Abstract
Sugar alcohols (polyols) exist widely in nature. While some specific sugar alcohol phosphatases are known, there is no known phosphatase for some important sugar alcohols (e.g., sorbitol-6-phosphate). Using liquid chromatography-mass spectrometry-based metabolomics, we screened yeast strains with putative phosphatases of unknown function deleted. We show that the yeast gene YNL010W, which has close homologues in all fungi species and some plants, encodes a sugar alcohol phosphatase. We term this enzyme, which hydrolyzes sorbitol-6-phosphate, ribitol-5-phosphate, and (D)-glycerol-3-phosphate, polyol phosphatase 1 or PYP1. Polyol phosphates are structural analogs of the enediol intermediate of phosphoglucose isomerase (Pgi). We find that sorbitol-6-phosphate and ribitol-5-phosphate inhibit Pgi and that Pyp1 activity is important for yeast to maintain Pgi activity in the presence of environmental sugar alcohols. Pyp1 expression is strongly positively correlated with yeast growth rate, presumably because faster growth requires greater glycolytic and accordingly Pgi flux. Thus, yeast express the previously uncharacterized enzyme Pyp1 to prevent inhibition of glycolysis by sugar alcohol phosphates. Pyp1 may be useful for engineering sugar alcohol production.
Graphical Abstract
Introduction
Polyols, the reduced forms of sugars, are an important natural family of carbohydrates 1, 2 Glycerol, the simplest polyol, is the backbone of phospholipids and is excreted by many microbes in response to stress 3, 4 Longer chain polyols include erythritol, ribitol, xylitol, arabitol, sorbitol, and mannitol, all of which exist only in the (D)-form in nature and are usually found in plants. Due to the inability of most organisms to assimilate long chain polyols into glycolysis, they are often regarded as inert solutes with unclear physiological functions. While the biological function of polyols has remained obscure, they have gained commercial interest in the last decade due to their increased usage as a sugar substitute by the food industry 5
Many fungi species are able to produce polyols via reduction of the corresponding sugar 6 To engineer polyol production, the standard approach is to first dephosphorylate the sugar phosphate and then reduce the resulting sugar to the polyol 7–10. This engineering approach differs from the natural glycerol production pathway, where (L)-glycerol-3-phosphate (a.k.a. sn-glycerol-3-phosphate) is first made from the reduction of dihydroxyacetone phosphate, followed by the dephosphorylation of the phosphorylated polyol 11, 12. This natural approach avoids accumulation of the potentially toxic metabolite dihydroxyacetone (DHA). Moreover, it avoids making a dephosphorylated sugar that might escape from the cell. However, this series of reactions has not been feasible as an engineering strategy due to lack of a suitable polyol phosphatase.
Because many enzymes remain unannotated even in the best studied organisms such as Baker’s yeast 13 it is possible that various polyol phosphate phosphatases exist but have yet to be characterized. One approach to metabolic enzyme annotation is to knock out the corresponding gene and assess the metabolome of the resulting strain. In general, substrates of the enzyme are likely to increase and products to decrease. This approach has now been successfully used to assign function to a variety of enzymes14–22.
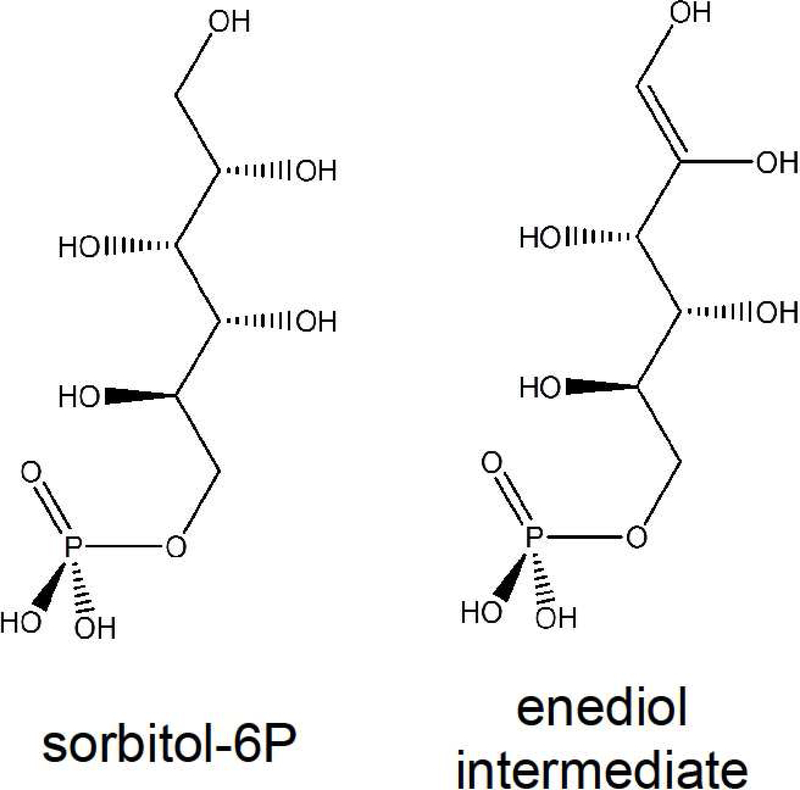
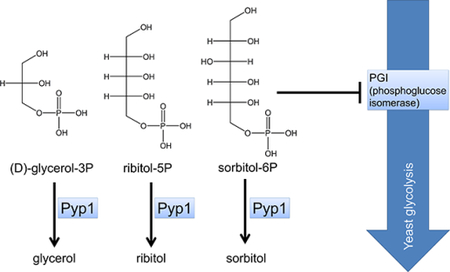
Using this approach and subsequent biochemical studies, here we show that YNL010W, a gene conserved across all fungi species and some plants, encodes a polyol phosphatase (Pyp1). We further show that the enzyme prevents polyol phosphate accumulation in yeast, and that this is physiologically important due to polyols being Pgi transition state analogues (Scheme 1). Thus, through assigning function to this previously unannotated yeast gene, we identify both a new glycolytic regulatory mechanism and a promising new enzyme for polyol metabolic engineering.
Scheme 1.
Results
A yeast Pyp1 deletion mutant accumulates octulose-8-phosphate and polyol phosphates
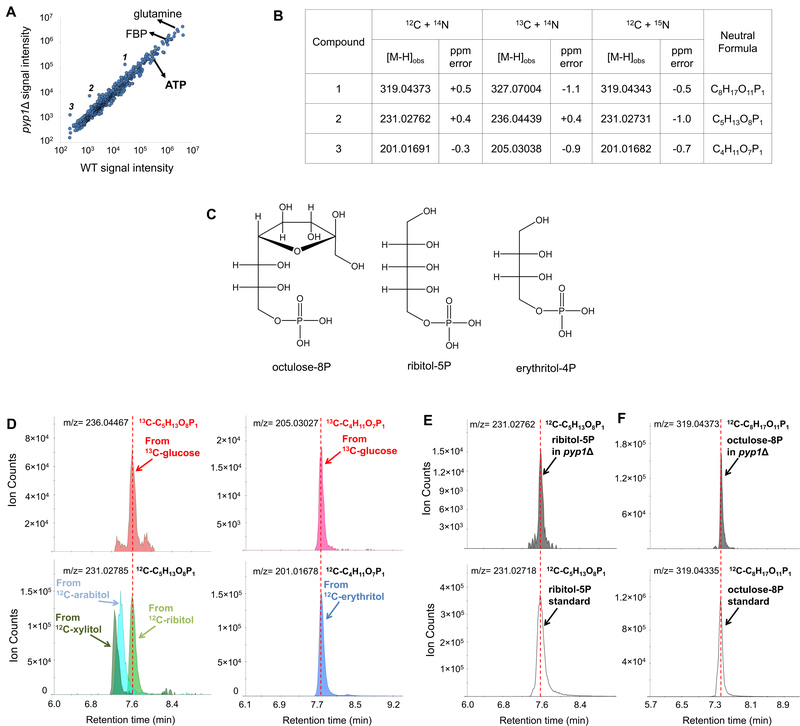
We screened yeast strains, with putative phosphatases of unknown function deleted, for changes in metabolite concentrations. Yeast were grown in glucose minimal media, and metabolites were extracted into 40:40:20 methanol: acetonitrile: water, followed by metabolome analysis by reversed-phase ion-pairing liquid chromatography – high resolution mass spectrometry (LC-MS). We found that the deletion of PYP1, formerly known as the uncharacterized gene YNL010W, while not significantly altering the concentrations of most metabolites, led to the statistically significant (false discovery rate < 0.05) accumulation of three compounds in negative ion mode (Figure 1A). The metabolites’ formulae were obtained by labeling cells with 13C and 15N and observing no shift from nitrogen labeling and a shift of +8, +5, or +4 daltons from carbon labeling (Figure 1B)23 Exact masses of these compounds matched putative formulae of C8H17O11P1, C5H13O8P1 and C4H11O7P1. Searching for these formulae in the KEGG database returned the metabolites octulose-8-phosphate (C8H17O11P1); ribitol-5-phosphate, arabitol-5-phosphate, and xylitol-5-phosphate (C5H13O8P1); and erythritol-4-phosphate (C4H11O7P1) (Figure 1C).
Figure 1. Metabolomic phenotype of pyplΔ cells.
(A). Metabolite profiles from glucose-grown wild type and pyp1Δ yeast cells detected by untargeted negative ion mode LC-high resolution MS. 1, 2, and 3 represent compounds with significantly different signal intensity between wild type and pyp1Δ cells. The x-axis represents the average of signal intensity in WT cells (N = 3). The y-axis represents the average of signal intensity in pyp1Δ cells (N = 3). Adduct and isotopic peaks were excluded. (B). Impact of 13C- and 15N-labeling on peaks of interest. Compounds 1, 2 and 3 were assigned formula as C8H17O11P1, C5H13O8P1 and C4H11O7P1 respectively. (C). Chemical structures of octulose-8P, ribitol-5P and erythritol-4P. (D). Extracted ion chromatograms of polyol phosphates produced from 13C-glucose (above) or by phosphorylation of 12C-polyols fed to the pyp1Δ cells (below). pyp1Δ cells growing exponentially on 2% 13C-glucose was switched to 2% 12C- polyols. Yeast metabolome right before the switch and four hours after the switch were analyzed by LC- MS. Retention time identifies the glucose-derived five carbon polyol phosphate as ribitol-5-phosphate and the four carbon polyol phosphate as erythritol-4P. (E and F). Extracted ion chromatogram for endogenous ribitol-5P (E) and octulose-8P (F) compared to synthetic standards.
To identify the five- and four-carbon polyol phosphates, we performed an isotope labeling experiment involving switching pyp1Δ cells from U-13C-glucose to unlabeled ribitol, arabitol, xylitol or erythritol. Although baker’s yeast cannot effectively utilize these polyols as carbon sources, in the absence of glucose, they slowly transport and phosphorylate them. We found that feeding ribitol and erythritol resulted in the build-up of intracellular metabolites with exact mass and LC retention time matching the C5H13O8P1 and C4H11O7P1 that accumulated with Pyp1 deletion (Figure 1D). Feeding of arabitol and xylitol resulted in the accumulation of polyol phosphates with different retention times (Figure 1D). Based on these results, we synthesized ribitol-5P and confirmed that exact mass and retention time matched to the endogenous 5-carbon sugar alcohol (Figure 1E). Octulose-8P has been reported in yeast previously18, and the chromatographic retention time of the accumulated C8H17O11P1 exactly matched the synthetic octulose-8P standard (Figure 1F). Thus, the compounds that accumulate in the pyp1Δ strain are octulose-8P, ribitol-5P and erythritol-4P.
Ribitol-5P and erythritol-4P are polyol phosphates, but octulose-8P is a sugar phosphate. We were curious why Pyp1 deletion resulted in accumulation of metabolites from these different structural families. The most common conformer of the seven carbon sugar sedoheptulose and its derivatives in solution is β-furanose 24. Octulose-8P likely has a similar β-furanose ring structure (Figure 1C), which has three carbons (6’−8’) of octulose out of the ring, forming a tail that resembles (D)-glycerol-3P, a polyol phosphate. Note that (D)-glycerol-3P is the same compound as (L)-glycerol-IP, which is the enantiomer of the common central carbon metabolite (L)-glycerol-3P (a.k.a. sn-glycerol-3P). We use the (D)-glycerol-3P nomenclature to emphasize the structural similarity to longer (D)-polyol phosphates, including ribitol-5P and sorbitol-6P (Figure 1C).
Recombinant Pyp1 dephosphorylates ribitol-5-phosphate, sorbitol-6-phosphate, and (D)- glycerol-3-phosphate
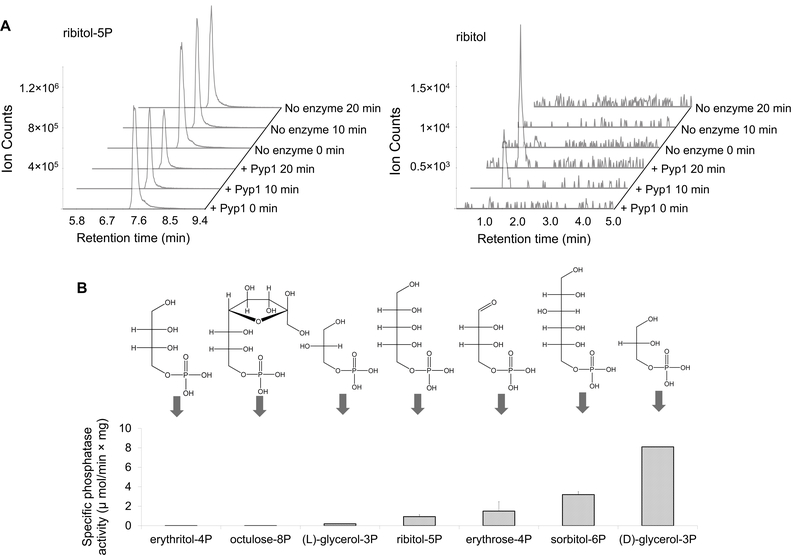
Although polyols are excretion products of fungi and plants, polyol phosphates were not previously thought to be an intermediate in this pathway (sugar phosphate → sugar → polyol). To determine if the accumulated compounds were indeed the substrates of Pyp1, we analyzed the biochemical activity of the purified recombinant Pyp1 on these compounds. Incubation with Pyp1 led to the depletion of ribitol- 5P and the accumulation of ribitol (Figure 2A), whereas no detectable phosphatase activity was found against octulose-8P or erythritol-4P (Figure 2B). Because flux through octulose-8P and erythritol-4P is very low in cells, minimal Pyp1 activity may nevertheless be sufficient to alter the cellular concentration 18. Alternatively, Pyp1 may need to bind an activator or other substrate in order to hydrolyze these species, or the accumulation of octulose-8P and erythritol-4P may be an indirect consequence of Pyp1 loss (e.g. occurring secondary to build-up of a more preferred substrate).
Figure 2. Pyp1 hydrolyzes (D)-polyol phosphates.
(A). Purified Pypl’s phosphatase activity was assayed against 0.5 mM ribitol-5P by LC-MS. Incubation with Pyp1 led to the depletion of ribitol-5P (left panel) and the accumulation of ribitol (right panel). (B). Pyp1’s (D)-polyol phosphatase activity was measured in the presence of 0.5 mM substrate and 5 mM Mg2+ (pH = 7.0, 30°C) by monitoring the appearance the polyol. See Experimental Procedures for details. The y-axis represents specific phosphatase activity (μmoles product per min per mg of protein) (mean ± range, N = 2).
To better define the range of substrates of Pyp1, we also measured its activity against erythrose-4P, sorbitol-6P, racemic glycerol-3P, and (L)-glycerol-3P. Incubation with Pyp1 led to the hydrolysis of erythrose-4P, sorbitol-6P and racemic glycerol-3P, but not (L)-glycerol-3P (Figure 2B). Thus, unlike the known glycerol-3-phosphatases (Hor2 and Rhr2) in the glycerol biosynthetic pathway, which act on both (L) and (D)-glycerol-3P 12, Pyp1 is specific to (D)-glycerol-3P. No other phosphatase activity was found for other common phosphorylated metabolites (see Supplementary Table 1). The best biochemical substrates for Pyp1 do not perfectly match those that accumulated in cells, likely because cellular accumulation depends on the absence of other routes of metabolizing the compounds.
Source of ribitol-5P in glucose-grown yeast
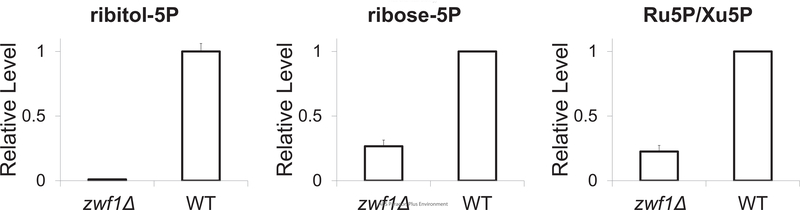
Although polyol phosphates have not been previously described in Baker’s yeast, they have been noted in other fungi 25. There are two likely routes for their cellular biosynthesis. The first involves the reduction of the corresponding sugar phosphate and the second involves phosphorylation of polyols. Both activities have been described in fungi but the responsible genes remain unknown25. In Baker’s yeast grown on glucose, the only known polyol is glycerol, and thus polyol phosphates are likely made from the reduction of sugar phosphates (as in Haemophilus influenza, Staphylococcus aureus26, and Escherichia coli27). Consistent with this, deletion of Zwf1, the first step in the oxidative pentose phosphate pathway (PPP), eliminated the endogenous peak for ribitol-5P, presumably due to decreased levels of ribose-5P and ribulose-5P (Figure 3). This is consistent with ribitol-5P being made via the reduction of ribose-5P or ribulose-5P in yeast grown on glucose.
Figure 3. Intracellular levels of ribitol-5-phosphate and ribulose-5-phosphate/xylulose-5-phosphate in wild type and zwf1Δ strain.
The metabolomes of wild type and zwf1Δ strains growing on 2% glucose were measured by LC-MS. The y-axis represents the relative level for select metabolites (mean ± range, N = 2).
Growth on sorbitol requires Pyp1
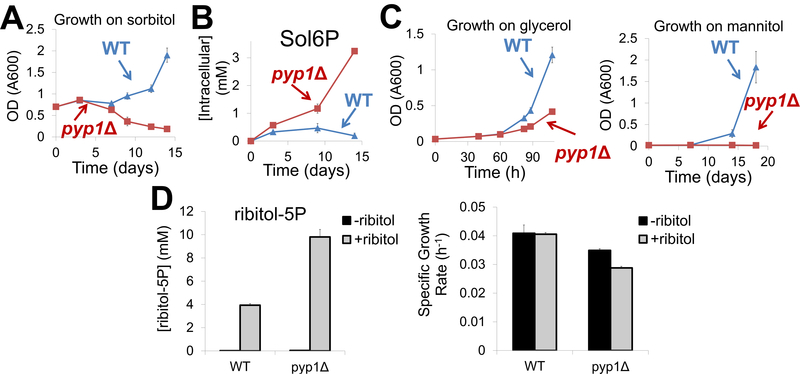
When polyols are present in the growth environment, polyol phosphates can be made by their phosphorylation. Of the natural long-chain polyols, the only one known to support yeast growth as the sole carbon source is sorbitol 28.Catabolism of sorbitol begins with its dehydrogenation to fructose, which then enters central metabolism.. Due to the structural similarity of sorbitol to glucose, we hypothesized that yeast might sometimes erroneously phosphorylate sorbitol into sorbitol-6P, which could be toxic in excess. To explore this possibility, we fed both wild type and pyp1Δ strains minimal media containing sorbitol as the sole carbon source. While wild type cells grew to saturation after a long lag phase, pyp1Δ cells never grew (Figure 4A). Metabolome profiling revealed that sorbitol-6P levels in the pyp1Δ strain were much higher than the wild type strain (Figure 4B). These results are consistent with Pyp1 being required to dephosphorylate sorbitol-6P, which otherwise accumulates to toxic levels, thereby precluding growth on sorbitol.
Figure 4. Pyp1 accelerates yeast growth on or in the presence of polyols.
(A). Growth of wild type and pyp1Δ yeast on sorbitol. Yeast cells grown on YPD medium were switched to minimal media containing 2% sorbitol as the carbon source. The x-axis represents days after the switch and the y-axis represents optical density (A600). (B). Absolute concentration of sorbitol-e-phosphate in wild type and pyp1Δ cells in the experiment shown in (A). The x-axis represents days after the switch and the y-axis represents absolute intracellular concentration (mean ± range, N = 2). (C). Growth of wild type and pyp1Δ cells on glycerol and mannitol. Yeast cells grown on YPD medium were switched to medium containing complete supplement mixture (CSM) and 3% glycerol as the carbon source or minimal medium containing 2% mannitol as the carbon source. The x-axis represents time after the switch and the y-axis represents optical density (A600). (D). Impact of ribitol in the presence of trehalose as the carbon source. Absolute concentration of ribitol-5P in wild type and pyp1Δ cells (left panel) and their growth rates (right panel) on minimal media containing 1% trehalose +/− 1% ribitol.
To investigate whether Pyp1 is important for growth on other polyol substrates, we fed pyp1Δ yeast glycerol or mannitol. The pyp1Δ cells grew poorly on glycerol. While wild type cells managed to reach saturation after a long lag phase on mannitol, pyp1Δ· cells were unable to grow on mannitol (Figure 4C). Thus, Pyp1 contributes to polyol phosphate detoxification, and such detoxification is required for growth on diverse polyol substrates.
To more directly ascertain whether the impaired growth was a result of polyol phosphate toxicity, we fed yeast trehalose (a dimer of glucose that can be slowly utilized by yeast resulting in carbon-limited growth29, 30) with or without addition of ribitol. In wild type yeast, addition of ribitol to the medium resulted in accumulation of intracellular ribitol-5P, without impacting growth rate (Figure 4D). In pyp1 yeast, the ribitol-5P levels rose yet higher, resulting in a ~20% decrease in the growth rate (Figure 4D). Thus, buildup of polyol phosphate compounds impairs yeast growth, including on substrates other than polyols.
Polyol phosphates are inhibitors of phosphoglucose isomerase (Pgi)
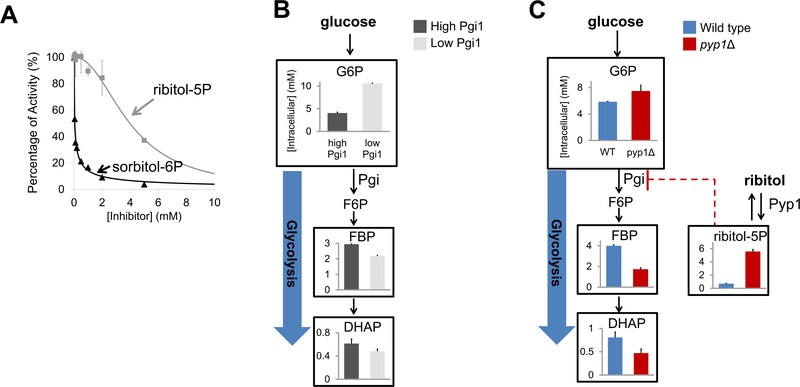
One potential mechanism by which polyol phosphates could impair cell growth is through metabolic enzyme inhibition. Phosphoglucose isomerase (Pgi1) catalyzes the interconversion between glucose-6P and fructose-6P in glycolysis and is essential for yeast to grow on either glucose or fructose as the sole carbon source 31. Sorbitol-6P is structurally similar to the enediol transition state of the Pgi reaction (Scheme 1) and sorbitol-6P and other structural mimics of the enediol intermediate are known Pgi inhibitors 32. We confirmed that sorbitol-6P and less potently ribitol-5P inhibit Pgi (Figure 5A). Motivated by these observations, we sought to determine whether polyol phosphates significantly and selectively inhibit Pgi in yeast cells. We hypothesized that two metabolic hallmarks of Pgi inhibition would be increased glucose-6P and decreased glycolytic intermediates downstream of fructose-6P, with fructose-1, 6-bisphosphate (FBP) a convenient marker compound. To evaluate whether decreased Pgi activity indeed induces these metabolic changes, we constructed a yeast strain with PGI under control of an estradiol-inducible promoter. In the low induction condition (1 nM estradiol, Pgi protein level ~1/7 of WT cells), we observed both increased glucose-6P and decreased FBP (Figure 5B).
Figure 5. Polyol phosphates inhibit phosphoglucose isomerase in vitro and in growing yeast.
(A). Activity of the purified Pgi1 was assayed in the presence of 0.6 mM fructose-6-phosphate (substrate), 5 mM Mg2+ (pH = 7.0, 30°C) and a range of 0.05–5 mM of sorbitol-6P or ribitol-5P by monitoring the appearance of glucose-6-phosphate. The x-axis represents polyol phosphate concentration and the y-axis represents relative Pgi1 activity (mean ± range of N = 2 replicates). (B). Metabolome profiling of cells with high (100 nM estradiol, comparable to WT protein level, shown in black) and low (1 nM estradiol, ~1/7 of WT protein level, shown in grey) Pgi1 induction. This experiment is used to define a characteristic low-Pgi metabolome. (C). Metabolome profiling of pyp1Δ (dark red) and wild type (blue) cells at t = 1 h after switching from trehalose + ribitol to glucose + ribitol. This experiment shows that PYP1 deletion results, in the presence of ribitol, in a characteristic low-Pgi metabolome.
We then tested whether ribitol-5P accumulation results in these metabolic hallmarks of Pgi deficiency. Cells were initially grown in trehalose + ribitol and then switched to glucose + ribitol to enhance glycolytic flux. Relative to wild-type yeast, the pyp1Δ strain accumulated dramatically more ribitol-5P (Figure 5C). Critically, the yeast lacking Pyp1 also manifested both hallmarks of physiological Pgi inhibition, increased glucose-6P and decreased FBP (Figure 5C).
Discussion
Sorbitol-6-phosphatase activity has been found in apple leaves 33 and silk worms 34. The gene encoding this enzyme, however, has not been discovered. In engineered fungi, sorbitol production has been achieved by expressing bacterial sorbitol-6P dehydrogenase, but again the gene encoding the required phosphatase activity remained missing 35. Here we identify the previously unannotated yeast gene YNL010W, which we now term PYP1, as a polyol phosphate phosphatase. Pyp1 dephosphorylates a variety of compounds with the common structural motif of a (D)-glycerol-3-phosphate tail, including (D)- glycerol-3P, erythrose-4P, ribitol-5P, and sorbitol-6P (Figure 2B). It is likely that Pyp1 would also dephosphorylate erythritol-4P, arabitol-5P, and xylitol-5P. The ability of Pyp1 to hydrolyze a variety of sugar alcohol phosphates gives rise to an intriguing opportunity to employ Pyp1 in the production of diverse sugar alcohol consumables.
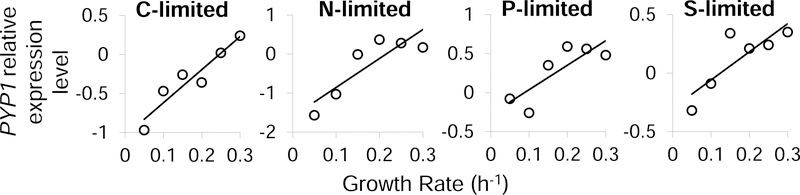
One functional role of Pyp1 is to limit the polyol phosphate concentrations in cells. High levels of polyol phosphates impair yeast growth, at least in part by inhibition of the upper glycolytic enzyme phosphoglucose isomerase (Pgi), for which sorbitol-6-phosphate is a transition state analogue. Other sugar phosphate isomerases, such as triose-phosphate isomerase and ribose-5-phosphate isomerase, have similar enediol reaction intermediates 36,37, and thus may also be inhibited by polyol phosphates. Similar to strong inhibition of Pgi by sorbitol-6 phosphate, triose-phosphate isomerase may be inhibited by (D)-glycerol-3P, and ribose-5-phosphate isomerase by ribitol-5P or xylitol-5P. Flux through each of these enzymes tends to increase with faster yeast growth rate. For example, ribose-5-phosphate isomerase is required to feed ribosome biogenesis. Thus rapidly growing yeast cells may be particularly sensitive to enzyme inhibition by polyol phosphates. To determine whether Pyp1 function is associated with growth rate, we analyzed its expression as a function of growth rate across 25 different chemostat conditions 38. Interestingly, Pypl’s expression was strongly positively correlated with growth rate, regardless of the limiting nutrient used (top 6% of all transcripts in genome) (Figure 6). Thus, unlike most protective or detoxification genes (e.g. against heat, osmolarity or oxidative stress), which are highly expressed under slower growth conditions 38,39, PYP1 is a fast-growth gene that maintains high Pgi flux by dephosphorylating polyol phosphates.
Figure 6. Pyp1 is highly expressed and functionally important in rapid growth.
Relative expression level of PYP1 at different growth rates in C-, N-, P- and S-limited chemostats 38. The x-axis represents different growth rates and the y-axis represents relative expression level of PYP1.
The transition state inhibition by polyol phosphates and their derivatives is not limited to sugar phosphate isomerase. In plants, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) also has an enediol intermediate and is known to be inhibited by 2-carboxy-D-arabitol-1-phosphate (CA1P) 40. The corresponding 2-carboxy-D-arabitol-1-phosphatase is activated by light and controls cellular level of CA1P. In darkness, RuBisCO is inhibited and also protected by CA1P from proteolysis 41.
Pyp1 is conserved across fungi species and some plants and bacteria, including Ricinus communis, Dehalococcoides ethenogenes, and Bacillus anthracis42. The highly conserved nature of Pyp1 in fungi indicates the importance of clearing polyol phosphates. It is likely that the homologous enzymes play the same role in bacteria and plants. Although we have not identified a Pyp1 homolog in mammals, nor are linear polyol phosphates longer than three carbons known mammalian metabolites, polyols are also produced in humans and can accumulate in disease43. For example, during hyperglycemia (e.g., due to diabetes), sorbitol accumulates in a number of organs due to the action of aldose reductase on glucose, which may contribute to diabetic complications including diabetic retinopathy, peripheral neuropathy and diabetic kidney disease44–46. It is unclear whether such sorbitol sometimes becomes phosphorylated to sorbitol-6P and, if so, whether sorbitol-6P contributes to disease pathology.
Beyond their potential cellular toxicity, polyol phosphates may also have a regulatory role in central carbon metabolism. Pgi sits at the branch point between glycolysis and the oxidative pentose phosphate pathway. Despite being ideally situated to regulate the branching ratio between these pathways (Figure 5), Pgi is not an allosteric enzyme, with no physiological regulators known32, 47. It is tempting to speculate that polyol phosphates might serve as endogenous Pgi regulators, with Pgi an enzyme controlled by active site competition48–53. Further investigation is merited to see whether polyol phosphates contribute to physiological metabolic flux control.
Experimental Procedures
Chemicals and reagents
Most chemicals, reagents and media components in the study, including (D)-sorbitol-6-phosphate (S1753), (D)-ribose-5-phosphate (83875), (D)-erythritol-4-phosphate (08897), (D)-erythrose-4- phosphate (E0377), (DL)-glycerol-3-phosphate (61668), (L)-glycerol-3P (94124), (D)-glycerol-3P (92034), NaBH4 (452882), (D)-sorbitol (S1876), ribitol (adonitol, A5502), (D)-arabitol (A3381), xylitol (X3375), erythritol (PHR1479), tributylamine (90780), and acetic acid (695092), were obtained from Sigma-Aldrich (St. Louis, MO). LCMS-grade acetonitrile (A955), methanol (A456), water (W6) were obtained from Fisher Scientific (Waltham, MA). 13C6-glucose (CLM-1396) was from Cambridge Isotope Laboratories (Tewksbury, MA).
Yeast strains and media
Yeast strains were derived from prototrophic S288C. Prototrophic deletions were created by homologous recombination using the allele amplified by PCR from the synthetic genetic analysis (SGA) deletion set 54. The Pgi1 inducible strain was generated as described:55 uman estradiol was used to induce PGI1 via insertion of a synthetic promoter involving chimeric transcription factor-estrogen receptor in front of PGI1. 1 nM and 5 nM estradiol were added to achieve low and high expression levels of yeast Pgi1, resulting in a 2.5-fold difference in Pgi1 protein level 55.
Cells were grown in minimal media comprising 6.7g/L Difco Yeast Nitrogen Base without amino acids plus 2% (w/v) glucose, sorbitol or mannitol. Glycerol medium was composed of 6.7g/L Difco Yeast Nitrogen Base without amino acids, 0.79 g/L Complete Supplement Mixture (Sunrise Science, San Diego, CA) and 3% (v/v) glycerol. Trehalose minimal medium was prepared by mixing 6.7g/L Difco Yeast Nitrogen Base without amino acids and 1% (w/v) trehalose, and adjusting pH to 4.8 by adding succinic acid.
Yeast culture conditions and extraction
The metabolome of batch culture Saccharomyces cerevisiae was characterized as described previously 56: Saturated overnight cultures were diluted 1:30 and grown in liquid media in a shaking flask to A600 of ~ 0.6. A portion of the cells (3 mL) were filtered onto a 50 mm nylon membrane filter (Millipore, Billerica, MA), which was immediately transferred into −20 °C extraction solvent (40:40:20 acetonitrile/methanol/water). For carbon upshift, 100 mL of cell culture grown on trehalose at A600 of ~0.6 were poured onto a 100 mm cellulose acetate membrane filter (Sterlitech, Kent, WA) resting on a vacuum filter holder with a 1000 mL funnel (Kimble Chase, Vineland, NJ) and were washed with 100 mL pre-warmed (30°C) glucose minimal medium. Immediately after the wash media went through, the filter was taken fromthe holder and the cells were washed into a new flask containing 100 mL pre-warmed (30 °C) glucose minimal medium. Samples were then taken at the indicated time points after the switch, and filtered and quenched as described above.
LC-MS metabolite measurement
Cell extracts were analyzed by reversed phase ion-pairing liquid chromatography (LC) coupled by electrospray ionization (ESI) (negative mode) to a high-resolution, high-accuracy mass spectrometer (Exactive; Thermo Fisher Scientific, Waltham, Massachusetts) operating in negative ion mode scanning m/z 70–1000with 100,000 resolution at m/z 200. Liquid chromatography separation was achieved on a Synergy Hydro-RP column (100 mm × 2 mm, 2.5 μm particle size, Phenomenex, Torrance, CA) with the following gradient: 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; 25 min, 0% B. Solvent A is 97:3 water/methanol with 10 mM tributylamine and 15 mM acetic acid; solvent B is methanol. Other LC parameters are autosampler temperature 4 °C, injection volume 10 μL, and column temperature 25 °C. Peaks differing between wild-type and pyplΔ strain were determined using the in-house developed, open-source software MAVEN 57,58. Compounds’ identities were verified by mass and retention time match to authenticated standards. Differences in metabolome between wild type and pyp1Δ strains were tested for significance using Student’s T-test. The resulting P-values were then corrected using the Benjamini-Yekutieli False Discovery Rate (FDR) model 59.
The number of C and N atoms in each accumulated compound was determined by the method described 23. Yeast batch cultures were grown with uniformly labeled glucose or ammonium sulfate (Cambridge Isotopes, Andover, MA) for > 20 generations to ensure complete labeling of the metabolome.
Absolute intracellular metabolite concentrations in steadily growing wild type and pyplΔ cells were determined as described previously 60. Metabolite concentrations after perturbations were computed based on fold-change in ion counts relative to steadily-growing cells (grown and analyzed in parallel) multiplied by the known absolute concentration in the steadily growing cells, as determined using an isotope ratio-based approach 61.
Synthesis of ribitol-5-phosphate and octulose-8-phosphate
Synthesis of ribitol-5-phosphate was performed as described previously through the reduction of ribose-5-phosphate using NaBH4 62. Synthesis of (D)-glycero-D-altro-octulose-8-phosphate was performed enzymatically as described 63.
Protein purification and enzymatic assays
For Pyp1’s polyol phosphatase activity assay, a yeast Open Reading Frame (ORF) strain with an expression vector containing C-terminal His-tagged PYP1 (Open Biosystems, Thermo Fisher Scientific, San Jose, CA) was grown on galactose to induce Pyp1 expression. The resulting cells were lysed using glass beads and His-tagged Pyp1 was purified using Qiagen Ni-NTA spin columns according to the manufacturer’s instructions. Phosphatase activity against ribitol-5-phosphate, sorbitol-6-phosphate, erythritol-4-phosphate and octulose-8-phosphate was determined by monitoring the increase of ribitol, sorbitol, erythritol or octulose using LC-MS. The reaction mixture contained 100 mM Tris-HCl at the pH 8.0, 10 mM MgCl2 and a range of substrate concentrations (0.01 to 5 mM).
Purified Pyp1 was also assayed against other compounds with similar structures and 90 common phosphorylated metabolites (see Supplementary Table for the list of compounds). Briefly, the gene encoding Pyp1 was amplified by PCR using S. cerevisiae genomic DNA. The amplified fragments were cloned into a modified pET15b vector (Novagen, Darmstadt, Germany) and overexpressed in the E. coli BL21 (DE3) Gold strain (Stratagene, La Jolla, California) as previously described 64. The recombinant protein was purified using metal ion affinity chromatography on nickel chelate resin (Qiagen, Hilden, Germany) to high homogeneity and stored at −80 °C. Purified Pyp1 was then screened for phosphatase activity as described previously65. Due to the difficulty of obtaining (D)-glycerol-3P, the activity of Pyp1 on (D)-glycerol-3P was determined using racemic glycerol-3P as the substrate. Because Pyp1 has minimal activity on (L)-glycerol-3P, its activity on racemic glycerol-3P was taken as the activity on (D)-glycerol-3P. Compounds with specific activity higher than 0.1 μmol/mg/min, as well as erythritol-4P, are shown in Figure 2B.
To measure the inhibition of Pgi by sorbitol-6-phosphate and ribitol-5-hosphate, yeast Pgi1 was purchased from Sigma Aldrich (St. Louis, MO). Phosphoglucose isomerase activity was determined by adding fructose-6-phosphate and monitoring the appearance of glucose-6-phosphate using LC/MS. We found such LC/MS based assay is consistently more sensitive and accurate than the more typical colorimetric-based assay, which involves coupling the Pgi1 activity with glucose-6-phosphate dehydrogenase activity and monitoring the appearance of NADH. The reaction mixture contained 100 mM Tris-HCl at pH 8.0, 10 mM MgCl2, 0.6 mM fructose-6-phosphate (the physiological concentration in cells grown exponentially on glucose), and a range of sorbitol-6-phosphate and ribitol-5-phosphate concentrations (0.05 to 5 mM). The resulting data were fitted to the Hill equation using the GraphPad Prism Software.
Supplementary Material
Acknowledgments
We thank S. McIsaac, S. Silverman, S. Hackett and X. Su for helpful discussions. This research was funded by NSF CAREER award MCB-0643859, Joint DOE-AFOSR Award DOE DE-SC0002077 - AFOSR FA9550–09-1 −0580, NSF grant CBET-0941143, NIH R01 grant CA163591, and DOE Center for Advanced Bioenergy and Bioproducts Innovation (CABBI) DE-SC0018420 to J.D. R. and CA211437 to W.L., with additional support from the Princeton University Center for Quantitative Biology (P50 GM071508) and from the Government of Canada through Genome Canada, Ontario Genomics Institute (2009-OGI-ABC-1405), and Ontario Research Fund (ORF-GL2–01-004), and from NSF grant OCE-1233964.
References:
- 1.Teo G, Suzuki Y, Uratsu SL, Lampinen B, Ormonde N, Hu WK, DeJong TM, and Dandekar AM (2006) Silencing leaf sorbitol synthesis alters long-distance partitioning and apple fruit quality, Proc Natl Acad Sci U S A 103, 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shindou T, Sasaki Y, Miki H, Eguchi T, Hagiwara K, and Ichikawa T (1988) Determination of Erythritol in Fermented Foods by High-Performance Liquid-Chromatography, J Food Hyg Soc Jpn 29, 419–422. [Google Scholar]
- 3.Pahlman AK, Granath K, Ansell R, Hohmann S, and Adler L (2001) The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress, J Biol Chem 276, 3555–3563. [DOI] [PubMed] [Google Scholar]
- 4.Nevoigt E, and Stahl U (1997) Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae, FEMS Microbiol Rev 21, 231–241. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw DJ, and Marsh PD (1994) Effect of Sugar Alcohols on the Composition and Metabolism of a Mixed Culture of Oral Bacteria Grown in a Chemostat, Caries Res 28, 251–256. [DOI] [PubMed] [Google Scholar]
- 6.Chang Q, Griest TA, Harter TM, and Petrash JM (2007) Functional studies of aldo-keto reductases in Saccharomyces cerevisiae, Bba-Mol Cell Res 1773, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon HJ, Jeya M, Kim IW, and Lee JK (2010) Biotechnological production of erythritol and its applications, Applied Microbiology and Biotechnology 86, 1017–1025. [DOI] [PubMed] [Google Scholar]
- 8.Povelainen M, and Miasnikov AN (2006) Production of D-arabitol by a metabolic engineered strain of Bacillus subtilis, Biotechnology journal 1, 214–219. [DOI] [PubMed] [Google Scholar]
- 9.Povelainen M, and Miasnikov AN (2007) Production of xylitol by metabolically engineered strains of Bacillus subtilis, Journal of biotechnology 128, 24–31. [DOI] [PubMed] [Google Scholar]
- 10.Toivari MH, Maaheimo H, Penttila M, and Ruohonen L (2010) Enhancing the flux of D-glucose to the pentose phosphate pathway in Saccharomyces cerevisiae for the production of D-ribose and ribitol, Appl Microbiol Biotechnol 85, 731–739. [DOI] [PubMed] [Google Scholar]
- 11.Albertyn J, Hohmann S, Thevelein JM, and Prior BA (1994) GPD1, which encodes glycerol- 3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway, Mol Cell Biol 14, 4135–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norbeck J, Pahlman AK, Akhtar N, Blomberg A, and Adler L (1996) Purification and characterization of two isoenzymes of DL-glycerol-3-phosphatase from Saccharomyces cerevisiae - Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway, Journal of Biological Chemistry 271, 13875–13881. [DOI] [PubMed] [Google Scholar]
- 13.Ellens KW, Christian N, Singh C, Satagopam VP, May P, and Linster CL (2017) Confronting the catalytic dark matter encoded by sequenced genomes, Nucleic Acids Res 45, 11495–11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, and Willmitzer L (2000) Metabolite profiling for plant functional genomics, Nat Biotechnol 18, 1157–1161. [DOI] [PubMed] [Google Scholar]
- 15.Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, and Oliver SG (2001) A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations, Nat Biotechnol 19, 45–50. [DOI] [PubMed] [Google Scholar]
- 16.Saghatelian A, and Cravatt BF (2005) Discovery metabolite profiling--forging functional connections between the proteome and metabolome, Life sciences 77, 1759–1766. [DOI] [PubMed] [Google Scholar]
- 17.Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, and Saito K (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis, The Plant cell 20, 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clasquin MF, Melamud E, Singer A, Gooding JR, Xu X, Dong A, Cui H, Campagna SR, Savchenko A, Yakunin AF, Rabinowitz JD, and Caudy AA (2011) Riboneogenesis in yeast, Cell 145, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clasquin MF, Melamud E, and Rabinowitz JD (2012) LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine, Current protocols in bioinformatics /editoral board, Baxevanis Andreas D. … [et al. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quanbeck SM, Brachova L, Campbell AA, Guan X, Perera A, He K, Rhee SY, Bais P, Dickerson JA, Dixon P, Wohlgemuth G, Fiehn O, Barkan L, Lange I, Lange BM, Lee I, Cortes D, Salazar C, Shuman J, Shulaev V, Huhman DV, Sumner LW, Roth MR, Welti R, Ilarslan H, Wurtele ES, and Nikolau BJ (2012) Metabolomics as a Hypothesis-Generating Functional Genomics Tool for the Annotation of Arabidopsis thaliana Genes of “Unknown Function”, Frontiers in plant science 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su X, Chen W, Lee W, Jiang H, Zhang S, and Lin H (2012) YBR246W is required for the third step of diphthamide biosynthesis, J Am Chem Soc 134, 773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu YF, Letisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, and Rabinowitz JD (2013) Nucleotide degradation and ribose salvage in yeast, Mol Syst Biol 9, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegeman AD, Schulte CF, Cui Q, Lewis IA, Huttlin EL, Eghbalnia H, Harms AC, Ulrich EL, Markley JL, and Sussman MR (2007) Stable isotope assisted assignment of elemental compositions for metabolomics, Anal Chem 79, 6912–6921. [DOI] [PubMed] [Google Scholar]
- 24.Kuchel PW, Berthon HA, Bubb WA, Mcintyre LM, Nygh NK, and Thorburn DR (1990) C-13 and P-31 Nmr-Studies of the Pentose-Phosphate Pathway in Human Erythrocytes, Biomed Biochim Acta 49, S105–S110. [PubMed] [Google Scholar]
- 25.Jennings DH (1984) Polyol Metabolism in Fungi, Adv Microb Physiol 25, 149–193. [DOI] [PubMed] [Google Scholar]
- 26.Pereira MP, and Brown ED (2004) Bifunctional catalysis by CDP-ribitol synthase: Convergent recruitment of reductase and cytidylyltransferase activities in Haemophilus influenzae and Staphylococcus aureus, Biochemistry 43, 11802–11812. [DOI] [PubMed] [Google Scholar]
- 27.Novotny MJ, Reizer J, Esch F, and Saier MH (1984) Purification and Properties of D-Mannitol-1-Phosphate Dehydrogenase and D-Glucitol-6-Phosphate Dehydrogenase from Escherichia-Coli, Journal of Bacteriology 159, 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarthy AV, Schopp C, and Idler KB (1994) Cloning and Sequence Determination of the Gene Encoding Sorbitol Dehydrogenase from Saccharomyces-Cerevisiae, Gene 140, 121–126. [DOI] [PubMed] [Google Scholar]
- 29.Jules M, Guillou V, Francois J, and Parrou JL (2004) Two distinct pathways for trehalose assimilation in the yeast Saccharomyces cerevisiae, Appl Environ Microbiol 70, 2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walther T, Novo M, Rossger K, Letisse F, Loret MO, Portais JC, and Francois JM (2010) Control of ATP homeostasis during the respiro-fermentative transition in yeast, Mol Syst Biol 6, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguilera A (1986) Deletion of the phosphoglucose isomerase structural gene makes growth and sporulation glucose dependent in Saccharomyces cerevisiae, Mol Gen Genet 204, 310–316. [DOI] [PubMed] [Google Scholar]
- 32.Milewski S, Janiak A, and Wojciechowski M (2006) Structural analogues of reactive intermediates as inhibitors of glucosamine-6-phosphate synthase and phosphoglucose isomerase, Archives of Biochemistry and Biophysics 450, 39–49. [DOI] [PubMed] [Google Scholar]
- 33.Zhou R, Cheng LL, and Wayne R (2003) Purification and characterization of sorbitol-6-phosphate phosphatase from apple leaves, Plant Sci 165, 227–232. [Google Scholar]
- 34.Oda Y, Iwami M, and Sakurai S (2005) Membrane-bound sorbitol 6-phosphatase in fat body cells controls the dynamics of sorbitol 6-phosphate, a major hemolymph sugar in the silkworm, Insect Biochem Molec 35, 1284–1292. [DOI] [PubMed] [Google Scholar]
- 35.Ladero V, Ramos A, Wiersma A, Goffin P, Schanck A, Kleerebezem M, Hugenholtz J, Smid EJ, and Hols P (2007) High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering, APPLIED AND ENVIRONMENTAL MICROBIOLOGY 73, 1864–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R, Andersson CE, Savchenko A, Skarina T, Evdokimova E, Beasley S, Arrowsmith CH, Edwards AM, Joachimiak A, and Mowbray SL (2003) Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle, Structure 11, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komives EA, Chang LC, Lolis E, Tilton RF, Petsko GA, and Knowles JR (1991) Electrophilic catalysis in triosephosphate isomerase: the role of histidine-95, Biochemistry 30, 3011–3019. [DOI] [PubMed] [Google Scholar]
- 38.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, and Botstein D (2008) Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast, Molecular biology of the cell 19, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, and Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes, Molecular biology of the cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andralojc PJ, Dawson GW, Parry MA, and Keys AJ (1994) Incorporation of carbon from photosynthetic products into 2-carboxyarabinitol-1-phosphate and 2-carboxyarabinitol, The Biochemical journal 304 ( Pt 3), 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan S, Andralojc PJ, Lea PJ, and Parry MA (1999) 2’-carboxy-D-arabitinol 1-phosphate protects ribulose 1, 5-bisphosphate carboxylase/oxygenase against proteolytic breakdown, European journal of biochemistry / FEBS 266, 840–847. [DOI] [PubMed] [Google Scholar]
- 42.Gibney PA, Hickman MJ, Bradley PH, Matese JC, and Botstein D (2013) Phylogenetic Portrait of the Saccharomyces cerevisiae Functional Genome, G3 (Bethesda) 3, 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee AYW, Chung SK, and Chung SSM (1995) Demonstration That Polyol Accumulation Is Responsible for Diabetic Cataract by the Use of Transgenic Mice Expressing the Aldose Reductase Gene in the Lens, Proceedings of the National Academy of Sciences of the United States of America 92, 2780–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, Giardino I, and Brownlee M (2000) Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage, Nature 404, 787–790. [DOI] [PubMed] [Google Scholar]
- 45.Brownlee M (2001) Biochemistry and molecular cell biology of diabetic complications, Nature 414, 813–820. [DOI] [PubMed] [Google Scholar]
- 46.Schrijvers BF, De Vriese AS, and Flyvbjerg A (2004) From hyperglycemia to diabetic kidney disease: the role of metabolic, hemodynamic, intracellular factors and growth factors/cytokines, Endocrine reviews 25, 971–1010. [DOI] [PubMed] [Google Scholar]
- 47.Noltmann EA (1972) Aldose-ketose isomerases, Academic Press. [Google Scholar]
- 48.Goyal S, Yuan J, Chen T, Rabinowitz JD, and Wingreen NS (2010) Achieving optimal growth through product feedback inhibition in metabolism, PLoS computational biology 6, e1000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinrich R, and Rapoport TA (1974) Linear Steady-State Treatment of Enzymatic Chains - General Properties, Control and Effector Strength, European Journal of Biochemistry 42, 89–95. [DOI] [PubMed] [Google Scholar]
- 50.Fell D (1997) Understanding the Control of Metabolism, Portland Press. [Google Scholar]
- 51.Kacser H, Burns JA, and Fell DA (1995) The Control of Flux, Biochemical Society Transactions 23, 341–366. [DOI] [PubMed] [Google Scholar]
- 52.Kell DB, and Westerhoff HV (1986) Metabolic Control-Theory - Its Role in Microbiology and Biotechnology, Fems Microbiology Reviews 39, 305–320. [Google Scholar]
- 53.Hofmeyr JHS, and Cornishbowden A (1991) Quantitative Assessment of Regulation in Metabolic Systems, European Journal of Biochemistry 200, 223–236. [DOI] [PubMed] [Google Scholar]
- 54.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, and Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants, Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 55.McIsaac RS, Oakes BL, Wang X, Dummit KA, Botstein D, and Noyes MB (2013) Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast, Nucleic Acids Res 41, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu YF, Zhao X, Glass DS, Absalan F, Perlman DH, Broach JR, and Rabinowitz JD (2012) Regulation of yeast pyruvate kinase by ultrasensitive allostery independent of phosphorylation, Molecular cell 48, 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu WY, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, and Rabinowitz JD (2010) Metabolomic Analysis via Reversed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer, Analytical Chemistry 82, 3212–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melamud E, Vastag L, and Rabinowitz JD (2010) Metabolomic Analysis and Visualization Engine for LC-MS Data, Analytical Chemistry 82, 9818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamini YY, Daniel. (2001) The control of the false discovery rate in multiple testing under dependency, Ann Stat 20, 1165–1188. [Google Scholar]
- 60.Bennett BD, Yuan J, Kimball EH, and Rabinowitz JD (2008) Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach, Nature Protocols 3, 1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, and Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli, Nature Chemical Biology 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Egan W, Schneerson R, Werner KE, and Zon G (1982) Structural Studies and Chemistry of Bacterial Capsular Polysaccharides - Investigations of Phosphodiester-Linked Capsular Polysaccharides Isolated from Hemophilus-Influenzae Types a, B, C, and F - Nmr Spectroscopic Identification and Chemical Modification of End Groups and the Nature of Base-Catalyzed Hydrolytic Depolymerizationlu, Journal of the American Chemical Society 104, 2898–2910. [Google Scholar]
- 63.Kapuscinski M, Franke FP, Flanigan I, Macleod JK, and Williams JF (1985) Improved Methods for the Enzymic Preparation and Chromatography of Octulose Phosphates, Carbohydrate research 140, 69–79. [Google Scholar]
- 64.Kuznetsova E, Xu L, Singer A, Brown G, Dong A, Flick R, Cui H, Cuff M, Joachimiak A, Savchenko A, and Yakunin AF (2010) Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae, J Biol Chem 285, 21049–21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, and Yakunin AF (2006) Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family, J Biol Chem 281, 36149–36161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.