes (2.47 fold; p-value < 0.05) (Figures 5A and S4 in the Supporting Information). To strengthen our data, we tested in THP1 cells, a monocytic cell line that expresses the luciferase gene under the control of interferon regulatory factor (IRF) pathway, the effect of STDO, LSTDO, and liposomes. IRF pathway is under the regulation of the TLRs family. We found that only STDO stimulated the IRF pathway highlighting, even in vitro, the ability of LSTDO to attenuate the activation of inflammatory pathways (>3.13 fold; p-value < 0.01) (Figure 5B,C).
Figure 5.
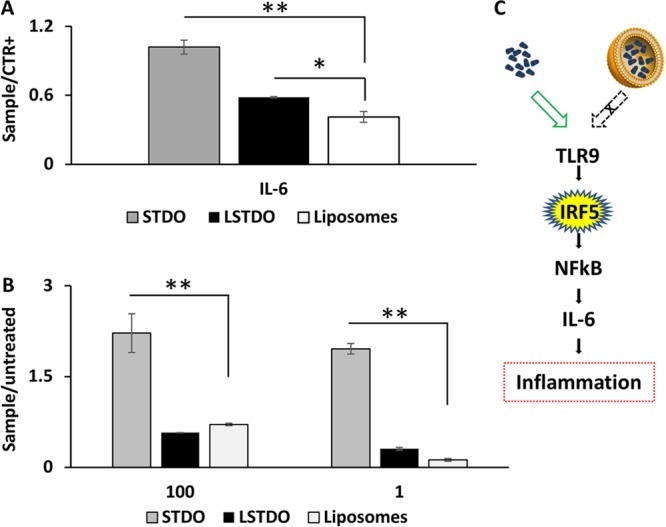
LSTDO attenuates the inflammation induced by DNA origami. (A) ELISA test on FVB/N mice. Mice (two groups of four mice) were treated with STDO, LSTDO, and liposomes to detect acute inflammation. After 6 h, blood was collected and analyzed: IL-6 increased in mice treated with STDO (*p-value < 0.05). (B) Evaluation of interferon activation in THP1 monocytic cell line. THP1 cell line was modified in order to secrete a luciferase under the control of interferon pathway. Cells were treated with 10 and 100 μg/mL of STDO, LSTDO, and liposomes. Interferon production was enhanced by the treatment with STDO (**p-value < 0.01; * <0.05). (C) Schematic representation of the TLR9 stimulation by STDO and LSTDO.
In the present work, we have addressed key limitations in the bio-application of DNA origami, i.e., their short in vivo stability and immune-activation, and seed the basis for a new drug remote loading concept. We show that the internalization of a short tube DNA origami inside a liposome prevents the inflammatory response and allows the loading of doxo with high efficiency, resulting into a high drug/lipid ratio that is essential for clinical application. The system possesses a pH-dependent controlled release with faster kinetics at pH 5.5, a tumor-like condition. Since many drugs are able to interact with DNA and STDO could be easily customizable, we strongly believe that our DDS could be helpful for the development of new liposomal biomimetic drug formulations to obtain a efficient and safe multistage systems.
Acknowledgments
C.R.S., S.P., I.C., and F.Ri. are thankful to Associazione Italiana per la Ricerca sul Cancro (My First AIRC no. 1569).
Glossary
ABBREVIATIONS
- AGE
agarose gel electrophoresis
- BSA
bovine serum albumin
- DDS
drug delivery system
- DLS
dynamic light scattering
- doxo
doxorubicin
- DPPE-PEG
1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine-PEG
- DSPC
1,2-distearoyl-sn-glycero-3-phosphocholine
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- IFN
interferon
- i.p.
intraperitoneally
- i.v.
intravenously
- LSTDO
liposome short tube DNA origami
- PEG
polyethylene glycol
- PBS
phosphate buffered saline
- PI
polydispersity index
- QD
quantum dot
- RES
reticuloendothelial system
- RT
room temperature
- STDO
short tube DNA origami
- TEM
transmission electron microscopy
- TrDO
triangular DNA origami
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00557.
Experimental section and additional figures and table(PDF)
Author Contributions
S.P., V.K., and F.Ri. conceived the study, interpreted the data, and wrote the manuscript. S.P., M.H., C.R.S., and S.B. performed experiments and analyzed the data. I.C. supported the development of in vitro experiments. G.C. and F.L.R. performed the biodistribution analysis by LC–MS/MS. V.C. performed histopathology analyses. F.Ro. interpreted data on DNA origami stability. M.A. revised the manuscript. F.Ri. supervised all the phases of the project. All authors read and commented the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Chabner B. A.; Roberts T. G. Timeline: Chemotherapy and the War on Cancer. Nat. Rev. Cancer 2005, 5 (1), 65–72. 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- Min Y.; Caster J. M.; Eblan M. J.; Wang A. Z. Clinical Translation of Nanomedicine. Chem. Rev. 2015, 115 (19), 11147–11190. 10.1021/acs.chemrev.5b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. J. W.; Semple S. C.; Klimuk S. K.; Ansell S.; Maurer N.; Cullis P. R. Characterization of the Drug Retention and Pharmacokinetic Properties of Liposomal Nanoparticles Containing Dihydrosphingomyelin. Biochim. Biophys. Acta, Biomembr. 2007, 1768 (5), 1121–1127. 10.1016/j.bbamem.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Hofheinz R.-D.; Gnad-Vogt S. U.; Beyer U.; Hochhaus A. Liposomal Encapsulated Anti-Cancer Drugs. Anti-Cancer Drugs 2005, 16 (7), 691–707. 10.1097/01.cad.0000167902.53039.5a. [DOI] [PubMed] [Google Scholar]
- Barenholz Y. Doxil®--the First FDA-Approved Nano-Drug: Lessons Learned. J. Controlled Release 2012, 160 (2), 117–134. 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440 (7082), 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Langecker M.; Arnaut V.; Martin T. G.; List J.; Renner S.; Mayer M.; Dietz H.; Simmel F. C. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 338 (6109), 932–936. 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmi D. N.; Adamson R. J.; Attrill H.; Goddard A. D.; Gilbert R. J. C.; Watts A.; Turberfield A. J. DNA-Templated Protein Arrays for Single-Molecule Imaging. Nano Lett. 2011, 11 (2), 657–660. 10.1021/nl1037769. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S.; Subramaniam S.; Stewart A. F.; Grundmeier G.; Keller A. Regular Nanoscale Protein Patterns via Directed Adsorption through Self-Assembled DNA Origami Masks. ACS Appl. Mater. Interfaces 2016, 8 (45), 31239–31247. 10.1021/acsami.6b10535. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Wang J.; Shigematsu H.; Xu W.; Shih W. M.; Rothman J. E.; Lin C. Self-Assembly of Size-Controlled Liposomes on DNA Nanotemplates. Nat. Chem. 2016, 8 (5), 476–483. 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave N.; Liu J. Programmable Assembly of DNA-Functionalized Liposomes by DNA. ACS Nano 2011, 5 (2), 1304–1312. 10.1021/nn1030093. [DOI] [PubMed] [Google Scholar]
- Li P.; Liu D.; Miao L.; Liu C.; Sun X.; Liu Y.; Zhang N. A PH-Sensitive Multifunctional Gene Carrier Assembled via Layer-by-Layer Technique for Efficient Gene Delivery. Int. J. Nanomed. 2012, 7, 925–939. 10.2147/IJN.S26955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Sun Y.; Wang L.; Wang D.; Zhou T.; Yang Z.; Chen Z.; Wang Q.; Fan Q.; Liu D. Frame-Guided Assembly of Vesicles with Programmed Geometry and Dimensions. Angew. Chem., Int. Ed. 2014, 53 (10), 2607–2610. 10.1002/anie.201310715. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335 (6070), 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Ora A.; Järvihaavisto E.; Zhang H.; Auvinen H.; Santos H. A.; Kostiainen M. A.; Linko V.; Mortensen M. R.; Gothelf K. V.; Kjems J.; Peng C. G.; Charisse K.; Borodovsky A.; Manoharan M.; Donahoe J. S.; Truelove J.; Nahrendorf M.; Langer R.; Anderson D. G. Cellular Delivery of Enzyme-Loaded DNA Origami. Chem. Commun. 2016, 52 (98), 14161–14164. 10.1039/C6CC08197E. [DOI] [PubMed] [Google Scholar]
- Tasciotti E. Smart Cancer Therapy with DNA Origami. Nat. Biotechnol. 2018, 36 (3), 234–235. 10.1038/nbt.4095. [DOI] [PubMed] [Google Scholar]
- Jiang Q.; Song C.; Nangreave J.; Liu X.; Lin L.; Qiu D.; Wang Z.-G.; Zou G.; Liang X.; Yan H.; Ding B. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 2012, 134 (32), 13396–13403. 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- Zhao Y.-X.; Shaw A.; Zeng X.; Benson E.; Nyström A. M.; Högberg B. DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties. ACS Nano 2012, 6 (10), 8684–8691. 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Jiang Q.; Li N.; Dai L.; Liu Q.; Song L.; Wang J.; Li Y.; Tian J.; Ding B.; Du Y. DNA Origami as an in Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8 (7), 6633–6643. 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- Halley P. D.; Lucas C. R.; McWilliams E. M.; Webber M. J.; Patton R. A.; Kural C.; Lucas D. M.; Byrd J. C.; Castro C. E. Daunorubicin-Loaded DNA Origami Nanostructures Circumvent Drug-Resistance Mechanisms in a Leukemia Model. Small 2016, 12 (3), 308–320. 10.1002/smll.201502118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Lytton-Jean A. K. R.; Chen Y.; Love K. T.; Park A. I.; Karagiannis E. D.; Sehgal A.; Querbes W.; Zurenko C. S.; Jayaraman M.; Peng C. G.; Charisse K.; Borodovsky A.; Manoharan M.; Donahoe J. S.; Truelove J.; Nahrendorf M.; Langer R.; Anderson D. G. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo SiRNA Delivery. Nat. Nanotechnol. 2012, 7 (6), 389–393. 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.; Bayda S.; Hadla M.; Caligiuri I.; Spena C. R.; Palazzolo S.; Kempter S.; Corona G.; Toffoli G.; Rizzolio F. Enhanced Chemotherapeutic Behavior of Open-Caged DNA@Doxorubicin Nanostructures for Cancer Cells. J. Cell. Physiol. 2016, 231 (1), 106–110. 10.1002/jcp.25057. [DOI] [PubMed] [Google Scholar]
- Chopra A.; Krishnan S.; Simmel F. C. Electrotransfection of Polyamine Folded DNA Origami Structures. Nano Lett. 2016, 16 (10), 6683–6690. 10.1021/acs.nanolett.6b03586. [DOI] [PubMed] [Google Scholar]
- Kiviaho J. K.; Linko V.; Ora A.; Tiainen T.; Järvihaavisto E.; Mikkilä J.; Tenhu H.; Nonappa; Kostiainen M. A. Cationic Polymers for DNA Origami Coating - Examining Their Binding Efficiency and Tuning the Enzymatic Reaction Rates. Nanoscale 2016, 8 (22), 11674–11680. 10.1039/C5NR08355A. [DOI] [PubMed] [Google Scholar]
- Lacroix A.; Edwardson T. G. W.; Hancock M. A.; Dore M. D.; Sleiman H. F. Development of DNA Nanostructures for High-Affinity Binding to Human Serum Albumin. J. Am. Chem. Soc. 2017, 139 (21), 7355–7362. 10.1021/jacs.7b02917. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy N.; Bastings M. M. C.; Nathwani B.; Ryu J. H.; Chou L. Y. T.; Vinther M.; Li W. A.; Anastassacos F. M.; Mooney D. J.; Shih W. M. Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation. Nat. Commun. 2017, 8, 15654. 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault S. D.; Shih W. M. Virus-Inspired Membrane Encapsulation of DNA Nanostructures to Achieve in Vivo Stability. ACS Nano 2014, 8 (5), 5132–5140. 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvinen H.; Zhang H.; Nonappa; Kopilow A.; Niemelä E. H.; Nummelin S.; Correia A.; Santos H. A.; Linko V.; Kostiainen M. A. Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv. Healthcare Mater. 2017, 6 (18), 1700692. 10.1002/adhm.201700692. [DOI] [PubMed] [Google Scholar]
- Stahl E.; Martin T. G.; Praetorius F.; Dietz H. Facile and Scalable Preparation of Pure and Dense DNA Origami Solutions. Angew. Chem., Int. Ed. 2014, 53 (47), 12735–40. 10.1002/anie.201405991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayda S.; Hadla M.; Palazzolo S.; Kumar V.; Caligiuri I.; Ambrosi E.; Pontoglio E.; Agostini M.; Tuccinardi T.; Benedetti A.; Riello P.; Canzonieri V.; Corona G.; Toffoli G.; Rizzolio F. Bottom-up Synthesis of Carbon Nanoparticles with Higher Doxorubicin Efficacy. J. Controlled Release 2017, 248, 144–152. 10.1016/j.jconrel.2017.01.022. [DOI] [PubMed] [Google Scholar]
- Kim H.; Surwade S. P.; Powell A.; O’Donnell C.; Liu H. Stability of DNA Origami Nanostructure under Diverse Chemical Environments. Chem. Mater. 2014, 26 (18), 5265–5273. 10.1021/cm5019663. [DOI] [Google Scholar]
- Tamkovich S. N.; Cherepanova A. V.; Kolesnikova E. V.; Rykova E. Y.; Pyshnyi D. V.; Vlassov V. V.; Laktionov P. P. Circulating DNA and DNase Activity in Human Blood. Ann. N. Y. Acad. Sci. 2006, 1075, 191–196. 10.1196/annals.1368.026. [DOI] [PubMed] [Google Scholar]
- Samejima K.; Earnshaw W. C. Trashing the Genome: The Role of Nucleases during Apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6 (9), 677–688. 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- Paludan S. R.; Bowie A. G. Immune Sensing of DNA. Immunity 2013, 38 (5), 870–880. 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


