Abstract
Determination mitochondrial DNA (mtDNA) sequences from extremely small amounts of DNA extracted from tissue of limited amounts and/or degraded samples is frequently employed in medical, forensic, and anthropologic studies. Polymerase chain reaction (PCR) amplification followed by DNA cloning is a routine method, especially to examine heteroplasmy of mtDNA mutations. In this review, we compare the mtDNA mutation patterns detected by three different sequencing strategies. Cloning and sequencing methods that are based on PCR amplification of DNA extracted from either single cells or pooled cells yield a high frequency of mutations, partly due to the artifacts introduced by PCR and/or the DNA cloning process. Direct sequencing of PCR product which has been amplified from DNA in individual cells is able to detect the low levels of mtDNA mutations present within a cell. We further summarize the findings in our recent studies that utilized this single cell method to assay mtDNA mutation patterns in different human blood cells. Our data show that many somatic mutations observed in the end-stage differentiated cells are found in hematopoietic stem cells (HSCs) and progenitors within the CD34+ cell compartment. Accumulation of mtDNA variations in the individual CD34+ cells is affected by both aging and family genetic background. Granulocytes harbor higher numbers of mutations compared with the other cells, such as CD34+ cells and lymphocytes. Serial assessment of mtDNA mutations in a population of single CD34+ cells obtained from the same donor over time suggests stability of some somatic mutations. CD34+ cell clones from a donor marked by specific mtDNA somatic mutations can be found in the recipient after transplantation. The significance of these findings is discussed in terms of the lineage tracing of HSCs, aging effect on accumulation of mtDNA mutations and the usage of mtDNA sequence in forensic identification.
Keywords: MtDNA, Single cell analysis, Mutation, Hematopoietic stem cells
1. Introduction
Mitochondrial DNA, a 16.6kb, circular and double-stranded molecule that is located in mitochondria, encodes 13 respiratory chain subunits, 2 ribosomal RNAs, and 22 tRNAs [1]. Each cell contains hundreds to thousands of copies of mtDNA, which replicate independent of the nuclear genomic DNA. In comparison to nuclear DNA, mtDNA has limited repair capacity and is proximate to the sites of reactive oxygen species generation; it is thus more vulnerable to mutations [2]. Because of its abundance and specific characters, such as maternal inheritance, absence of recombination, and a high mutation rate, mtDNA has been widely used in forensic science and anthropology [2–15]. In medicine, hundreds of mtDNA mutations have been reported to cause or associate with a variety of degenerative diseases (refer to MITOMAP at www.mitomap.org for more details). Within the past decade, a high frequency of somatic mtDNA mutation has been observed in tumor and aging tissues, leading to the hypothesis that mtDNA mutations play an active role in tumorigenesis and aging [2,16–23]. However, there are numerous technical problems inherent in the assay of specimens and/or clinical samples that are of limited quantity or poor quality; degraded DNA and/or an extremely low quantity of DNA, as from a single cell or a few mtDNA copies are frequently encountered [24]. In such cases, to generate an amplicon, avoid contamination, and obtain an authentic sequence are daunting tasks. Fortunately, methods and caveats have recently been elucidated that help to optimize the analysis of ancient DNA [25,26]. A posterior check of data according to mtDNA phylogenetic information is also helpful to identify potential problems [10,27–33]. Some mtDNA disease-associated mutations are present in a heteroplasmic status (co-existence of mutant and wild-type mtDNA) and show a threshold effect [34]. One routine method to determine the heteroplasmic mutation load is PCR amplification (one-step or two-step PCR, depending on the quality and amount of initial template DNA) and cloning, followed by physical isolation of sufficient positive clones (usually 5–20) for sequencing. This method has been widely employed in ancient DNA studies [8,35] and in medical research [36–38].
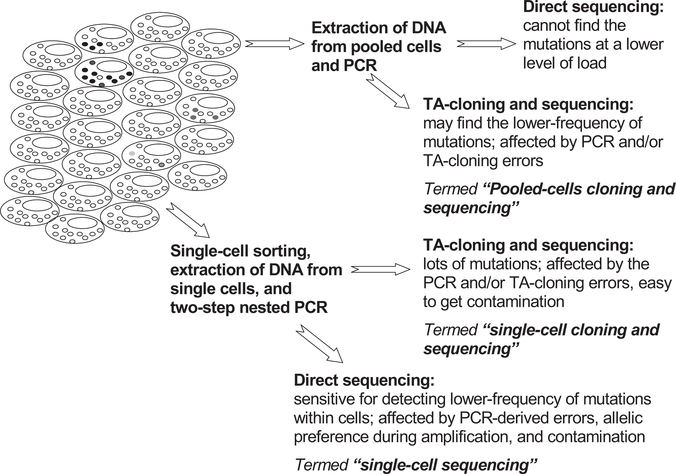
In recent years, we and others have designed methods for determining mtDNA mutations in single cells and have analyzed the mutation spectra of individual hematopoietic cells [19,21,39–42], single epithelial cells [43] and postmitotic cardiomyocytes [44,45], single muscle fibers [46–50], single neuron and glia cells [51–54], single cells of different cell lines [55], as well as DNA from minimal amounts of tissues [56,57]. Single-cell sequencing analysis is capable of detecting the lower levels of mtDNA mutations within individual cells [21,58]. We describe here the sensitivity and shortcoming of different sequencing approaches based on PCR amplification of DNA extracted from single cells and/or pooled cells (Fig. 1), and we focus on our own new and reported data in hematopoietic cells to discuss mtDNA somatic mutation patterns and process.
Fig. 1.
Strategies for determining mtDNA mutations within and among cells. Heteroplasmy of mtDNA mutations can exist within a cell and among different cells. Different mutation detection strategies have different sensitivity to pick up the lower levels of mutations and/or artifacts.
2. mtDNA mutation status in cells
Each cell usually has hundreds to thousands of mitochondria. Within each mitochondrion, there are many copies of mtDNA which are localized to numerous intramitochondrial nucleoprotein complexes (nucleoid) and are compartmentalized within mitochondrion [59]. Because of the unique compartmentalized structure of mitochondrion, and the abundance of mitochondria and mtDNA copy per cell, mutations always occur in a heteroplasmic status within a population of cells or tissues: the existence of wild type and mutant mtDNA will be present within cells (intracell heteroplasmy) and among cells (inter-cell heteroplasmy); the mutation load within a cell or tissue may vary from 0% to 100% (Fig. 1). Due to clonal expansion and genetic drift, cells with a certain mutation can become prevalent in a tissue [44,60,61]. However, in most cases, random genetic drift has been proposed to be the main factor that results in fixation of a somatic mutation in a cell or tissue [62,63]. However, recent studies based on next generation sequencing technologies showed that mtDNA heteroplasmy is extensive in tissues and positive selection acting on some somatic mtDNA mutations [20,22,23,64]. Meanwhile, mtDNA heteroplasmy at the single-cell level is affected by both genetic background and aging [19,65] and may be facilitated by intercellular exchange of mtDNA [55].
3. “Single-cell sequencing” versus “pooled-cells cloning and sequencing”
To evaluate different methods of detecting the low level of mtDNA mutation in cells, we first performed a comparison between two mutation detection methods in a leukemia patient (sample UPN16 in Yao et al. [21]). The first method, “single-cell sequencing”, has been fully described by us [19,21,41,42]. In brief, single cells sorted by flow-cytometry are lysed, then subjected to direct two-step nested PCR amplification, followed by direct sequencing of the purified PCR products. Mutations are scored according to the sequencing chromatograms, following the same approach in our previous study [21]. The second method, “pooled-cells cloning and sequencing”, includes one-step PCR amplification using DNA extracted from a homogenate of many blast cells, followed by TA-cloning and sequencing of the plasmids from positive clones (Supplementary method).We used the network method [66] to display mutations observed in single cells and single plasmids. Due to the limit of mutation scoring based on sequencing chromatograms, we only score mutations that appear with a heteroplasmy of at least 10% according to the sequencing chromatograms [21]; this conservative mutation scoring underestimates the total mutation events in a population of cells, especially when the mutations are present at a very marginal level. In a previous study, Song et al. [67] claimed that they could accurately detect mutations with a heteroplasmic level as low as 3% based on the sequencing chromatograms, by using the program Mutation Quantifier. However, in our own experience, we are unable to reliably distinguish background noise from true signal if the mutation load was under 10%.
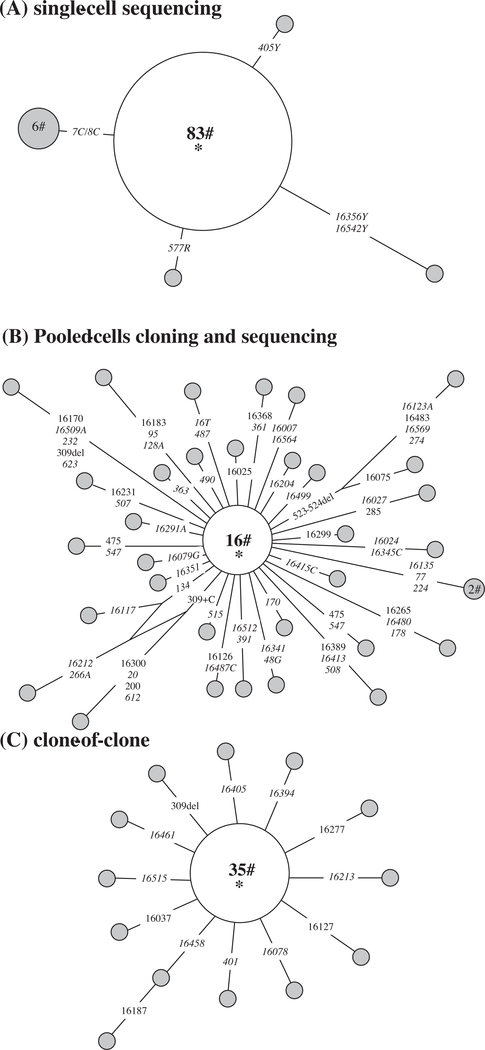
As shown in Fig. 2A, five haplotypes were found among a total of 92 individual leukemia blast cells from UPN16 that had been sorted by flow cytometry and analyzed for 1.2 kb fragment covering the entire mtDNA control region (data were initially reported in Yao et al. [21]). When we extracted DNA from pooled blast cells from this sample and performed PCR amplification (using 100 ng of genomic DNA), TA-cloning and sequencing, we observed a high frequency of mutations: 34 haplotypes were identified in a total of 50 plasmid DNAs with the 1.2 kb mtDNA insert isolated from positive Escherichia coli clones. Moreover, the sequences of some plasmid clones differed from each other in as many as ten separate mutations (Fig. 2B; new data in this study). Such a high frequency of mutations would not be simply attributable to artifacts introduced by PCR and/or TA-cloning, as we did not find such a high mutation pattern in a “clone-of-clone” assay (Supplementary method) to quantify the errors introduced by this method; in brief, we picked up a plasmid which contains an inserted sequence identical to the main haplotype sequence in Fig. 2B, and then performed PCR amplification, TA-cloning, and sequencing. Among the 48 plasmids sequenced, we observed 14 haplotypes (including the main haplotype that occurred in 35 plasmids; Supplementary Table S1). Most of the haplotypes differed from the main haplotype by only one mutation (Fig. 2C; new data in this study). It is important to note that the consensus sequences of mtDNA fragments determined in single cells and cloned plasmids are identical, justifying the use in forensics or in ancient DNA studies. The observed mutations in the pooled-cells cloning and sequencing method are thus composed of real somatic mutations and errors derived from PCR and/or TA-cloning.
Fig. 2.
MtDNA mutations detected by using the “single-cell sequencing” method (A), “pooled-cells cloning and sequencing” method (B), and “clone-of-clone” method (C) to show the artifacts. The relationship of haplotypes in single cells and plasmid clones are presented in a network profile. The order of mutations on the branch is arbitrary. Each circle represents one mtDNA haplotype, with area of the circle proportional to its frequency, and is further specified by the number of individual cells or plasmid clones sharing that haplotype. The main haplotype, which is located at the center of each network and denoted by a star (*), contains the consensus sequence (16069–16126-73–185-228–263-295–315+C-462–482-489) of all single cells or plasmid clones. The mutations observed in single cells are all heteroplasmic and are represented with all the status, e.g. 405Y means site 405 has both C and T, 7C/8C means heteroplasmy of two length mutations of C-tract (7C and 8C) in region 303–309. In all plasmid clones, we did not observe heteroplasmic mutations; mutations are thus listed as site for transitions and transversions are highlighted by adding suffixes A, G, C or T. Insertion and deletion are demonstrated by “+ inserted base” and “del”, respectively.
We tentatively estimated the point mutation frequency observed in the three experiments shown in Fig. 2. Mutations detected multiple times were counted only once. The clone-of-clone method shows a mutation frequency of 2.08 × 10−4 substitutions/bp, which may represent the error rate introduced by the PCR and/or the TA-cloning method and serves as the background noise of the technique (Supplementary Table S1). The pooled-cells cloning and sequencing method has a mutation frequency of 1.05 × 10−3 substitutions/base pair (bp), much higher than that observed by the single-cell sequencing method (3.62 × 10−5 substitutions/bp, which is estimated based on data showing in Fig. 2A). The extremely high mutation frequency estimated by using the pooled-cells cloning and sequencing method contradicted to previous studies that reported a much lower frequency ~1–5 × 10−5 [22,23] and should be received with caution, as all methods involved cloning and sequencing are highly affected by artifacts and any non-reproducible mutations must be rejected as potential artifacts. For single-cell sequencing method, according to our previous experience [19], we could consistently reproduce the majority of mutations (~80%) in single cells among duplicates in independent amplifications of single cells, which suggested that the mutation frequency estimated by this approach should be more reliable. The mutation frequency that we estimated from our single-cell sequencing method was similar to those reported by others [22,23,64], albeit different methods were used in the latter studies. Intriguingly, studies in flies [68] and mice [69] also showed similar mutation frequency as well.
4. Mutations in single cells determined by “single-cell cloning and sequencing” method
As mentioned above, each cell has hundreds to thousands of mtDNA copies; differences among these mtDNA copies constitute intracellular heteroplasmy. The single-cell sequencing method might not be able to sample the trivial difference among these copies. A sensitive method to detect these minor changes among copies of mtDNA within a cell, by “single-cell cloning and sequencing”, relies on the DNA cloning approach (Supplementary method): amplification of the target mtDNA fragment by a two-step nested PCR, TA-cloning for the PCR product, and sequencing of the positive clones are required. A potential difficulty inherent in these manipulations is increased PCR-derived artifacts from the two-step PCR, which would falsely introduce nucleotide changes that could not be distinguished from true alterations in mtDNA sequences. This method has been used to identify mtDNA somatic mutations in single neurons and glia cells from post-mortem human substantia nigra [51]: Cantuti-Castelvetri and coworkers reported that neurons had a higher frequency of somatic mutations as compared with glia cells, and remarkably many mutations were transversions [51]. Based on a counting for the number of transversions in their mutation list (disregarding the frequency of each transversion), we found that approximately 34% of mutations in glia cells and 26% of mutations in neurons are transversions. This mutation pattern obviously contrasts to the estimated transition/transversion ratio based on population-derived data [70,71], the mutation spectra of single cell analysis [19], as well as tissue-specific mutation pattern revealed by high-throughput sequencing [20,22,23]. Notably, there was an enrichment of G> T and C >A transversions in both glia cells (57% (12/21) of transversions is G> T and 33% (7/21) of transversion is C >A) and neurons (34% (13/38) of transversions is G>T and 29% (11/38) of transversion is C >A) [51]. However, this pattern was not found in other studies and there was no evidence for a striking accumulation of G> T and C >A transversions with age in brain tissues according to recent studies using the next generation sequencing technology [23] and the ultra-sensitive Duplex Sequencing methodology [22]. Therefore, the mutation pattern reported by Cantuti-Castelvetri et al. [51] may either reflect the elevated level of oxidative stress encountered in these cells, or tissue/cell specific pattern, or simply be affected by a technical artifact.
To clarify the potential technical artifact of the “single-cell cloning and sequencing” approach, we determined the mutation patterns in three single CD34+ cells and one granulocyte from one healthy donor (Supplementary Table S1). For each cell, at least 80 E. coli positive clones were isolated for sequencing. As expected, we observed a very high number of nucleotide changes in the 1.2 kb plasmid inserts sequenced. Some haplotypes differed from each other by as many as 12 mutations. We failed to confirm in single hematopoietic cells the remarkable increase in transversions and enrichment of G> T and C >A as described in single neuron and glia by Cantuti-Castelvetri et al. [51], suggesting potential cell-specific pattern or lab-specific pattern. We summarized the mutation information in the plasmid inserts picked from each of the four single cells (Supplementary Table S1). About 16% of cloned plasmids had an inserted mtDNA sequence identical to the consensus sequence of all the clones, also identical to the sequence that was determined by directly sequencing the PCR product of the single cells. Note that there were some differences in the percentage of plasmids harboring consensus sequence among the three single CD34+ cells (10%, 11%, and 19%, respectively), which might reflect the real mutation status among these cells and/or be generated randomly during the DNA amplification and plasmid physical isolation. The overall mutation frequency estimated from the four single cells determined by using the single-cell cloning and sequencing method (Supplementary Table S1) showed a mean value of 1.40 × 10−3 substitutions/bp, a value slightly higher than the mutation frequency (1.05 × 10−3 substitutions/bp) determined by the pooled-cells cloning and sequencing method, in which DNA from a cell homogenate in the leukemia patient and one step PCR amplification were used. This biased mutation frequency difference might reflect the different technical procedures that were used in the analyses, rather than of potentially biological significance. Again, we cannot take this mutation frequency if we did not discard all non-reproducible mutations.
5. mtDNA alterations in different type of blood cells from the same donor
Different tissues are composed of different types of cells and have tissue-specific gene expression patterns. In the hematopoietic system, blood cell production or hematopoiesis occurs through a highly organized system: mature erythrocytes, leucocytes, and platelets are derived from their respective intermediate progenitor cells and are replenished continuously. Committed progenitors are ultimately originated from a limited number of primitive hematopoietic stem cells [72,73]. Due to this hierarchical structure, mtDNA somatic mutations that occurred in or are fixed in HSCs should be found in the differentiated mature cells that were derived from more primitive cells of the lineage. Tracing this sentinel somatic mutation(s) might help estimate the active number of the HSCs during hemotopiesis, as well as to discern the aging effect on hematopoiesis. Mutations that occur in the committed progenitor cells or in mature cells would be obviously restricted to these cells and/or their immediate progeny and might not be present in the HSCs (disregarding parallel mutations).
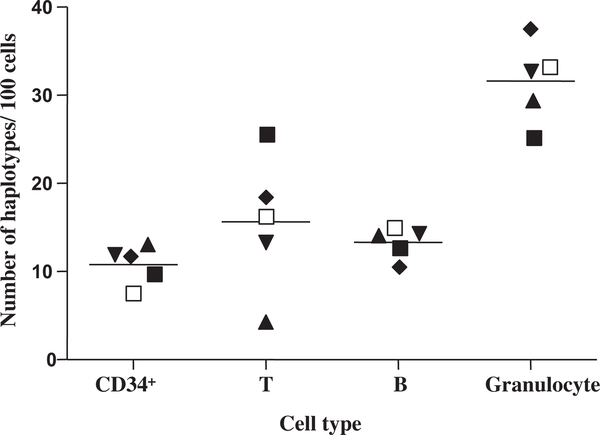
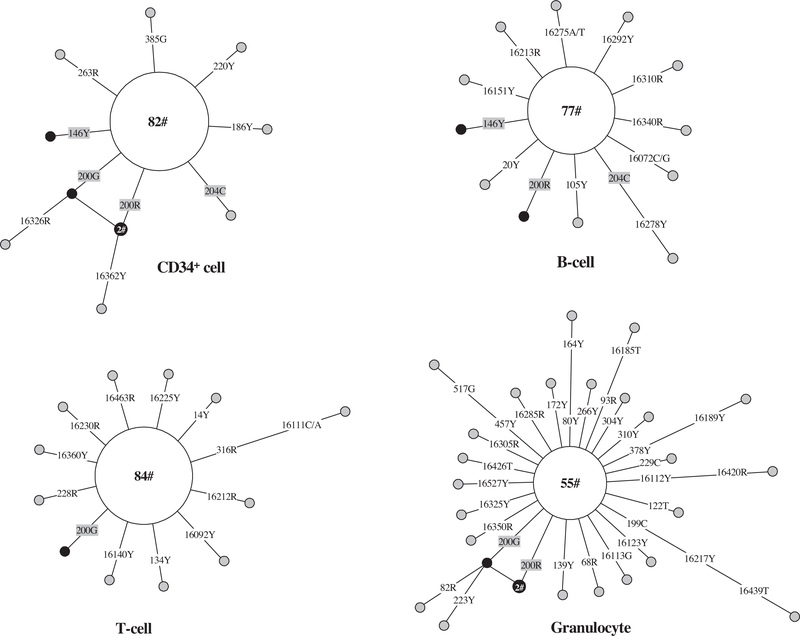
Using the single-cell sequencing method, we compared mutation patterns in single CD34+ cells (containing HSCs and progenitors), B cells, T cells, and granulocytes from five donors [41]. In general, the granulocytes showed higher intracellular heterogeneity compared with CD34+ cells, T cells, and B cells (Fig. 3). In a study aimed at quantifying the mtDNA common deletion (4977 bp) in blood cells, Mohamed et al. [74] also found that granulocytes had a higher level of a common deletion as compared to T and B cells. The higher number of mutations in granulocytes could be explained by their function and especially the high level of reactive oxygen species in these cells. Some of the mutations observed in differentiated cells could be traced to the CD34+ cells, such as 146T > Y (Y means heterogeneous for both C and T), 204T > C, and 200A >G or R (R means heterogeneous for both A and G) in one donor shown in Fig. 4, suggesting that these mutations occurred relatively early in blood cell development.
Fig. 3.
Total number of haplotypes defined by nucleotide substitutions relative to the aggregate or consensus sequence in a population of single CD34+, T, and B cells, and granulocytes. The data are taken from Ogasawara et al. [41]. Different donors are demonstrated by different shapes. The horizontal lines denote the mean values.
Fig. 4.
Network profiles of mtDNA mutations in single CD34+, T, and B cells, and granulocytes from the same donor. The original data for this donor (donor 5) were previously reported by Ogasawara et al. [41]. Only haplotypes defined by nucleotide substitutions relative to the aggregate sequence are considered. The aggregate sequence / the main haplotype contains sequence variations 16270–16292-16362–73-150–263-315 + C-517. The order of mutations on the branch is arbitrary. Each circle represents one mtDNA haplotype, with the area of the circle proportional to the frequency of the haplotype in a population of cells, and is further specified by the number of cells sharing that haplotype. Mutations shared between CD34+ and differentiated cells were marked in gray shade; the CD34+ and differentiated cell clones with identical sequence (not including the aggregate sequence) were marked in black.
6. Longitudinal assessment of mtDNA mutation in the same type of cells from same donor
In an organism’s lifespan, HSCs are mitotically quiescent and undergo a carefully regulated process of balanced self-renewal; both conditions are required in order to maintain the stem cell pool and allow periodically regulated commitment to differentiate to mature progeny [72,75,76]. Genomic instability and transformation of HSCs likely cause leukemia and other malignant hematological diseases [73]. Because of the rapid replenishment of fully differentiated end-stage blood cells in normal hematopoiesis, those mtDNA somatic mutations that arose in mature cells and/or committed progenitors at a later differentiation stage would be eliminated from the blood and should not be detectable over time. Only mutations that arose in HSCs and multipotential progenitors would be observed over an extended period time. In addition, with aging and other environmental effects, new mutations might arise in these cells.
To discern the dynamic process of HSC clones marked by mtDNA somatic mutations, we collected peripheral blood samples from one healthy donor (donor CABO) during an eight-year interval and compared the mtDNA mutation pattern in single CD34+ cells in our previous study [77]. Many of the somatic mutations observed in single CD34+ cells from the same donor at the two different time points were different. However, five CD34+ cell clones, characterized by nucleotide transitions at sites 227, 309, 541, 16221, and 16272, respectively, could be easily discerned in the serial assessment of mtDNA mutation in a population of single CD34+ cells (cf. Fig. 1C in Yao et al. [77]). This result confirms the idea that HSCs are quiescent in vivo, and suggests that primitive stem cells and somatic mutations in the stem cell compartment might persistent over time [77]. Further evidence for a quiescent status of primitive CD34+ cells marked by specific mtDNA variant(s) could be demonstrated by the analysis of CD34+ cells from mononuclear cells with different storage and transport [42]. We were able to identify CD34+ cells marked by certain mtDNA variant in two batches of CD34+ cells sampled at different time points and subjected to different storage and transport. To name a few, cells with 16131T > Y in donor #1, cells with 16129G > R in donor #2, cells with 182T > Y in donor #3, and cells with 207T > C in donor #4, were observed at both time points of sample collection [42].
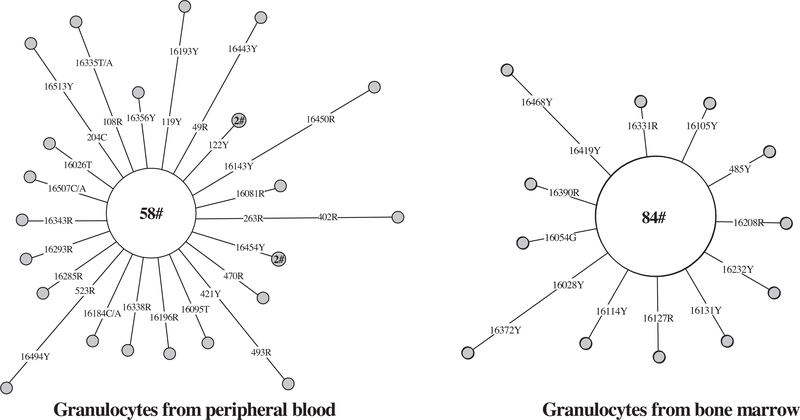
In contrast, for end-stage differentiated cells, it would be difficult to detect identical mutations in cells collected over months to year, unless these mutations arose in the HSCs and pluripotential progenitors, or were recurrent. In one donor that we analyzed, mtDNA mutations in single granulocytes from peripheral blood [41] and bone marrow [21] during a two-year interval showed no overlapping somatic mutations (Fig. 5). The number of haplotypes defined by substitutions per 100 granulocytes from bone marrow (13/96) was lower than that of granulocytes from peripheral blood (23/82) collected almost two years earlier. Unresolved is whether the difference of mutation levels in granulocytes from bone marrow and peripheral blood is secondary to tissue origin. Nevertheless, this result suggests a quick change of somatic mutations in the end-stage mature cells, consistent with the fast replenishment of these cells in blood.
Fig. 5.
Longitudinal assessment of mtDNA mutations in single granulocytes over almost two years. The original data for single granulocytes from peripheral blood and bone marrow were taken from Ogasawara et al. [41] and Yao et al. [21], respectively. The aggregate sequence of the single granulocytes /main haplotype harbors 16223–16278-16294–16390-73–146-152–195-263–315 + C- (523–524) delAC. For additional information regarding the network, see legend of Fig. 4.
7. Donor-specific somatic mutations in CD34+ cells can be transplanted to a recipient
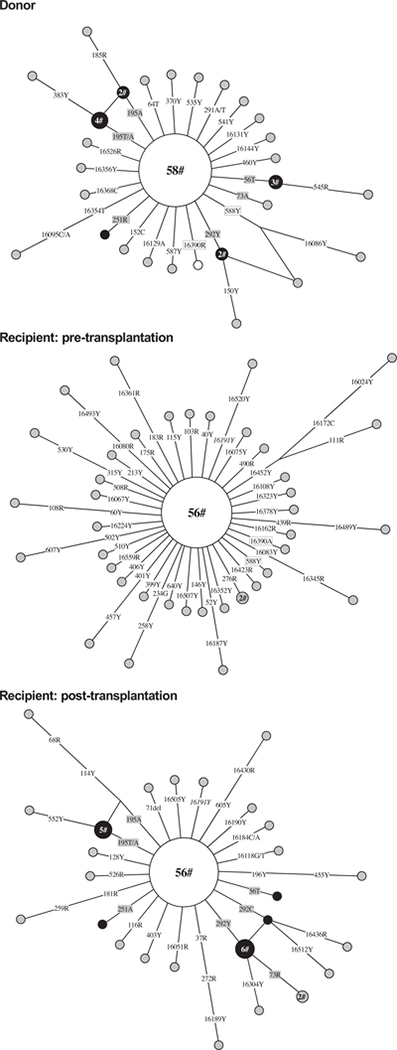
Hematopoietic stem cell transplantation is effective therapy for hematologic and lymphoid tumors as well as many other rare diseases [78]. Allogeneic transplantation, especially when HSCs from an unrelated donor are used, could produce a dilemma for forensic identification by using the blood sample from the recipient after transplantation, as the whole blood system is essentially reconstructed by the donor’s stem cells. Moreover, HSCs may also incorporate and differentiate into mature cells in various tissues [79,80]. Somatic mutations that arise in HSCs may be stable for a long time (as discussed above), and donor HSCs containing these mutations repopulate the recipient. We observed mtDNA mutations in a patient with aplastic anemia who received a HSC transplant from his brother (sample BURDC in Yao et al. [77]); there was clear evidence that CD34+ cells with specific mtDNA mutations from the donor had equal potentiality to repopulate the recipient (Fig. 6).
Fig. 6.
Network profile of mtDNA mutations observed in single CD34+ cell populations from a donor and a recipient before and after the nonmyeloidablative allogeneic stem cell transplantation. The original data were from Yao et al. [77] and we only considered the nucleotide substitutions. The main haplotype harbors sequence variations 16111–16189-16224–16256-16311–16519-56 + C-58–73-263–315 + C-497-(523–524) insAC. The CD34+ cell clones recognized by donor-specific somatic mutations (in gray shade) that repopulated the recipient eight months after transplantation were marked in black filled circles. Mutations at sites 16390 and 588 were found in both donor and recipient before transplantation and were in box; mutation at site 16191 found in the recipient before transplantation was still detected after transplantation and was in italic. For additional information regarding the network, see legend of Fig. 4.
Since the overall mtDNA sequences of the donor and recipient are largely the same (as they are maternally inherited), we focused on donor- and recipient-specific somatic mutations observed in CD34+ cells in order to distinguish the source. Fig. 6 shows the variants observed in single CD34+ cells from a donor and recipient pair before and after transplantation based on the original data published in our previous study [77]. Subsequently, the donor-originated nucleotide substitutions at sites 56, 195, 251, and 292 were detected in CD34+ cells from the recipient eight months later after transplant. Nearly all of the nucleotide substitutions present in CD34+ cells from the recipient before transplantation were not seen in the patient after transplantation, likely eliminated through the graft versus host effect [81]. This case indicates that mtDNA somatic mutations in stem cells from the donor can be transplanted to the recipient and can be used as a useful marker to trace the single HSC clone.
8. Family-specific pattern of mtDNA mutations
The inheritance of lower level of heteroplasmic mtDNA received wide concerns in the past decades. Recent studies using the next generation sequencing technologies showed that nearly half of identified mtDNA heteroplasmy is likely to be inherited, and the identified mtDNA mutations have a tissue-related, allele-related and age-related pattern [20]. Similarly, Greaves and coworker showed that neutral mtDNA mutations occur at early or may be transmitted through the germline [64]. In our recent study [19], we analyzed mtDNA mutations in 5071 single CD34+ cells from 49 individuals (including 31 maternally related members from four families and 18 unrelated donors), and we found that CD34+ cells from members of the same family shared several family-unique mtDNA variants, albeit at very low frequency, suggesting a pedigree-specific occurrence of these mutations (if these mutations were not recurrent) [19]. For instance, single CD34+ cells from different members of Family A had mutations at sites 16192, 204, 16079, 16129, 215, 217 etc, whereas single CD34+ cells from members of Family B shared mutations at sites 146, 204, 16129, 16131, 16093, 16150, 16187 etc [19]. The appearance of such a pedigree-specific mutation pattern raised more unanswered questions: If these mutations were inherited, how they were maintained at the very low level of heteroplasmy? What is the driving force behind this process? Suppose these variants occurred randomly, how did they target to the same positions in different members? Note that not all these family-specific mutations were hypervariable sites according to human population data [19].
Along with the occurrence of family-specific mtDNA mutations, we found that the age-related accumulation of mtDNA mutations in CD34+ cells varied in different families, suggesting an influence by family genetic background [19]. The different genetic background effects on mtDNA mutation accumulation in single HSCs were also evident in our previous mouse experiments, in that we showed that B6 mice has an obvious age-related increase of mtDNA mutations, but not for BALB mice [65]. Taken all these lines of evidence together, it seems probable that highly frequent mutations in single HSCs do not occur randomly during the aging process.
9. Occurrence of mtDNA somatic mutations in single leukemia cells
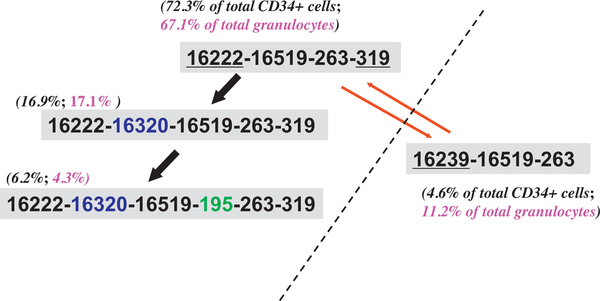
The marked level of mtDNA mutations observed in single cells is not unexpected, especially when we consider the different microenvironments of the cells from different tissues and the dynamics of the cell population. In leukemia, the increased generation of reactive oxygen species and other intrinsic aspects of leukemia pathophysiology would contribute to mtDNA genetic alterations, leading to a high level of mtDNA somatic mutations. Conversely, rapid expansion of a clonal leukemic cell population would act to either dilute mutations or to fix specific mtDNA variants by genetic drift, resulting in lower overall heterogeneity. We tested this hypothesis by measuring the level of mtDNA mutations in single leukemic cells and we found that the pattern was more complex than we had thought [21]. The single leukemic cells from the patients presented a higher variance of mtDNA heterogeneity than that of normal controls. In particular, there is no association of the level of mtDNA mutations in single cells with clinical leukemia types. Some relapsed patients presented a complex shift of major haplotypes in single leukemic cells, which might be related to chemotherapy effects or other factors [21]. For instance, in the relapsed acute myeloid leukemia patient UPN22 reported in our previous study [21], we saw a quick shift of single cells with step-wise mutations at sites 16320 and 195 based on the consensus haplotype 16222–16519-263–319, and a possible introduced / contaminated haplotype 16239–16519-263 caused by a platelet transfusion (Fig. 7) [21]. Apparently, the overall pattern of mtDNA heterogeneity in single blast cells from different patients had no uniform pattern but bore individual-specific feature [21]. This result differed from the finding by Ericson et al. [82], who found a decrease of the frequency of mtDNA mutations in colorectal cancer relative to normal tissues, possibly a result of the decrease in reactive oxygen species-mediated mtDNA damage due to the change of glucose metabolism in neoplastic tissues.
Fig. 7.
Diverse mtDNA haplotypes identified in a relapsed patient with acute myeloid leukemia. This patient (UPN22) received a platelet transfusion proximate to blood collection, and contamination by donor blood cells might account for the presence of haplotype 16239–16519-263, which differed from the main haplotype 16222–16519-263–319 by three variants that were underlined. The percentage of cells sharing each haplotype was marked in parentheses besides the haplotype. The step-wise occurrence of mutations 16320 and 195 based on haplotype 16222–16519-263–319 was marked by arrows. The original data were from Yao et al. [21].
10. Summary
We conclude from our data that mtDNA somatic mutations are frequent and common in blood cells; their detection and quantification depend on the methods employed. Different techniques have different sensitivity to identify true mutations and to generate methodological artifacts introduced by PCR amplification and DNA cloning. Single cell analysis for mutational spectrum and frequency is the best approach to trace the somatic mutations in single cells. However, this approach is also prone to error, unless one limits the analyses to intracellular clonally expanded mutations. If a mutation is not expanded in a cell (e.g. is present in one copy) then there is no good way to distinguish true mutations from artifacts, without the use of a high-resolution mutation detection assay, such as the Duplex Sequencing methodology [22]. Intracellular clones, in contrast, can be easily recognized and confirmed by repeated PCR-sequencing from the same cell (clonally expanded mutation will be repeatedly detected in the same cell by this procedure, which undoubtedly confirms their existence). It should also be mentioned that when amplifying from individual cells, there is a chance to fall into a situation where PCR may start with a single copy, and it is crucial to present proof that that single PCR originated from multiple copies, otherwise repeated PCR should be performed to consistently reproduce the mutation(s). Our results are also of importance inference to routine procedures employed in forensic laboratories, where only minimal amounts of often degraded DNA samples may be retrieved from crime scenes: the correct sequence of a specimen can be determined from consensus sequence of 10 or more single cells or cloned mtDNA in plasmids, filtering out both somatic mutations and/or technical artifacts.
The single-cell sequencing analysis has the ability to trace certain HSC clone and to uncover the mutational pattern of hematopoietic cells at the single cell level. We showed that different hematopoietic cell types may have different mutation spectra. End-stage, fully differentiated granulocytes have more mutations compared with other HSCs and progenitors; many somatic mutations in differentiated cells can be traced to stem cells and progenitors. Somatic mutations arising in stem cells and progenitors appear relatively stable for long periods of time and may be transplanted to another individual; accumulation of mtDNA mutations in HSCs is affected by genetic background and the aging effect varies in different families. These findings add new implications for the occurrence of mtDNA mutations in HSCs and understanding their putative role in ageing. Can we use mtDNA mutations as a marker to reconstruct the dynamics of normal human hematopoiesis, as had been demonstrated by recent studies using genetically manipulated labels in situ in mice [75,76], or to estimate the number of HSC clones active in white cell production? Considering the high mutation rate and complex intrinsic features of mtDNA mutations, and inherent defect of artifacts in a method aiming at achieving a high sensitivity, we would not be very optimistic about this attempt although it may deserve a try in principal. On the other hand, rapid expansion effect, chemotherapy effect, as well as intrinsic aspects of leukemia pathophysiology, would affect mtDNA mutations in leukemic cells. With the use of new cutting-edge technologies with a high sensitive and reliability, we will be able to depict the overall picture of low level of heteroplasmic mtDNA mutations in cells and tissues, and will finally resolve the fundamental properties regarding the nature of mtDNA heterogeneity.
Supplementary Material
Acknowledgements
We thank the two anonymous reviewers for their critical comments on the early version of the manuscript. Y.-G.Y. was supported by the MOST of China (2011CB910902) and the National Natural Science Foundation of China (31171225 and 30925021).
Footnotes
Conflict of interest
The authors declare no conflict in interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.mrfmmm.2015.06.009
References
- [1].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG, Sequence and organization of the human mitochondrial genome, Nature 290 (1981) 457–465. [DOI] [PubMed] [Google Scholar]
- [2].Wallace DC, A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine, Annu. Rev. Genet 39 (2005) 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cann RL, Stoneking M, Wilson AC, Mitochondrial DNA and human evolution, Nature 325 (1987) 31–36. [DOI] [PubMed] [Google Scholar]
- [4].Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC, African populations and the evolution of human mitochondrial DNA, Science 253 (1991) 1503–1507. [DOI] [PubMed] [Google Scholar]
- [5].Ivanov PL, Wadhams MJ, Roby RK, Holland MM, Weedn VW, Parsons TJ, Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II, Nat. Genet 12 (1996) 417–420. [DOI] [PubMed] [Google Scholar]
- [6].Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP, Phylogeographic differentiation of mitochondrial DNA in Han Chinese, Am. J. Hum. Genet 70 (2002) 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao YG, Bravi CM, Bandelt HJ, A call for mtDNA data quality control in forensic science, Forensic Sci. Int 141 (2004) 1–6. [DOI] [PubMed] [Google Scholar]
- [8].Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Paabo S, No evidence of Neandertal mtDNA contribution to early modern humans, PLoS Biol. 2 (2004) e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ, Harvesting the fruit of the human mtDNA tree, Trends Genet. 22 (2006) 339–345. [DOI] [PubMed] [Google Scholar]
- [10].Salas A, Carracedo A, Macaulay V, Richards M, Bandelt HJ, A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics, Biochem. Biophys. Res. Commun 335 (2005) 891–899. [DOI] [PubMed] [Google Scholar]
- [11].Salas A, Bandelt HJ, Macaulay V, Richards MB, Phylogeographic investigations: the role of trees in forensic genetics, Forensic Sci. Int 168 (2007) 1–13. [DOI] [PubMed] [Google Scholar]
- [12].Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, Torroni A, Richards MB, The archaeogenetics of Europe, Curr. Biol 20 (2010) R174–183. [DOI] [PubMed] [Google Scholar]
- [13].Achilli A, Perego UA, Lancioni H, Olivieri A, Gandini F, Hooshiar Kashani B, Battaglia V, Grugni V, Angerhofer N, Rogers MP, Herrera RJ, Woodward SR, Labuda D, Smith DG, Cybulski JS, Semino O, Malhi RS, Torroni A, Reconciling migration models to the Americas with the variation of North American native mitogenomes, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 14308–14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford Orlando TWL Jr., Metspalu E, Karmin M, Tambets K, Rootsi S, Magi R, Campos PF, Balanovska E, Balanovsky O, Khusnutdinova E, Litvinov S, Osipova LP, Fedorova SA, Voevoda MI, DeGiorgio M, Sicheritz-Ponten T, Brunak S, Demeshchenko S, Kivisild T, Villems R, Nielsen R, Jakobsson M, Willerslev E, Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans, Nature 505 (2014) 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tian JY, Wang HW, Li YC, Zhang W, Yao YG, van Straten J, Richards MB, Kong QP, A genetic contribution from the Far East into Ashkenazi Jews via the ancient Silk Road, Sci. Rep 5 (2015) 8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chinnery PF, Samuels DC, Elson J, Turnbull DM, Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism, Lancet 360 (2002) 1323–1325. [DOI] [PubMed] [Google Scholar]
- [17].Taylor RW, Turnbull DM, Mitochondrial DNA mutations in human disease, Nat. Rev. Genet 6 (2005) 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lu J, Sharma LK, Bai Y, Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis, Cell Res. 19 (2009) 802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yao YG, Kajigaya S, Feng X, Samsel L, McCoy JP Jr., Torelli G, Young NS, Accumulation of mtDNA variations in human single CD34+ cells from maternally related individuals: effects of aging and family genetic background, Stem Cell Res. 10 (2013) 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li M, Schroder R, Ni S, Madea B, Stoneking M, Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yao YG, Ogasawara Y, Kajigaya S, Molldrem JJ, Falcao RP, Pintao MC, McCoy Rizzatti JPEG Jr., Young NS, Mitochondrial DNA sequence variation in single cells from leukemia patients, Blood 109 (2007) 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kennedy SR, Salk JJ, Schmitt MW, Loeb LA, Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage, PLoS Genet. 9 (2013) e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Williams SL, Mash DC, Zuchner S, Moraes CT, Somatic mtDNA mutation spectra in the aging human putamen, PLoS Genet. 9 (2013) e1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yao YG, Bandelt HJ, Young NS, External contamination in single cell mtDNA analysis, PLoS One 2 (2007) e681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gilbert MT, Bandelt HJ, Hofreiter M, Barnes I, Assessing ancient DNA studies, Trends Ecol. Evol. 20 (2005) 541–544. [DOI] [PubMed] [Google Scholar]
- [26].Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M, Genetic analyses from ancient DNA, Annu. Rev. Genet 38 (2004) 645–679. [DOI] [PubMed] [Google Scholar]
- [27].Bandelt HJ, Lahermo P, Richards M, Macaulay V, Detecting errors in mtDNA data by phylogenetic analysis, Int. J. Legal Med 115 (2001) 64–69. [DOI] [PubMed] [Google Scholar]
- [28].Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V, The fingerprint of phantom mutations in mitochondrial DNA data, Am. J. Hum. Genet 71 (2002) 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bandelt HJ, Salas A, Contamination and sample mix-up can best explain some patterns of mtDNA instabilities in buccal cells and oral squamous cell carcinoma, BMC Cancer 9 (2009) 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yao YG, Macauley V, Kivisild T, Zhang YP, Bandelt HJ, To trust or not to trust an idiosyncratic mitochondrial data set, Am. J. Hum. Genet 72 (2003) 1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yao YG, Kong QP, Salas A, Bandelt HJ, Pseudomitochondrial genome haunts disease studies, J. Med. Genet 45 (2008) 769–772. [DOI] [PubMed] [Google Scholar]
- [32].Yao YG, Salas A, Logan I, Bandelt HJ, mtDNA data mining in GenBank needs surveying, Am. J. Hum. Genet. 85 (2009) 929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Salas A, Yao YG, Macaulay V, Vega A, Carracedo A, Bandelt HJ, A critical reassessment of the role of mitochondria in tumorigenesis, PLoS Med. 2 (2005) e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].DiMauro S, Schon EA, Mitochondrial respiratory-chain diseases, N. Engl. J. Med 348 (2003) 2656–2668. [DOI] [PubMed] [Google Scholar]
- [35].Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Paabo S, DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA, Nucleic Acids Res. 29 (2001) 4793–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang J, Asin-Cayuela J, Fish J, Michikawa Y, Bonafe M, Olivieri F, Passarino G, De Benedictis G, Franceschi C, Attardi G, Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes, Proc. Natl. Acad. U. S. A. Sci 100 (2003) 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wardell TM, Ferguson E, Chinnery PF, Borthwick GM, Taylor RW, Jackson G, Craft A, Lightowlers RN, Howell N, Turnbull DM, Changes in the human mitochondrial genome after treatment of malignant disease, Mutat. Res. 525 (2003) 19–27. [DOI] [PubMed] [Google Scholar]
- [38].Howell N, Smejkal CB, Mackey DA, Chinnery PF, Turnbull DM, Herrnstadt C, The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates, Am. J. Hum. Genet 72 (2003) 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shin MG, Kajigaya S, McCoy Levin JPBC Jr., Young NS, Marked mitochondrial DNA sequence heterogeneity in single CD34+ cell clones from normal adult bone marrow, Blood 103 (2004) 553–561. [DOI] [PubMed] [Google Scholar]
- [40].Shin MG, Kajigaya S, Tarnowka M, McCoy Levin JPBC Jr., Young NS, Mitochondrial DNA sequence heterogeneity in circulating normal human CD34 cells and granulocytes, Blood 103 (2004) 4466–4477. [DOI] [PubMed] [Google Scholar]
- [41].Ogasawara Y, Nakayama K, Tarnowka M, McCoy JP Jr., Kajigaya S, Levin BC, Young NS, Mitochondrial DNA spectra of single human CD34+ cells, T, cells, B cells, and granulocytes, Blood 106 (2005) 3271–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yao YG, Kajigaya S, Samsel L, McCoy JP Jr., Torelli G, Young NS, Apparent mtDNA sequence heterogeneity in single human blood CD34+ cells is markedly affected by storage and transport, Mutat. Res 751–752 (2013) 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG, High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection, Nat. Genet 28 (2001) 147–150. [DOI] [PubMed] [Google Scholar]
- [44].Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K, Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues, Proc. Natl. Acad. Sci. U. S. A. 99 (2002) 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Khrapko K, Bodyak N, Thilly WG, van Orsouw NJ, Zhang X, Coller HA, Perls TT, Upton M, Vijg J, Wei JY, Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions, Nucleic Acids Res. 27 (1999) 2434–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM, Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR, Nucleic Acids Res. 30 (2002) e68e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Del Bo R, Crimi M, Sciacco M, Malferrari G, Bordoni A, Napoli L, Prelle A, Biunno I, Moggio M, Bresolin N, Scarlato G, Pietro Comi G, High mutational burden in the mtDNA control region from aged muscles: a single-fiber study, Neurobiol. Aging 24 (2003) 829–838. [DOI] [PubMed] [Google Scholar]
- [48].Cormio A, Milella F, Vecchiet J, Felzani G, Gadaleta MN, Cantatore P, Mitochondrial DNA mutations in RRF of healthy subjects of different age, Neurobiol. Aging 26 (2005) 655–664. [DOI] [PubMed] [Google Scholar]
- [49].Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM, Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers, Am. J. Hum. Genet 79 (2006) 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Payne BA, Cree L, Chinnery PF, Single-cell analysis of mitochondrial DNA, Methods Mol. Biol 1264 (2015) 67–76. [DOI] [PubMed] [Google Scholar]
- [51].Cantuti-Castelvetri I, Lin MT, Zheng K, Keller-McGandy CE, Betensky RA, Johns DR, Beal MF, Standaert DG, Simon DK, Somatic mitochondrial DNA mutations in single neurons and glia, Neurobiol. Aging 26 (2005) 1343–1355. [DOI] [PubMed] [Google Scholar]
- [52].Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K, Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons, Nat. Genet 38 (2006) 518–520. [DOI] [PubMed] [Google Scholar]
- [53].Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM, High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease, Nat. Genet 38 (2006) 515–517. [DOI] [PubMed] [Google Scholar]
- [54].Reeve A, Meagher M, Lax N, Simcox E, Hepplewhite P, Jaros E, Turnbull D, The impact of pathogenic mitochondrial DNA mutations on substantia nigra neurons, J. Neurosci 33 (2013) 10790–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jayaprakash AD, Benson EK, Gone S, Liang R, Shim J, Lambertini L, Toloue MM, Wigler M, Aaronson SA, Sachidanandam R, Stable heteroplasmy at the single-cell level is facilitated by intercellular exchange of mtDNA, Nucleic Acids Res. 43 (2015) 2177–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, Leedham SJ, Deheragoda M, Sasieni P, Novelli MR, Jankowski JA, Turnbull DM, Wright NA, McDonald SA, Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Greaves LC, Beadle NE, Taylor GA, Commane D, Mathers JC, Khrapko K, Turnbull DM, Quantification of mitochondrial DNA mutation load, Aging Cell 8 (2009) 566–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kraytsberg Y, Nicholas A, Khrapko K, Are somatic mitochondrial DNA mutations relevant to our health? A challenge for mutation analysis techniques, Expert Opin. Med. Diagn 1 (2007) 109–116. [DOI] [PubMed] [Google Scholar]
- [59].Malka F, Lombes A, Rojo M, Organization, dynamics and transmission of mitochondrial DNA: focus on vertebrate nucleoids, Biochim. Biophys. Acta 1763 (2006) 463–472. [DOI] [PubMed] [Google Scholar]
- [60].Khrapko K, Nekhaeva E, Kraytsberg Y, Kunz W, Clonal expansions of mitochondrial genomes: implications for in vivo mutational spectra, Mutat. Res 522 (2003) 13–19. [DOI] [PubMed] [Google Scholar]
- [61].McDonald SA, Preston SL, Greaves LC, Leedham SJ, Lovell MA, Jankowski JA, Turnbull DM, Wright NA, Clonal expansion in the human gut: mitochondrial DNA mutations show us the way, Cell Cycle 5 (2006) 808–811. [DOI] [PubMed] [Google Scholar]
- [62].Elson JL, Samuels DC, Turnbull DM, Chinnery PF, Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age, Am. J. Hum. Genet 68 (2001) 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chinnery PF, Taylor GA, Howell N, Brown DT, Parsons TJ, Turnbull DM, Point mutations of the mtDNA control region in normal and neurodegenerative human brains, Am. J. Hum. Genet 68 (2001) 529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Greaves LC, Nooteboom M, Elson JL, Tuppen HA, Taylor GA, Commane DM, Arasaradnam RP, Khrapko K, Taylor RW, Kirkwood TB, Mathers JC, Turnbull DM, Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing, PLoS Genet. 10 (2014) e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yao YG, Ellison FM, McCoy JP, Chen J, Young NS, Age-dependent accumulation of mtDNA mutations in murine hematopoietic stem cells is modulated by the nuclear genetic background, Hum. Mol. Genet 16 (2007) 286–294. [DOI] [PubMed] [Google Scholar]
- [66].Bandelt HJ, Macaulay V, Richards M, Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA, Mol. Phylogenet. Evol 16 (2000) 8–28. [DOI] [PubMed] [Google Scholar]
- [67].Song X, Deng JH, Liu CJ, Bai Y, Specific point mutations may not accumulate with aging in the mouse mitochondrial DNA control region, Gene 350 (2005) 193–199. [DOI] [PubMed] [Google Scholar]
- [68].Itsara LS, Kennedy SR, Fox EJ, Yu S, Hewitt JJ, Sanchez-Contreras M, Cardozo-Pelaez F, Pallanck LJ, Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations, PLoS Genet. 10 (2014) e1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA, Mitochondrial point mutations do not limit the natural lifespan of mice, Nat. Genet 39 (2007) 540–543. [DOI] [PubMed] [Google Scholar]
- [70].Soares P, Ermini L, Thomson N, Mormina M, Rito T, Rohl A, Salas A, Oppenheimer S, Macaulay V, Richards MB, Correcting for purifying selection: an improved human mitochondrial molecular clock, Am. J. Hum. Genet 84 (2009) 740–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bandelt H-J, Kong Q-P, Richards M, Macaulay V, Estimation of mutation rates and coalescence times: some caveats, in: Bandelt H-J, Macaulay V, Richards M (Eds.), Human Mitochondrial DNA and the Evolution of Homo sapiens, Springer Verlag, Berlin, Heidelberg, 2006, pp. 47–92. [Google Scholar]
- [72].Dick JE, Lapidot T, Biology of normal and acute myeloid leukemia stem cells, Int. J. Hematol 82 (2005) 389–396. [DOI] [PubMed] [Google Scholar]
- [73].Wang JC, Dick JE, Cancer stem cells: lessons from leukemia, Trends Cell Biol. 15 (2005) 494–501. [DOI] [PubMed] [Google Scholar]
- [74].Mohamed SA, Wesch D, Blumenthal A, Bruse P, Windler K, Ernst M, Kabelitz D, Oehmichen M, Meissner C, Detection of the 4977 deletion of mitochondrial DNA in different human blood cells, Exp. Gerontol 39 (2004) 181–188. [DOI] [PubMed] [Google Scholar]
- [75].Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Hofer T, Rodewald HR, Fundamental properties of unperturbed haematopoiesis from stem cells in vivo, Nature 518 (2015) 542–546. [DOI] [PubMed] [Google Scholar]
- [76].Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD, Clonal dynamics of native haematopoiesis, Nature 514 (2014) 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yao YG, Childs RW, Kajigaya S, McCoy JP Jr., Young NS, Mitochondrial DNA sequence heterogeneity of single CD34+ cells after nonmyeloablative allogeneic stem cell transplantation, Stem Cells 25 (2007) 2670–2676. [DOI] [PubMed] [Google Scholar]
- [78].Copelan EA, Hematopoietic stem-cell transplantation, N. Engl. J. Med 354 (2006) 1813–1826. [DOI] [PubMed] [Google Scholar]
- [79].Körbling M, Katz RL, Khanna A, Ruifrok AC, Rondon G, Albitar M, Champlin RE, Estrov Z, Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem cells, N. Engl. J. Med 346 (2002) 738–746. [DOI] [PubMed] [Google Scholar]
- [80].Grove JE, Bruscia E, Krause DS, Plasticity of bone marrow-derived stem cells, Stem Cells 22 (2004) 487–500. [DOI] [PubMed] [Google Scholar]
- [81].Childs R, Clave E, Contentin N, Jayasekera D, Hensel N, Leitman S, Read EJ, Carter C, Bahceci E, Young NS, Barrett AJ, Engraftment kinetics after nonmyeloablative allogeneic peripheral blood stem cell transplantation: full donor T-cell chimerism precedes alloimmune responses, Blood 94 (1999) 3234–3241. [PubMed] [Google Scholar]
- [82].Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O’Sullivan J, Salk JJ, Bielas JH, Decreased mitochondrial DNA mutagenesis in human colorectal cancer, PLoS Genet. 8 (2012) e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.