Abstract
Thioredoxins (Trxs) play an important role in defending against oxidative stress and keeping disulfide bonding correct to maintain protein function. Edwardsiella piscicida, a severe fish pathogen, has been shown to encode several thioredoxins including TrxA, TrxC, and TrxH, but their biological roles remain unknown. In this study, we characterized TrxH of E. piscicida (named TrxHEp) and examined its expression and function. TrxHEp is composed of 125 residues and possesses typical thioredoxin H motifs. Expression of trxHEp was upregulated under conditions of oxidative stress, iron starvation, low pH, and during infection of host cells. trxHEp expression was also regulated by ferric uptake regulator (Fur), an important global regulatory of E. piscicida. Compared to the wild type TX01, a markerless trxHEp in-frame mutant strain TX01∆trxH exhibited markedly compromised tolerance of the pathogen to hydrogen peroxide, acid stress, and iron deficiency. Deletion of trxHEp significantly retarded bacterial biofilm growth and decreased resistance against serum killing. Pathogenicity analysis shows that the inactivation of trxHEp significantly impaired the ability of E. piscicida to invade host cells, reproduce in macrophages, and infect host tissues. Introduction of a trans-expressed trxH gene restored the lost virulence of TX01∆trxH. There is likely to be a complex relationship of functional complementation or expression regulation between TrxH and another two thioredoxins, TrxA and TrxC, of E. piscicida. This is the first functional report of TrxH in fish pathogens, and the findings suggest that TrxHEp is essential for coping with adverse circumstances and contributes to host infection of E. piscicida.
Introduction
Edwardsiella piscicida (formerly included in E. tarda) [1, 2], one family member of Enterobacteriaceae, is a Gram-negative, motile, rod-shaped bacterium. It is a serious fish pathogen and has a broad host range that includes many species of economically important fish such as Japanese eel, flounder, turbot, red sea bream, tilapia, and channel catfish [3]. Edwardsiellosis, which is caused by E. piscicida, is one of the most severe diseases in aquaculture with freshwater as well as marine water supply, and has led to great economic losses in the aquaculture industry worldwide [4–7]. In addition to fish, E. piscicida is also pathogenic to humans and can cause gastrointestinal disorders and other diseases [8]. Since E. piscicida is an important pathogen of aquatic animals, more and more studies about E. piscicida are reported. A large number of virulence factors/systems, such as type III (T3SS) and type VI (T6SS) secretion systems, a two-component regulatory system, hemolysin, LuxS/AI-2 quorum sensing system, molecular chaperons and RNA-binding protein Hfq, ferric uptake regulator, lysozyme inhibitors, and so on, are known to be involved in E. piscicida stress resistance or pathogenicity [9–13]. Accumulating evidence has made clear that E. piscicida is an intracellular pathogen with the capacity to evade host immune defense [11, 14]. Recently, a study on intracellular trafficking pathways indicates a clear preference of E. piscicida for certain endocytic pathways and an involvement of endosomes, lysosomes, and cytoskeletons in the infection process [15]. Cheng et al. reported that this bacteria uses the tricarboxylic acid cycle to evade complement-mediated killing [16]. Another study showed that Sip2 in E. piscicida contributes to serum survival, acid resistance, intracellular replication, and host infection [17]. However, studies on the tolerance of E. piscicida to oxidative stress are limited.
Oxidative stress is a problem often encountered by most microorganisms. Reactive oxygen species (ROS) are abundantly generated in various environments including in host environments, leading microorganisms to develop multiple strategies to cope with oxidative stress [18]. One of these strategies is the thioredoxin system. The thioredoxin system is widely distributed in several species, from archaea and bacteria to humans, and is composed of thioredoxin (Trx), thioredoxin reductase (TrxR), and nicotinamide adenosine dinucleotide phosphate (NADP/NADPH) [19]. This antioxidant system transfers electrons from NADPH through TrxR to Trx and then to a large range of proteins that play critical roles in DNA synthesis and oxidative stress [20]. The major function of the Trx system is to reduce disulfide bridges in target proteins and help organisms recover from ROS stress [21, 22]. In microbes, such as Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Helicobacter pylori, thioredoxins have been confirmed to involve in maintaining the intracellular redox homeostasis and play a key role in the protection of cells against ROS [23–25]. Moreover, the Trx system also provides the electron donor for many critical enzymes by its function of disulfide reductase and thus is related to a series of cellular functions [26].
Trx is a small (around 12 kDa) ubiquitous protein characterized by a conserved motif involving two redox-active Cys separated by a pair of amino acids (CxxC motif) [27]. Besides the oxidoreductase function, Trx has the function of regulating cell growth, inhibiting apoptosis and regulating gene transcription [28]. The E. coli genome encodes two Trx, Trx1 (TrxA), and Trx2 (TrxC). Trx1 is the first thioredoxin discovered in E. coli, and its function as a hydrogen donor in redox reactions was identified [29]. H. pylori also contains two genes that encode Trx, trxA and trxC. TrxA expression in H. pylori is induced by stress agents, and it is the electron donor for antioxidant enzymes [24, 30]. TrxC acts as a chaperone and transforms denatured arginase into its active form [31]. There are three thioredoxin in the E. piscicida genome, TrxA, TrxC, TrxH [32]. To our knowledge, there are no reports about the function study of TrxH in this pathogenic bacteria. In this study, we characterized TrxH in E. piscicida (named TrxHEp), examined its expression profiles under different conditions, and analyzed its role in adversity and infection. Our results provide the first insights into the biological function of E. piscicida TrxH.
Materials and methods
Bacteria and growth conditions
Escherichia coli BL21 (DE3) was purchased from TransGen (Beijing, China). E. coli S17-1λpir was purchased from Biomedal (Sevilla, Spain). E. piscicida TX01 was isolated from diseased fish [33]. Bacteria were cultured in Luria–Bertani broth (LB) at 37 °C (for E. coli) or 28 °C (for E. piscicida). Where indicated, chloramphenicol, tetracycline, polymyxin B, and 2,2′dipyridyl (Dp) were supplemented at the concentrations of 30 μg/mL, 15 μg/mL, 100 μg/mL, and 100 μM respectively.
Construction of the trxHEp mutation and its complementation
The primers used in this study are listed in Table 1. To construct the trxHEp knockout strain, TX01∆trxH, in-frame deletion of a 261 bp segment (residues 24 to 110) of trxHEp was performed by overlap extension PCR as follows: the first overlap PCR was performed with the primer pair TrxHF1/R1, the second overlap PCR was performed with the primer pair TrxHF2/R2, and the fusion PCR was performed with the primer pair TrxHF1/R2. The PCR products amplified by primer pair TrxHF1/R2 were inserted into the suicide plasmid pDM4 [34] at the Bgl II site, resulting in pDMTrxH. S17-1λpir was transformed with pDMTrxH, and the transformants were conjugated with TX01 as described previously [35]. The transconjugants were selected on LB agar plates supplemented with 10% sucrose. One of the colonies that were resistant to sucrose and sensitive to chloramphenicol (marker of pDM4) was analyzed by PCR, and the PCR products were subjected to DNA sequencing to confirm in-frame deletion. This strain was named TX01∆trxH. To construct the complementary strain TX01∆trxHC, trxHEp was amplified by PCR with primers TrxHF3/R3, and the following experimental operations were done as described previously [35].
Table 1.
Primers used in this study
| Primer | Sequences (5′ → 3′) |
|---|---|
| TrxhKOF1 | GGATCCCTCAACTTGCCAATACCAT (BamHI) |
| TrxhKOR1 | TCACCGATGCTAGCGGCCTGTACGG |
| TrxhKOF2 | CCGCTAGCATCGGTGAAACGTCGCCA |
| TrxhKOR1 | GGATCCATGCCGATCGCCATCAGC (BamHI) |
| TrxhF3 | GGATCCCTGAGTATTGCCGAGC (BamHI) |
| TrxhR3 | CATATGGCGGGTCAGAAAGTCAGAGA (HindIII) |
| TrxhRTF | AAACGGCAGGAGCGTGAA |
| TrxhRTR | GGGTGCTTTGCTGGCTGA |
| TrxARTF | GAGTATGCAGGGAAGCTGACCAT |
| TrxARTR | GGTTGGCATCCAGGAACTCTTT |
| TrxCRTF | TCCCGCCTGTCATACCACCA |
| TrxCRTR | GCGCCCAGAAGTCGATCAGTA |
| TrxhPF4 | GATATCATTCAGCCCGGCAAAGAC (EcoRV) |
| TrxhPR4 | GATATCATCCCACCTTTTTGTAATGTTC (EcoRV) |
Resistance to stressors (H2O2, iron deficiency and acidic pH) and to non-immune fish serum
TX01, TX01∆trxH and TX01∆trxHC were cultured in LB medium to exponential phase. To determine hydrogen peroxide and iron deficiency tolerance, LB agar plates (pH = 7) supplemented with or without 1 mM H2O2 or 50 μM 2,2′dipyridyl (Dp) were streaked with 5 μL TX01, TX01∆trxH and TX01∆trxHC, respectively. For acid resistance, LB agar plates with pH = 7 or pH = 5 were streaked with the three bacteria. The plate was incubated at 28 °C for 48 h, and bacterial growth was examined. For quantitative analysis, three strains were cultured in LB medium with three stress conditions for 24 h, then the populations of cultivated bacteria were counted by diluted plate. To determine hydrogen peroxide sensitivity, TX01 and TX01∆trxH were cultured in LB medium to exponential phase, then H2O2 was added to 5 mM, the bacterial growth was surveyed every hour. The experiment was performed three times.
TX01, TX01∆trxH and TX01∆trxHC were cultured in LB medium to exponential phase. Then the cells were washed with PBS and resuspended in PBS. Approximately 105 bacterial cells were mixed with 50 μL fish serum or PBS (control). After incubation with mild agitation at 28 °C for 60 min, the mixtures were serially diluted and plated in triplicate on LB agar plates. The plates were incubated at 28 °C for 48 h, and the colonies that appeared on the plates were enumerated. The survival rate was calculated as follows: [(number of serum-treated cells)/(number of control cells)] × 100%. The experiment was performed three times.
Biofilm assay
TX01, TX01∆trxH and TX01∆trxHC were cultured in LB medium to exponential phase and diluted to 105 CFU/mL. The diluted cells were transferred into a 96-well polystyrene plate (Nunc, Denmark) and incubated at 28 °C for 24 h without agitation. Then the wells were washed gently five times with PBS. The attached cells were treated with Bouin fixative for 1 h and stained with 1% crystal violet solution for 20 min. After the treatment, the unbound dye was removed by rinsing the plate several times with running water. The plate was air dried. The bound dye was eluted in ethanol, and the A570 of eluates was measured. The experiment was performed three times.
Invasion of eukaryotic cell lines
Interaction of bacteria with cultured fish cells
Examination of interactions between FG cells and E. piscicida was performed as described previously [36]. Briefly, TX01, TX01∆trxH and TX01∆trxHC were cultured in LB medium to exponential phase. FG cells were cultured in 96-well cell culture plates and 100 μL bacteria (1 × 107 CFU/mL) were added to FG cells at a multiplicity of infection (MOI) of 10:1. After incubation at 23 °C for 0, 2, 4, 8 h, the plates were washed five times with PBS. To determine the number of bacterial cells associated with FG cells, the washed FG cells were lysed with 200 μL of 1% (vol/vol) Triton X-100 in PBS, and the lysate was diluted serially and was plated on LB agar plates supplemented with polymyxin B. After incubation overnight at 28 °C, the numbers of colonies that appeared on the plates were counted. For microscopy observation, TX01 and TX01∆trxH were labeled with FITC (Solarbio, Beijing, China). FG cells were cultured in Glass Bottom cell Culture Dish and infected with FITC-labeled bacteria for 4 h, the plates were washed three times with PBS. The cells were fixed by polyformaldehyde, then the fluorescence of extracellular bacteria was quenched by adding trypan blue (2 mg/mL in PBS). After washing three times with PBS, the cells were observed with a confocal microscope (Olympus Fluoview FV1000).
Bacterial replication in macrophages
The experiment was performed as described by Sui et al. [15] with slight modification. RAW264.7, a murine monocyte-macrophage cell line, were cultured in Dulbecco’s minimal Eagle’s medium (DMEM) (Gibco, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA) at 37 °C in 5% CO2. Then 100 μL bacteria (1 × 107 CFU/mL) were added to RAW264.7 cells at a multiplicity of infection (MOI) of 10:1, followed by incubation at 28 °C for 2 h. Extracellular E. piscicida was killed by adding gentamicin (100 μg/mL) to the plate, followed by incubation at 28 °C for 1 h. The cells were washed three times with PBS and cultured in DMEM containing 10 μg/mL gentamicin for 0, 2, 4, and 8 h. At each time point, the number of bacteria in macrophage was detected as described above. The experiment was performed three times.
Fish and experimental challenges for bacterial dissemination in vivo
Clinically healthy Japanese flounder (Paralichthys olivaceus) (average 14.2 g) were purchased from a commercial fish farm in Shandong. The fish were maintained at ~22 °C in aerated seawater and fed daily with commercial dry pellets. Fish were acclimatized in the laboratory for 2 weeks before experimental manipulation. Before the experiment, fish were randomly sampled and examined for the presence of bacteria in the blood, liver, kidney, and spleen, and no bacteria were detected from the sampled fish. For tissue collection, fish were euthanized with an overdose of MS222 (tricaine methanesulfonate) (Sigma, USA) as previously described [37]. For tissue dissemination analysis, TX01, TX01∆trxH, and TX01∆trxHC were cultured in LB medium to an OD600 of 0.8. The cells were washed with PBS and resuspended in PBS to 106 CFU/mL. Japanese flounder were divided randomly into four groups and infected by intramuscular (i.m.) injection with 50 μL TX01, TX01∆trxH, TX01∆trxHC, or PBS. The kidney and spleen were taken aseptically from the fish at 24 h and 48 h post-infection (hpi). Bacterial recovery from the tissues was determined as reported previously [35]. The rest of the infected fish were monitored daily for mortality for 20 days. The experiment was performed in triplicate.
Quantitative real-time reverse transcriptase-PCR (RT-qPCR) analysis of trxHEp, trxA, and trxC expression in wild TX01 strain and mutants under different environmental conditions
To examine trxHEp expression under in vitro conditions, TX01 was grown in LB medium with different pH (pH 5 or 7) at 28 °C, grown in LB medium with or without 2,2′dipyridyl at 28 °C, and grown in LB medium with or without hydrogen peroxide at 28 °C. The bacteria were harvested by centrifugation and total RNA was extracted with HP Total RNA kit (Omega Bio-Tek, USA). The RNA was treated with DNase with the kit of RNase-Free DNase Set (Omega Bio-Tek, USA). One microgram of total RNA was used for cDNA synthesis with the Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). RT-qPCR were carried out as reported previously [35]. To examine trxHEp expression during infection of host cells, TX01 was cultured in LB medium to an OD600 of 0.8 and resuspended in PBS to 106 CFU/mL. The resuspension was used to infect FG cells, a cell line derived from flounder gill cells, as described above [38]. The cells were harvested and used for total RNA extraction and RT-qPCR analysis of trxHEp expression as described above. The experiment was performed three times.
A fur mutant strain of E. piscicida was obtained in another study (data not published). The wild type E. piscicida TX01 and fur mutant strain were cultured in LB medium to the early exponential phase, then bacteria were harvested and total RNA was extracted. The expression of trxHEp expression in two strains were examined by RT-qPCR as described above.
To examine the expression of trxA and trxC in wild type E. piscicida and trxHEp knockout strain, TX01 and TX01∆trxH were cultured in LB medium with or without H2O2, cultured in LB medium with pH = 7 or pH = 5, and incubated with or without FG cells. The bacteria were harvested and total RNA was extracted. The expression of trxA and trxC expression in two strains were examined by RT-qPCR as described above.
Transcriptional regulation of the trxHEp promoter by Fur
The speculative promoter of trxHEp (the 510 bp DNA upstream of trxHEp operon) was cloned by primers TrxhPF4/TrxhPR4 and was inserted into the SwaI site of pSC11, a promoter probe plasmid [39], resulting in pSH510. pSH510 was introduced into E. coli DH5α by transformation and cultured on X-gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside) plate. DH5α/pSH510 was then transformed with pT (control) and pTFur, which expressed the Fur and was constructed as described by Wang et al. [40], and cultured on X-gal plate. The transformants were subjected to β-galactosidase assay [40].
Statistical analysis
All statistical analyses were performed with SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). Data were analyzed with analysis of variance (ANOVA), and statistical significance was defined as P < 0.05.
Results
Characterization of the TrxHEp sequence
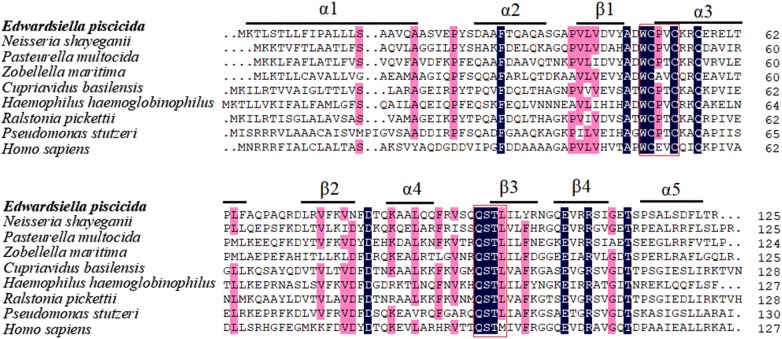
trxHEp codes for a putative protein of 125 amino acids, which contains a thioredoxin domain. According to BLAST sequence analyses, TrxHEp shares 49.6%–38.5% overall sequence identities with the thioredoxin homology of a number of species including Neisseria shayeganii, Pasteurella multocida, Zobellella maritima, Cupriavidus basilensis, Haemophilus haemoglobinophilus, Ralstonia pickettii, Pseudomonas stutzeri. Curiously, among the species with sequence identities of more than 30%, there are no common animal pathogenic bacteria even including E. coli (except Edwardsiella). TrxHEp also share sequence identity of 37.1% with thioredoxin of Homo sapiens. The WCXXC catalytic motif, the conserved active site of the TrxH family [41], exists in TrxHEp, suggesting that TrxHEp is one member of the TrxH protein family. Another conservative motif, QSTL/M, also exists in TrxHEp (Figure 1). Protein secondary structure prediction shows that the characteristic elements of the thioredoxin fold (α1, α2, α3, α4, α5, β1, β2, β3, and β4) are found in TrxHEp (Figure 1).
Figure 1.
Alignment of the amino acid sequences of TrxHEp homologues. Dots denote gaps introduced for maximum matching. The consensus residues are in black, the residues that are ≥ 75% identical among the aligned sequences are in pink. Conserved WCXXC catalytic motif and QSTL/M motif are boxed. The characteristic elements of the thioredoxin fold (α1, α2, α3, α4, α5, β1, β2, β3, and β4) are underlined.
Mutation of trxHEp has multiple effects
To examine its functional importance, the trxH gene of E. piscicida TX01 was knocked out by markerless in-frame deletion of a region encoding amino acid residues 24 to 110. The resulting mutant was named TX01∆trxH. Meantime, the complementary strain TX01∆trxHC, which is a genetic variant of TX01∆trxH that expresses trx in trans from a plasmid, was constructed.
Effect on bacterial resistance against stressors
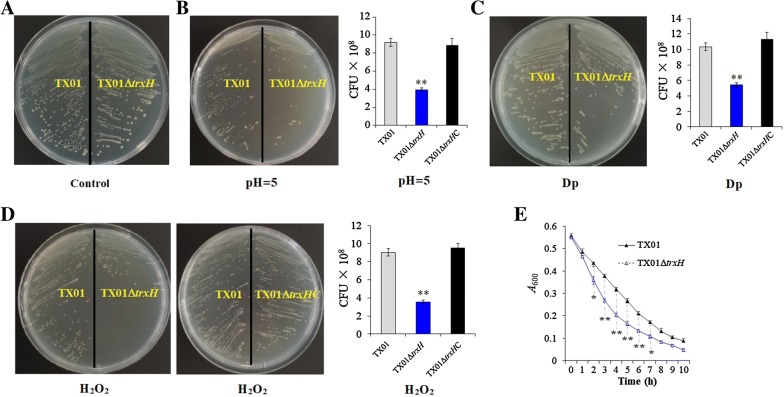
Growth analysis shows that when cultured in LB medium or LB agar medium, TX01∆trxH and TX01 exhibited a comparative growth rate (Figure 2A and data not shown). This indicates that the trxHEp mutation had no effect on growth of E. piscicida in normal conditions. However, when cultured in an LB agar plate containing H2O2, TX01 grew normally, whereas TX01∆trxH hardly grew, and the amount of TX01∆trxH in H2O2 stress was significantly lower than TX01 (Figure 2D). Consistently, when bacteria grew to early logarithmic phase in LB medium and hydrogen peroxide was added to the medium, growth analysis shows that TX01∆trxH exhibited a higher sensitivity to H2O2 than TX01 at a subsequent growth period (Figure 2E). When cultured under acidic condition, TX01∆trxH grew poorer than TX01 and the amount of TX01∆trxH was significantly lower than that of TX01 (Figure 2B). Similarly, when cultured under iron deficiency, the growth of TX01∆trxH was obviously retarded, compared to TX01 (Figure 2C). In contrast to TX01∆trxH, TX01∆trxHC and wild TX01 exhibited comparative capability of resistance against stresses caused by H2O2, acid, and Dp (Figure 2 and data not shown).
Figure 2.
Sensitivity of Edwardsiella piscicida to oxidative stress, iron deficiency, and acid stress. TX01, TX01∆trxH, and TX01∆trxHC were cultured in LB medium and LB agar plates with pH = 7 (normal A), with pH = 5 (B), with 80 μM 2,2′dipyridyl (Dp, C), or with 1 mM H2O2 (D) at 28 °C for 24–48 h. The populations of cultivated bacteria were counted by diluted plate. E TX01 and TX01∆trxH were cultured to early logarithmic phase in normal LB medium, then 5 mM H2O2 were added to the medium, and cell density was measured at different time points by determining absorbance at OD600. Data are the means of three independent experiments and presented as mean ± SEM (N = 3). N, the number of times the experiment was performed. *P < 0.05, **P < 0.01.
Effect on bacterial resistance to non-immune fish serum
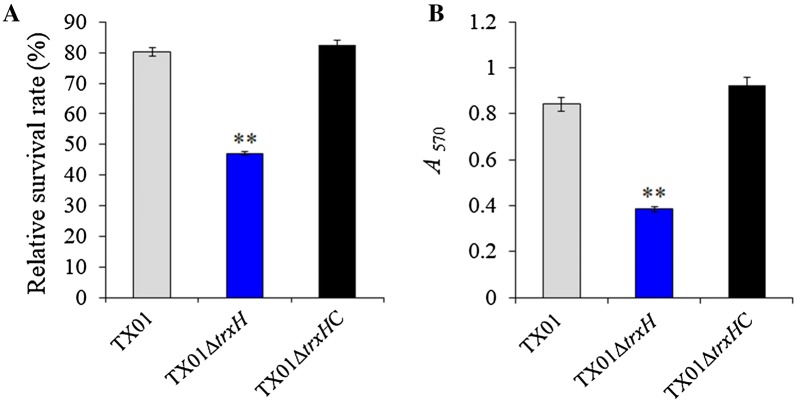
Resistance against host serum killing is an important part of the virulence mechanism in many pathogens, and E. piscicida is known to resist the bactericidal effect of fish serum [14]. To examine whether trxHEp mutation affects the ability of serum tolerance, TX01, TX01∆trxH, and TX01∆trxHC were incubated with non-immune flounder serum for 1 h, and the survival of bacteria was determined by plate count. The result shows that TX01 and TX01∆trxHC exhibit apparent serum resistance, as 80.3% and 82.6% of the cells survived after incubation with flounder serum, respectively. However, TX01∆trxH displayed only 47.1% of surviving cells after serum treatment, which is significantly lower than for TX01 (Figure 3A).
Figure 3.
Effects of trxHEp mutation on resistance to serum and biofilm growth. A Survival of E. piscicida in fish serum. TX01, TX01∆trxH, and TX01∆trxHC were incubated with non-immune flounder serum or PBS (control). After incubation, the survival of the bacteria was determined by plate counting. B The biofilm forming capacity of E. piscicida. TX01, TX01∆trxH, and TX01∆trxHC were incubated in polystyrene plate and biofilm formation was determined by measuring the the A570 of final eluates. Data are presented as mean ± SEM (N = 3). N, the number of times the experiment was performed. **P < 0.01.
Effect on biofilm formation
Since trxHEp mutations have no effect on planktonic growth of E. piscicida in normal conditions, we wanted to know whether bacterial biofilm formation was influenced by deletion of trxHEp. The biofilm forming capacity of E. piscicida were analyzed and the result shows that biofilm growth of TX01∆trxH was significantly slower (2.2-fold less) than that of TX01. And the biofilm growth of TX01∆trxHC was similar to that of TX01 (Figure 3B).
Effect on cell invasion in vitro
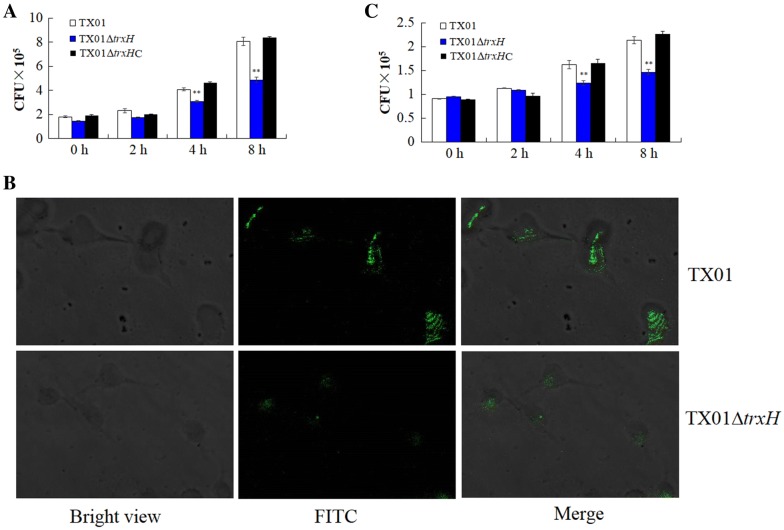
To examine whether trxHEp mutation played any role in the infectivity of TX01, cultured FG cells were incubated with the same dose of TX01, TX01∆trxH, or TX01∆trxHC. The bacterial cells associated with infection and those invading the host cells were enumerated. The results show that the amount of TX01∆trxH recovered from FG cells was significantly lower than that of TX01 at 4 and 8 hpi, and the amount of TX01∆trxHC was comparative to that of TX01 (Figure 4A). Consistently, microscopic analysis shows that fluorescence intensity of cells infected by FITC-labeled TX01∆trxH was significantly weaker than that of cells infected by FITC-labeled TX01 (Figure 4B), indicating that the ability to infect the host cell was declined when trxHEp was inactivated.
Figure 4.
Effect of trxHEp mutation on cellular infection and replication. A The invasion of Edwardsiella piscicida to flounder gill cells (FG cells). FG cells were infected with the same dose of TX01, TX01∆trxH, and TX01∆trxHC for various hours and washed with PBS. Then FG cells were lysed and the bacterial associated with and invaded into the host cells were determined. B FG cells were incubated with FITC-labeled E. piscicida TX01∆trxH or TX01, and the fluorescence of extracellular bacteria was quenched by adding trypan blue, then cells were observed with a confocal microscope. C Replication of E. piscicida in mouse macrophage cell RAW264.7. RAW264.7 cells were hatched with TX01, TX01∆trxH, and TX01∆trxHC for 2 h, then extracellular bacteria were killed. The cells were then incubated further for various hours, and the number of intracellular bacteria was determined by plate counting. Data are the means of three independent experiments and presented as mean ± SEM (N = 3). N, the number of times the experiment was performed. **P < 0.01.
It is known that E. piscicida is able to survive and replicate in the mouse macrophage cell line RAW264.7 [15]. To examine whether trxHEp mutation played any role in the intracellular survival of TX01, cultured RAW264.7 cells were incubated with the same dose of TX01 or TX01∆trxH, and extracellular bacteria were killed. The cells were then incubated further for various hours, and the number of intracellular bacteria was determined by plate count. The results show that the number of TX01∆trxH recovered from the intracellular RAW264.7 cells was significantly lower than those of TX01 at 4 and 8 hpi, and the amount of TX01∆trxHC was comparative to that of TX01 (Figure 4C).
Effect on bacterial dissemination in the fish tissues in vivo and general virulence
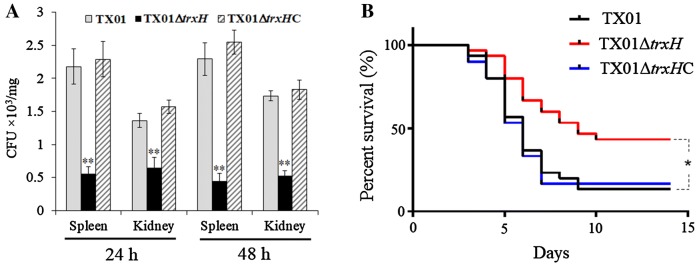
To examine the effect of trxHEp mutation on tissue infectivity, flounder were infected with the same dose of TX01, TX01∆trxH, or TX01∆trxHC, and bacterial recoveries from spleen and kidneys were determined at 24 and 48 hpi. The results show that bacterial recoveries from TX01∆trxH-infected fish were significantly lower than those from TX01-infected fish at 24 hpi and 48 hpi, and bacterial recoveries from TX01∆trxHC-infected fish were similar to those from TX01-infected fish (Figure 5A).
Figure 5.
In vivo infection of Edwardsiella piscicida in Japanese flounder. A Bacterial dissemination in the fish tissues. Flounder were infected with the same dose of TX01, TX01∆trxH, or TX01∆trxHC, bacterial recoveries from spleen and kidney were determined by plate count at 24 and 48 hpi. B Host mortality induced by E. piscicida. Flounder were infected with equivalent doses of TX01, TX01∆trxH, and TX01∆trxHC, and accumulated mortality were monitored for a period of 20 days (only 15 days are shown since no more deaths occurred after 15 days). Significance between the survivals of wild type and mutant infected fish was determined with logrank test. Data are presented as means ± SEM (N = 3). N, the number of times the experiment was performed. *P < 0.05, **P < 0.01.
To examine the effect of trxHEp mutation on the overall bacterial virulence, Japanese flounder infected with TX01, TX01∆trxH, or TX01∆trxHC were used to monitor for mortality. The results show that at the end of the monitored period (20 days), the survival rate of TX01∆trxH-infected fish was 43.3%, which was significantly higher than that of TX01-infected fish (13.3%) (Figure 5B).
Expression of trxHEp, trxA and trxC in wild TX01 strain and mutants under different environmental conditions
Expression of trxHEp is modulated by stress
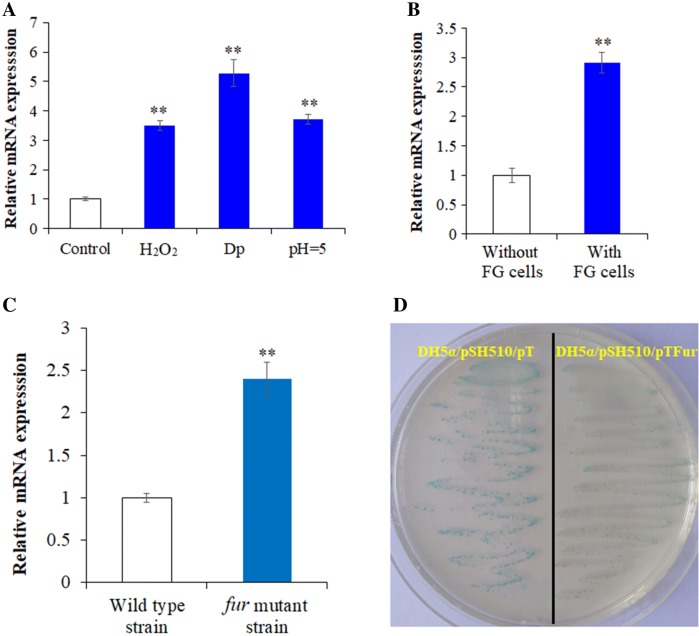
Since thioredoxin plays a role in maintaining the intracellular redox homeostasis, and since it may participate in response to adversity, we determined the expression of trxHEp under normal conditions (i.e., cultured in LB medium), adverse conditions (i.e., cultured in LB medium with H2O2, Dp, or cultured in low pH LB medium), and infecting host cell condition by RT-qPCR. The results show that the expression of trxHEp was significantly enhanced when bacteria faced hydrogen peroxide pressure, acid pressure, iron deficiency, and an intracellular environment, compared to the expression of trxHEp in normal condition (Figures 6A and B).
Figure 6.
Expression analysis of trxHEp. A Expression of trxHEp under different conditions. RT-qPCR was performed with total RNA extracted from Edwardsiella piscicida TX01 cultured in normal LB medium (control, pH = 7), in hydrogen peroxide condition (normal LB medium with H2O2), in acid condition (pH = 5), in iron limitation condition (with the iron chelator 2,2′dipyridyl, Dp), Expression level of trxHEp in normal LB medium was set as 1. B RT-qPCR was performed with total RNA extracted from TX01 incubated with or without (control) FG cells. Expression level of trxHEp without FG cells was set as 1. C RT-qPCR was performed with total RNA extracted from wild type TX01 and fur mutant strain cultured in normal LB medium. Expression level of trxHEp in wild type TX01 strain was set as 1. D DH5α/pSH510/pTFur and DH5α/pSH510/pT were streaked and cultured on LB plate with X-gal, kanamycin, and ampicillin. Data are presented as mean ± SEM (N = 3). N, the number of times the experiment was performed. **P < 0.01.
Expression of trxHEp is regulated by Fur (Ferric uptake regulator)
In a previous study of E. piscicida, we obtained a fur mutant train, which exhibited much higher virulence than wild type E. piscicida strain TX01 (data not published). Proteomic analysis shows that the expression of TrxH was significantly up-regulated in the fur mutant strain, compared to the wild type strain (data not shown). So we detected the expression of trxHEp at the mRNA level. RT-qPCR shows that the expression of trxHEp in the fur mutant strain was 2.4-fold higher than that of trxHEp in the wild type strain (Figure 6C). Subsequently, we detected the regulatory effect of Fur on promoter activity of trxHEp. The speculative promoter of trxHEp, H510, was cloned to a promoter probe plasmid pSC11 and introduced into DH5α by transformation, resulting in DH5α/pSH510. When DH5α/pSH510 was cultured in LB agar plate with X-gal, the bacterial colonies were blue, which indicates that H510 has promoter activity. DH5α/pSH510 was then transformed with pTFur and pT (control). On the X-gal plate, the blue of DH5α/pSH510/pTFur was obviously weak, compared with DH5α/pSH510/pT (Figure 6D). β-Galactosidase assays show that galactosidase activity produced by DH5α/pSH510/pTFur was significantly lower (3.1 fold) than that produced by DH5α/pSH510/pT. These results indicate that Fur negatively regulated transcription of trxHEp.
Expression of trxA and trxC in TX01∆trxH
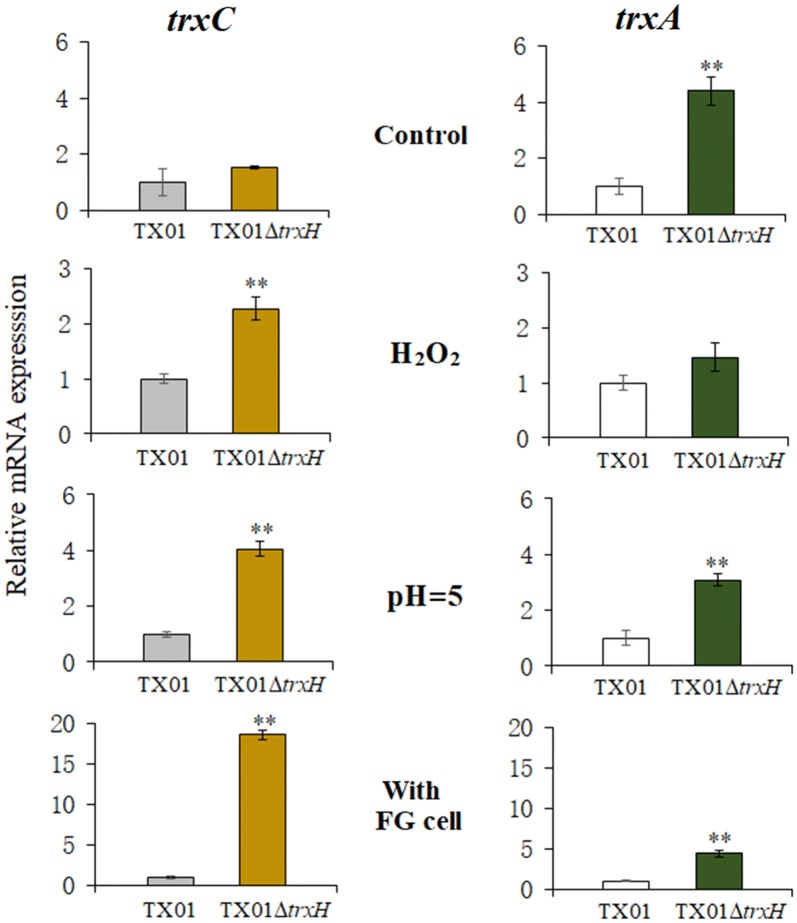
There are two other thioredoxins, TrxA and TrxC, in E. piscicida, and their functions are unknown. To detect whether TrxA and TrxC undertake more functions when the TrxH function was deactivated, we examined the expression of trxA and trxC in TX01∆trxH. The results show that the deletion of trxH has no effect on the expression of trxC in normal conditions. However, when facing oxidative stress, acid pressure, or infecting host cell, the expression of trxC in TX01∆trxH was significantly up-regulated, compared to that in TX01. On the contrary, the deletion of trxH significantly induced 4.4-fold expression of trxA in normal conditions, but trxA expression exhibited no difference between TX01 and TX01∆trxH in the presence of hydrogen peroxide. Under an acidic environment and infecting host cell, the mRNA level of trxA in TX01∆trxH was 3.1- and 4.3-fold high, compared to the level of trxA in TX01 (Figure 7). These findings suggest that there is likely a complex relationship of functional complementation or expression regulation between TrxH and another two thioredoxins, TrxA and TrxC, of E. piscicida.
Figure 7.
Expression of trxC and trxA in wild type Edwardsiella piscicida TX01 strain and TX01∆trxH strain. Total RNA was extracted from E. piscicida TX01 and TX01∆trxH cultured in normal LB medium (control, pH = 7), in hydrogen peroxide condition (with H2O2), in acid condition (pH = 5), or incubating with FG cells, respectively. Expressions of trxC and trxA were analyzed by RT-qPCR. Data are presented as mean ± SEM (N = 3). N, the number of times the experiment was performed. **P < 0.01.
Discussion
Thioredoxins play major roles in protection of cells against toxic oxygen species as well as maintaining the intracellular thio-disulfide balance in bacteria [42]. In E. piscicida, the function of thioredoxins is completely unknown. Moreover, we did not find any reports about the function of thioredoxin H in pathogen bacteria. In this study, a thioredoxin H homologue, TrxH, was identified and characterized from E. piscicida. TrxHEp contains an unusual motif (CPVC), not the typical Trx motif (CGPC), which is found in most Trx proteins. It is not known how this affects its activity or its roles.
It is reported that the expression of trx in many bacteria is induced by oxidative stress [20]. Similarly, in E. piscicida, the expression of trxHEp was significantly enhanced in the presence of H2O2. In other adverse conditions including acid stress and iron deficiency, trxHEp expression is upregulated. When bacteria infect host cells, the expression of trxHEp also increases. Under these conditions, the highest induction of trxHEp occured with iron deficiency, in which Fur exists predominately as a biologically inactive apoprotein [43]. Consistently, the expression of trxHEp in the fur mutant strain was higher than that in the wild type strain. As far as we know, this is the first report that Trx expression was regulated by Fur.
Besides TrxH, there are another two thioredoxin, TrxA and TrxC in E. piscicida. When trxHEp was inactivated, the expression of trxC did not change under normal conditions. However, under stress or infecting host cell conditions, trxC expression was significantly induced. As a chaperone, TrxC maybe undertook more roles in adverse conditions especially when trxH was inactivated in E. piscicida. On the contrary, the expression of trxA was directly or indirectly regulated by TrxH, which is sustained by the fact that Trx possesses the property of regulating gene expression [28]. However, in the presence of hydrogen peroxide trxA expression did not appear significantly different between wild TX01 and mutant TX01∆trxH. These findings show that there is likely to be a complex relationship between the three trx genes, and further research needs to be performed in the future.
Since the expression of trxHEp is induced by adverse conditions, it probably participates in stress resistance. Our results show that when trxHEp was inactive, the mutant TX01∆trxH exhibited a higher sensitivity to H2O2, which indicates that trxHEp plays an important role in tolerance to oxidative stress. Similar results were reported in other bacteria. In H. pylori, the ∆trxC mutant significantly decreased the ability of tolerance to oxygen exposure [24]. In Listeria monocytogenes, deletion of trxA markedly compromised tolerance of the pathogen to the thiol-specific oxidizing agent diamide [20]. Since Trx is involved in a series of cellular functions [26], we speculate that trxHEp participates in coping with other adversities, and our results confirm that trxHEp was involved in tolerance to acid stress and iron deficiency stress, which are similar to the adverse environment in the host.
It is generally known that Trx donates electrons to the disulfide bond system and helps isomerization of incorrectly paired disulfide bonds [20, 44]. Since many bacterial virulence factors require stable disulfide bonds for proper folding and function, Trx is closely related to infection of bacteria [45, 46]. It is reported that TrxA is essential for motility and contributes to host infection of L. monocytogenes [20]. In H. pylori, the ∆trxC mutant lacks mouse colonization ability, while the ability to colonize mouse stomachs is significantly reduced in the ∆trxA mutant [24]. In Magnaporthe oryzae, Trx1 and Trx2 also play important roles in bacterial pathogenesis [47]. In pathogenic fungus Paracoccidioides lutzii, Trx1 contributes to this organism’s virulence [19]. In this study, we also examined the effect of trxHEp on E. piscicida pathogenicity. It is known that biofilm formation and serum resistance are important parts of the virulence in multiple pathogens such as E. piscicida [17, 35] and H. pylori [48, 49]. In the latter, the inability to acquire iron from serum has been suggested to block H. pylori virulence by weakening its growth and survival in the serum [48]. Our results show that trxHEp mutation had no effect on growth of E. piscicida, but harmed bacterial biofilm growth. Similarly, the capability of serum resistance was significantly declined when trxHEp was inactivated. Consistently, upon infection with host cells, TX01∆trxH compared to TX01, shows an impaired ability to infect cells and to propagate in cells, indicating that trxH is required for bacterial invasion of host cells and host tissues. These observations suggest that trxHEp may play a role during bacterial invasion.
In conclusion, this study identified and characterized thioredoxin TrxHEp from fish pathogen E. piscicida. Our results indicate that TrxHEp possesses conservative thioredoxin motifs and that the trxHEp expression seems to be up-regulated under the pressure of several stressors, including iron deficiency. Regarding this fact, our results suggest the potential role of Fur for regulating trxHEp expression. TrxHEp not only played an important role in coping with adverse circumstances, but functioned as a factor that is essential to bacterial infection both at the cellular level and in a live fish model. This is the first study of TrxH in fish pathogen, and the results suggest that TrxHEp exerts pleiotropic effects on the pathogenesis of E. piscicida.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BYW performed the experiments and analyzed the data. HQH and SL were responsible for microbial cultivation and some data analysis. PT, HFD, JAX provided valuable assistance to sample preparation and research ideas. DMS and YHH designed the experiment and wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (41476138), the Financial Fund of the Ministry of Agriculture and Rural Affairs, P. R. of China (NFZX2018), and the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052018005).
Ethics approval and consent to participate
The study was approved by the ethics committee of Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences. Efforts were taken to ensure that all research animals received good care and humane treatment.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bi-ying Wang, Email: 757140358@qq.com.
Hui-qin Huang, Email: huanghuiqin@itbb.org.cn.
Shuang Li, Email: 1468664229@qq.com.
Ping Tang, Email: tp5257@126.com.
Hao-fu Dai, Email: daihaofu@itbb.org.cn.
Jian-an Xian, Email: xianjianan@itbb.org.cn.
Dong-mei Sun, Email: sdmlzw@126.com.
Yong-hua Hu, Email: huyonghua@itbb.org.cn.
References
- 1.Abayneh T, Colquhoun DJ, Sørum H. Edwardsiella piscicida sp. nov., a novel species pathogenic to fish. J Appl Microbiol. 2013;114:644–654. doi: 10.1111/jam.12080. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Zhao L, Yang M, Yin K, Zhou X, Leung KY, Liu Q, Zhang Y, Wang Q. Transcriptomic dissection of the horizontally acquired response regulator EsrB reveals its global regulatory roles in the physiological adaptation and activation of T3SS and the cognate effector repertoire in Edwardsiella piscicida during infection toward turbot. Virulence. 2017;8:1355–1377. doi: 10.1080/21505594.2017.1323157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanty BR, Sahoo PK. Edwardsiellosis in fish: a brief review. J Biosci. 2007;32:1331–1344. doi: 10.1007/s12038-007-0143-8. [DOI] [PubMed] [Google Scholar]
- 4.Xu TT, Zhang XH. Edwardsiella tarda: an intriguing problem in aquaculture. Aquaculture. 2014;431:129–135. doi: 10.1016/j.aquaculture.2013.12.001. [DOI] [Google Scholar]
- 5.Katharios P, Kokkari C, Dourala N, Smyrli M. First report of edwardsiellosis in cage-cultured sharpsnout sea bream, Diplodus puntazzo from the Mediterranean. BMC Vet Res. 2016;11:155. doi: 10.1186/s12917-015-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loch TP, Hawke J, Reichley S, Faisal M, Del Piero F, Griffin MJ. Outbreaks of edwardsiellosis caused by Edwardsiella piscicida and Edwardsiella tarda in farmed barramundi (Lates calcarifer) Aquaculture. 2017;481:202–210. doi: 10.1016/j.aquaculture.2017.09.005. [DOI] [Google Scholar]
- 7.Shafiei S, Viljamaa-Dirks S, Sundell K, Heinikainen S, Abayneh T, Wiklund T. Recovery of Edwardsiella piscicida from farmed whitefish Coregonus lavaretus (L.), in Finland. Aquaculture. 2016;454:19–26. doi: 10.1016/j.aquaculture.2015.12.011. [DOI] [Google Scholar]
- 8.Park SB, Aoki T, Jung TS. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res. 2012;43:67. doi: 10.1186/1297-9716-43-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty S, Sivaraman J, Leung KY, Mok YK. The two-component PhoBPhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J Biol Chem. 2011;286:39417–39430. doi: 10.1074/jbc.M111.295188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. Edwardsiella tarda—virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012;14:26–34. doi: 10.1016/j.micinf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Li MF, Wang C, Sun L. Edwardsiella tarda MliC, a lysozyme inhibitor that participates in pathogenesis in a manner that parallels Ivy. Infect Immun. 2015;83:583–590. doi: 10.1128/IAI.02473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin K, Guan Y, Ma R, Wei L, Liu B, Liu X, Zhou X, Ma Y, Zhang Y, Waldor MK, Wang Q. Critical role for a promoter discriminator in RpoS control of virulence in Edwardsiella piscicida. PLoS Pathog. 2018;14:e1007272. doi: 10.1371/journal.ppat.1007272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui S, Xiao J, Wang Q, Zhang Y. H-NS binding to evpB and evpC and repressing T6SS expression in fish pathogen Edwardsiella piscicida. Arch Microbiol. 2018;198:653–661. doi: 10.1007/s00203-016-1226-4. [DOI] [PubMed] [Google Scholar]
- 14.Li MF, Sun L, Li J. Edwardsiella tarda evades serum killing by preventing complement activation via the alternative pathway. Fish Shellfish Immunol. 2015;43:325–329. doi: 10.1016/j.fsi.2014.12.037. [DOI] [PubMed] [Google Scholar]
- 15.Sui ZH, Xu H, Wang H, Jiang S, Chi H, Sun L. Intracellular trafficking pathways of Edwardsiella tarda: from clathrin- and caveolin-mediated endocytosis to endosome and lysosome. Front Cell Infect Microbiol. 2017;7:400. doi: 10.3389/fcimb.2017.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng ZX, Gong QY, Wang Z, Chen ZG, Ye JZ, Li J, Wang J, Yang MJ, Ling XP, Peng B. Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front Immunol. 2017;8:1706. doi: 10.3389/fimmu.2017.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li MF, Sun L. Edwardsiella tarda Sip2: a serum-induced protein that is essential to serum survival, acid resistance, intracellular replication, and host infection. Front Microbiol. 2018;9:1084. doi: 10.3389/fmicb.2018.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staerck C, Gastebois A, Vandeputte P, Calenda A, Larcher G, Gillmann L, Papon N, Bouchara JP, Fleury MJJ. Microbial antioxidant defense enzymes. Microb Pathog. 2017;110:56–65. doi: 10.1016/j.micpath.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Cintra LC, Domingos FC, Lima YAR, Barbosa MS, Santos RS, Faria FP, Jesuíno RSA. Molecular cloning, expression and insulin reduction activity of a thioredoxin 1 homologue (TRX1) from the pathogenic fungus Paracoccidioides lutzii. Int J Biol Macromol. 2017;103:683–691. doi: 10.1016/j.ijbiomac.2017.05.114. [DOI] [PubMed] [Google Scholar]
- 20.Cheng C, Dong Z, Han X, Wang H, Jiang L, Sun J, Yang Y, Ma T, Shao C, Wang X, Chen Z, Fang W, Freitag NE, Huang H, Song H. Thioredoxin a is essential for motility and contributes to host infection of Listeria monocytogenes via redox interactions. Front Cell Infect Microbiol. 2017;7:287. doi: 10.3389/fcimb.2017.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 22.Yoshida T, Oka S, Masutani H, Nakamura H, Yodoi J. The role of thioredoxin in the aging process: involvement of oxidative stress. Antioxid Redox Signal. 2003;5:563–570. doi: 10.1089/152308603770310211. [DOI] [PubMed] [Google Scholar]
- 23.Peng H, Zhang Y, Trinidad JC, Giedroc DP. thioredoxin profiling of multiple thioredoxin-like proteins in Staphylococcus aureus. Front Microbiol. 2018;9:2385. doi: 10.3389/fmicb.2018.02385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhns LG, Wang G, Maier RJ. Comparative roles of the two Helicobacter pylori thioredoxins in preventing macromolecule damage. Infect Immun. 2015;83:2935–2943. doi: 10.1128/IAI.00232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao X, Yang F, Li H, So PK, Yao Z, Xia W, Sun H. Targeting the thioredoxin reductase-thioredoxin system from Staphylococcus aureus by silver ions. Inorg Chem. 2017;56:14823–14830. doi: 10.1021/acs.inorgchem.7b01904. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 27.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 28.Zeller T, Klug G. Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften. 2006;93:259–266. doi: 10.1007/s00114-006-0106-1. [DOI] [PubMed] [Google Scholar]
- 29.Laurent TC, Moore EC, Reichard P. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J Biol Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 30.Alamuri P, Maier RJ. Methionine sulfoxide reductase in Helicobacter pylori: interaction with methionine-rich proteins and stress induced expression. J Bacteriol. 2006;188:5839–5850. doi: 10.1128/JB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGee DJ, Kumar S, Viator RJ, Bolland JR, Ruiz J, Spadafora D, Testerman TL, Kelly DJ, Pannell LK, Windle HJ. Helicobacter pylori thioredoxin is an arginase chaperone and guardian against oxidative and nitrosative stresses. J Biol Chem. 2006;281:3290–3296. doi: 10.1074/jbc.M506139200. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Yang M, Xiao J, Wu H, Wang X, Lv Y, Xu L, Zheng H, Wang S, Zhao G, Liu Q, Zhang Y. Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLoS ONE. 2009;4(10):e7646. doi: 10.1371/journal.pone.0007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HL, Zhang M, Xiao ZZ, Sun L. Genetic mechanisms multi-antimicrobial resistance in a pathogenic Edwardsiella tarda strain. Aquaculture. 2009;289:134–139. doi: 10.1016/j.aquaculture.2009.01.002. [DOI] [Google Scholar]
- 34.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu YH, Zhou HZ, Jin QW, Zhang J. The serine protease autotransporter Tsh contributes to the virulence of Edwardsiella tarda. Vet Microbiol. 2016;189:68–74. doi: 10.1016/j.vetmic.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 36.Hu YH, Liu CS, Hou JH, Sun L. identification, characterization, and molecular application of a virulence-associated autotransporter from a pathogenic Pseudomonas fluorescens. Appl Environ Microbiol. 2009;75:4333–4340. doi: 10.1128/AEM.00159-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang GH, Wang JJ, Yue B, Du X, Du HH, Zhang M, Hu YH. High mobility group box 2 of black rockfish Sebastes schlegelii: gene cloning, immunoregulatory properties and antibacterial effect. Fish Shellfish Immunol. 2018;84:719–725. doi: 10.1016/j.fsi.2018.10.066. [DOI] [PubMed] [Google Scholar]
- 38.Hu YH, Dang W, Sun L. A TonB-dependent outer membrane receptor of Pseudomonas fluorescens: virulence and vaccine potential. Arch Microbiol. 2016;194:795–802. doi: 10.1007/s00203-012-0812-3. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Wang H, Zhang M, Sun L. Molecular analysis of the copper-responsive CopRSCD of a pathogenic Pseudomonas fluorescens strain. J Microbiol. 2009;47:277–286. doi: 10.1007/s12275-008-0278-9. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Cheng S, Sun K, Sun L. Molecular analysis of the fur (ferric uptake regulator) gene of a pathogenic Edwardsiella tarda strain. J Microbiol. 2008;46:350–355. doi: 10.1007/s12275-008-0038-x. [DOI] [PubMed] [Google Scholar]
- 41.Chen IH, Chen HT, Huang YP, Huang HC, Shenkwen LL, Hsu YH, Tsai CH. A thioredoxin NbTRXh2 from Nicotiana benthamiana negatively regulates the movement of Bamboo mosaic virus. Mol Plant Pathol. 2018;19:405–417. doi: 10.1111/mpp.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuzawa A. Thioredoxin and redox signaling: roles of the thioredoxin system in control of cell fate. Arch Biochem Biophys. 2017;617:101–105. doi: 10.1016/j.abb.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Troxell B, Hassan HM. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol. 2013;3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozhkova A, Stirnimann CU, Frei P, Grauschopf U, Brunisholz R, Grütter MG, Capitani G. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 2004;23:1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negrea A, Bjur E, Puiac S, Ygberg SE, Aslund F, Rhen M. Thioredoxin 1 participates in the activity of the Salmonella enterica serovar Typhimurium pathogenicity island 2 type III secretion system. J Bacteriol. 2009;191:6918–6927. doi: 10.1128/JB.00532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira DMS, Mendes SJF, Alawi K, Thakore P, Aubdool A, Sousa NCF, da Silva JFR, Castro JA, Jr, Pereira PIC, Silva LCN, Grisotto MAG, Monteiro-Neto V, Costa SKP, da Costa R, Calixto JB, Brain SD, Fernandes ES. Transient receptor potential canonical channels 4 and 5 mediate Escherichia coli-derived thioredoxin effects in lipopolysaccharide-injected mice. Oxid Med Cell Longev. 2018;2018:4904696. doi: 10.1155/2018/4904696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S, Jiang C, Zhang Q, Qi L, Li C, Xu JR. Thioredoxins are involved in the activation of the PMK1 MAP kinase pathway during appressorium penetration and invasive growth in Magnaporthe oryzae. Environ Microbiol. 2016;18:3768–3784. doi: 10.1111/1462-2920.13315. [DOI] [PubMed] [Google Scholar]
- 48.Hathroubi S, Servetas SL, Windham I, Merrell DS, Ottemann KM. Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol Mol Biol Rev. 2018;82:e00001–18. doi: 10.1128/MMBR.00001-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pich OQ, Merrell DS. The ferric uptake regulator of Helicobacter pylori: a critical player in the battle for iron and colonization of the stomach. Future Microbiol. 2013;8:725–738. doi: 10.2217/fmb.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]