Abstract
A critical step in the induction of apoptosis is the activation of the apoptotic initiator caspase 9. We show that at its normal physiological concentration, caspase 9 is primarily an inactive monomer (zymogen), and that activity is associated with a dimeric species. At the high concentrations used for crystal formation, caspase 9 is dimeric, and the structure reveals two very different active-site conformations within each dimer. One site closely resembles the catalytically competent sites of other caspases, whereas in the second, expulsion of the “activation loop” disrupts the catalytic machinery. We propose that the inactive domain resembles monomeric caspase 9. Activation is induced by dimerization, with interactions at the dimer interface promoting reorientation of the activation loop. These observations support a model in which recruitment by Apaf-1 creates high local concentrations of caspase 9 to provide a pathway for dimer-induced activation.
Keywords: apoptosis‖crystal structure‖zymogen activation
The protease caspase 9 is the central participant in a multicomponent complex known as the apoptosome, which controls cell deletion during development of the central nervous system and the apoptotic response to lethal cellular insults such as ionizing radiation or chemotherapeutic drugs (1–3). The task of caspase 9 is to generate the active forms of caspases 3 and 7 by limited proteolysis, and thereby transmit the apoptotic signal to the execution phase. Most proteases exist in their biologically relevant locations in a latent form, and caspase 9 is no exception. However, caspase 9 is unusual among its close relatives in that proteolysis between the large and small subunit is not sufficient for conversion of the latent zymogen to the catalytic form (4, 5). In common with other initiators of proteolytic pathways (6–8), caspase 9 requires association with specific cofactors for its activation, in this case an activated form of Apaf-1 within the apoptosome (1).
There are two fundamental ways in which protease zymogens are maintained in an inactive conformation. In one case, exemplified by the lysosomal cysteine proteases (9) and the matrix metalloproteinases (10), the catalytic site is in an active conformation, but access is blocked; limited proteolysis removes the blocking N-terminal propeptide. In the second case, such as the chymotrypsin family exemplified by the coagulation serine proteases, the catalytic site is not yet formed, and structural rearrangements are required (11). These structural rearrangements can occur by limited proteolysis, or by cofactor binding irrespective of proteolysis. Thus the executioner of the coagulation pathway, thrombin, is activated simply by proteolysis (12), but the initiator of the extrinsic branch of the coagulation pathway, factor VII, is inactive until bound by its cofactor (tissue factor; ref. 13).
The structure of active caspases 1, 3, 7, and 8 with peptidyl inhibitors occupying their catalytic sites has clarified the common mechanism of the enzymes (14–20), yet little was known of the molecular basis of activation of the latent forms, nor the molecular mechanism of maintenance of the inactive zymogens. Because proteolysis is not required for activation of caspase 9, there must exist another mechanism to achieve the catalytically competent form, and here we describe studies that illustrate such a mechanism.
Materials and Methods
Recombinant Proteins.
Full-length and ΔCARD (caspase recruitment domain) human caspase 9 (lacking the first 138 residues) were expressed in soluble form and purified from Escherichia coli (5). A cleavage-defective mutant of caspase 9 containing Asp/Ala substitutions at the underlined residues PEPDA and DQLDA within the interdomain linker segment was generated, expressed, and purified as described (5). Further mutations that abrogated cleavage of caspase 9 were generated by replacing the three acidic residues PEDES in the interdomain linker segment by Ala residues (21). The final noncleavable constructs contained all five Ala substitutions, and are designated as single-chain forms.
Reagents.
The polycaspase inhibitor benzoxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (Z-VAD-FMK) and the caspase 9 substrate acetyl-Leu-Glu-His-Asp-7-amino-4-trifluoromethyl coumarin (Ac-LEHD-AFC) were from Enzyme Systems Products, and the N-deblocked VAD-FMK was custom synthesized by Enzyme Systems Products (Livermore, CA). The irreversible inhibitor benzoxycarbonyl-Glu-Val-Asp-dichlorobenzylmethyl ketone (Z-EVD-Dcbmk) was a gift of Joe Wu, Idun Pharmaceuticals (San Diego).
Caspase 9 Assays.
Hydrolysis of Ac-LEHD-AFC by caspase 9 was followed on a Molecular Devices fmax plate reader at 37°C. The assay buffer was 10 mM Pipes/0.1 M NaCl/0.1 mM EDTA/10 mM DTT/10% sucrose/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), pH 7.2. Cleavage of the substrate as a function of time was followed by monitoring the emission at 510 nm on excitation at 405 nm, and the initial velocity was determined from the linear portion of progress curve. The enzymatically active concentration of caspase 9 was determined by titration with Z-VAD-FMK as described (22).
Gel Filtration.
Gel filtration of caspase 9 derivatives was performed on a Amersham Pharmacia Superdex 200 column in 20 mM Tris/100 mM NaCl buffer (pH 8.0) with a flow rate of 1 ml⋅min−1. The column dimensions were 30 × 1 cm, and 0.5-ml fractions were collected for analysis.
Caspase-9 Crosslinking.
Catalytically active ΔCARD caspase 9, and ΔCARD caspase 9 covalently inhibited with Z-VAD-FMK were dialyzed against 100 mM phosphate buffer (pH 7.5), and the protein concentration was adjusted to 2.4 μM. The crosslinking reaction was performed by incubating 50 μl of the protein solution with 50 μl of aqueous glutaraldehyde solutions at different concentrations (500 mM to 32 μM) for 5 min at 23°C. The reaction mixture was reduced for 30 min at 37°C with 20 μl of 2 M NaBH4. The crosslinked protein was precipitated by adding 50 μl of 80% trichloroacetic acid, washed, and run in SDS/PAGE in gels of 8–18% linear acrylamide gradient with a 2-amino-2-methyl-1,3-propanediol/glycine/HCl system (23). Samples for SDS/PAGE were boiled in SDS sample buffer containing 50 mM DTT for 5 min and then loaded onto the stacking gel.
N-Terminal Sequencing of Protein Samples.
For N-terminal sequencing, protein samples were resolved by SDS/PAGE or nondenaturing PAGE and transferred to Immobilon-P membrane (Millipore) by electroblotting (24). The membrane was briefly stained with Coomassie Brilliant Blue R250, destained, and washed with water. Appropriate bands were excised and sequenced by Edman degradation on a 476A protein sequencer (Applied Biosystems).
Crystallization.
Protein used for crystallization trials was further purified by gel filtration (Amersham Pharmacia Superdex 200). Initial crystallization trials were set up with 8–10 mg/ml uninhibited full-length caspase 9 (two chain) in 10 mM Tris/100 mM NaCl/1 mM DTT (pH 8.0), and commercially available screening kits from Hampton Research (Riverside, CA), and crystals where obtained from 0.1 M Mes (pH 6.5), 12% PEG 20,000. Two-chain ΔCARD caspase 9 inhibited with Z-EVD-Dcbmk crystallized under the same conditions and diffracted to 2.8 Å. The crystal symmetry and the unit cell dimensions where identical to those of the first crystal. Molecular replacement was performed with AMoRe (25) and caspase 3 as search model, map interpretation was done in MAIN (26), data processing and scaling were done in DENZO/SCALEPACK (27), and crystallographic refinement (simulated annealing, positional- and temperature-factor refinement) in Crystallography & NMR System (28). Noncrystallographic symmetry restraints were used between the two dimers in the asymmetric unit, but not between the catalytic in each dimer. In the Ramachandran plot of the 2.8-Å structure final model 90.2% of the ψ/φ angles fall in the core region and 9.8% in the additional allowed region.
Results and Discussion
Caspase 9 Conformation.
Caspases are initially single-chain proteins that can be processed to large and small chains (subunits). So far, only crystal structures of processed caspases have been reported, composed of two catalytic domains, each comprising a large and a small subunit (14–20). A catalytic domain consists of one large and one small subunit (whether processed or unprocessed), and we refer to this as a monomer. We also use the caspase-1 numbering convention throughout (17, 20).
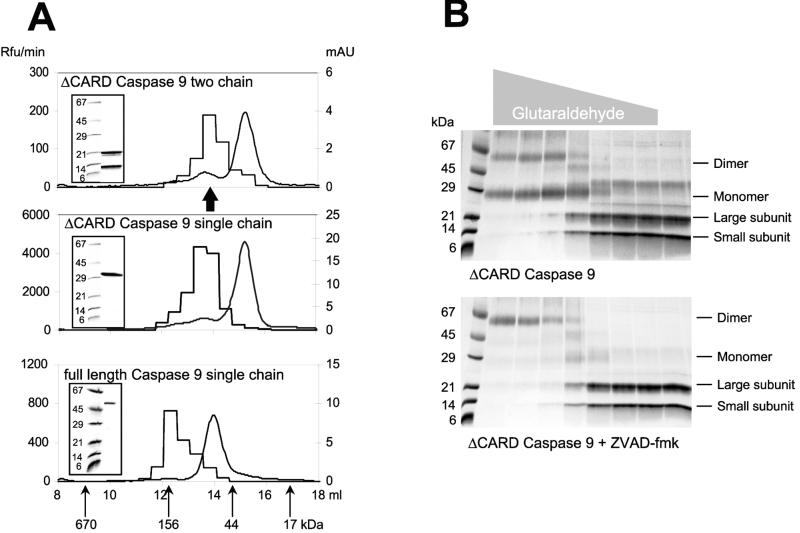
We produced full-length and CARD-domain-deleted forms of caspase 9 in E. coli (5), and during production both forms undergo autolytic processing. Processing indicates that both are active when expressed because of the very high concentrations achieved during expression and purification, a common property of caspases (29). To prevent processing we engineered mutants in which all of the potential cleavage sites in the interdomain linker are replaced by Ala. We assessed the conformation of ΔCARD caspase 9 (two chain), ΔCARD caspase 9 (single chain), and full length caspase 9 (single chain) by gel filtration (Fig. 1A). In each case the majority of protein eluted with a size equivalent to a single catalytic domain, irrespective of the presence of the CARD domain or processing status. Gel filtration demonstrates that the primary forms in solution are monomers at the concentrations used (1.2 μM), implying that the Kd is in the high micromolar range (Fig. 1). The monomers are inactive and catalytic activity requires dimerization. Chemical crosslinking of processed ΔCARD caspase 9 confirmed its monomeric status but, importantly, dimerization was favored when the protein was inactivated by the covalently reactive substrate analog Z-VAD-FMK (Fig. 1B). This is most likely because of trapping of the catalytic conformation of a transient dimer. In support of this possibility, Z-VAD-caspase 9 migrated as a dimer in gel filtration (Fig. 1A). We completely inhibited ΔCARD caspase 9 with N-terminal unblocked VAD-FMK, and sequenced the product by Edman degradation. Because the custom derivative contains a free amine, its complex with caspase 9 could be quantitatively sequenced to yield the absolute recovery of anilinothiazolinone derivatives of inhibitor and caspase in the first two cycles. Interestingly, this quantitation revealed a fractional yield of 0.48 (±0.05) for the VAD-FMK compared with the N terminus of ΔCARD caspase 9. Thus, in fully inhibited caspase 9 only ≈50% of the catalytic domains are labeled. Because VAD-FMK is a mechanism-based inactivator, requiring functional catalytic machinery for binding, caspase 9 must have only half of its potentially available catalytic sites in an active conformation.
Figure 1.
Identification of the active form of caspase 9. (A) Gel filtration of caspase 9 on a Amersham Pharmacia Superdex 200 column reveals that most of the protein for each form (smooth curge; right ordinate, mAU = milli-absorbance units at 280 nm) eluted as a monomer (≈35 kDa for the ΔCARD forms and ≈48 kDa for the full-length form), but the majority of activity against the substrate Ac-LEHD-AFC (step curve; left ordinate, Rfu = relative fluorescence units) is present in the fraction corresponding to dimers (≈70 kDa for the ΔCARD forms and ≈100 kDa for the full length form). Insets demonstrate the integrity of the purified recombinant proteins run in SDS/PAGE before gel filtration. The arrow marks the position of the protein peak for two chain caspase 9 inhibited by Z-VAD-FMK. (B) Crosslinking of ΔCARD caspase 9 by glutaraldehyde in the absence (Upper) or presence (Lower) of 1.0 μM Z-VAD-FMK. The gray triangle above the gels indicates the glutaraldehyde concentration (ranging from 500 to 0.032 mM). The majority of caspase 9 material is a monomer, and inhibitor binding forces the dimeric form. Thus, it appears that the inhibitor either drives or traps the dimeric form of caspase 9, indicating that the dimer is the active form of the enzyme.
These data indicate that caspase 9 is an inactive monomer at concentrations in the micromolar range, and that activity requires dimer formation. Moreover, we determined that full-length caspase 9 in vivo is also primarily a monomer, because gel filtration of cytosolic extracts from human embryonic kidney cells revealed a size of 45–50 kDa for the endogenous protein (not shown), consistent with the size for the natural protein observed previously (30).
Caspase 9 Structure.
To shed light on the structural basis of dimer-induced activation, we determined the crystal structure. Crystals obtained from two-chain caspase 9 diffracted to 3.5 Å, but the protein lacked the first 138 residues due to proteolytic removal of the CARD, most likely caused by a contaminating E. coli protease. We set up crystal trials with two-chain ΔCARD caspase 9, and determined the structure with a tripeptidyl inhibitor at a resolution of 2.8 Å (Table 1). The refined structures are nearly identical, with differences attributable to resolution, some terminal residues absent in the low-resolution structure, and the presence of the inhibitor. We describe the inhibitor-bound form because data were obtained at higher resolution.
Table 1.
Crystallographic data and refinement for ΔCARD caspase 9
| No inhibitor | Plus inhibitor | |
|---|---|---|
| Space group | C2 | C2 |
| Temperature, K | 100 | 100 |
| Beamline | NSLS X25 | SLAC 7–1 |
| Cell constants, Å | ||
| a | 144.3 | 143.8 |
| b | 81.5 | 81.8 |
| c | 125.2 | 125.4 |
| β, degrees | 111.8 | 111.8 |
| Measurements (total/unique) | 40,602/19,681 | 112,744/33,394 |
| Resolution limit, Å | 3.25 | 2.80 |
| Completeness (outermost shell), % | 88.2 (55.5) | 99.4 (93.4) |
| Rmerge (outermost shell), % | 8.2 (28.9) | 10.3 (28.3) |
| Number of atoms per asymmetric unit (occupancy > 0.00) | ||
| Non-hydrogen protein atoms | 6,570 | 7,084 |
| Solvent molecules | 0 | 274 |
| Inhibitor atoms | 0 | 45 |
| Resolution range, Å | 20–3.25 | 20–2.8 |
| Reflections used for refinement (working/test set) | 17,675/899 | 31,628/1,650 |
| R/Rfree, % | 22.1/28.7 | 23.3/27.6 |
| Root mean square standard deviation | ||
| Bond length, Å | 0.009 | 0.007 |
| Bond angles, degrees | 1.36 | 1.40 |
There are four catalytic domains and, in the case of the inhibitor bound structure, two inhibitor molecules per asymmetric unit. NSLS, National Synchrotron Light Source; SLAC, Stanford Linear Accellerator Center.
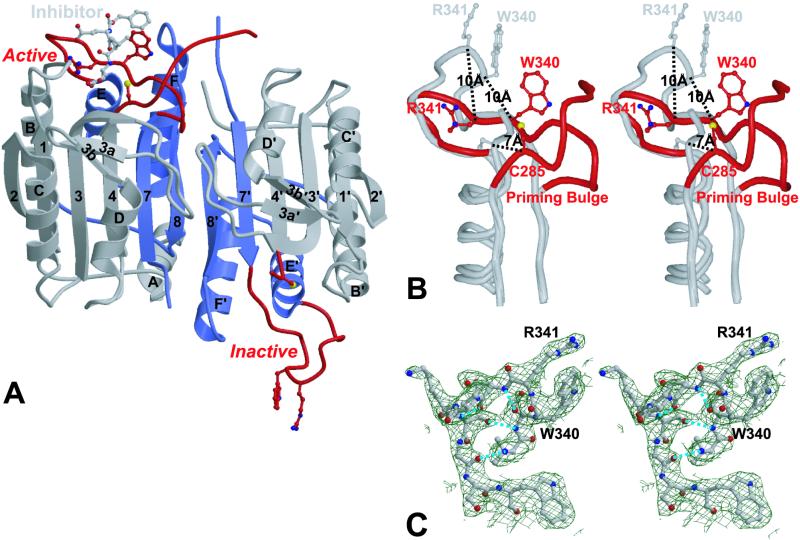
The crystals contain four catalytic domains, which comprise two caspase dimers, in the asymmetric unit. In common with all known caspase structures, each catalytic domain is composed of a large and a small subunit (Fig. 2), and a dimer is assembled by extending the central β-sheet. In the caspase structures published to date both catalytic domains are identical, and related by twofold symmetry. In contrast, the crystal structure of caspase 9 has captured two conformations of the catalytic domain, with one domain in a catalytically competent conformation, and the other catalytically incompetent. This explains the ≈50% labeling by VAD-FMK described above. Significantly, the structure is that of a caspase dimer. Although caspase 9 in solution is predominantly a monomer, the high protein concentrations used for crystal growth (approximately millimolar) forces dimerization.
Figure 2.
Schematic representation of ΔCARD caspase 9. (A) Caspase 9 is composed of two domains; the large subunit of each domain is colored gray and the small subunit is colored blue. The bound inhibitor molecule (gray), the side chains of the catalytic residue Cys-285 and the specificity determining residues Arg-341 and Trp-340 are in the highlighted ball and stick. Large deviations are seen in the environment of the catalytic and specificity-determining residues of the two domains (red). In the Left (active) domain, these residues adopt the conformations seen in other caspases, enabling binding of the inhibitor. In the Right (inactive) domain, the same residues are transposed from their catalytic conformation into a novel structure incapable of catalysis. The figure was made with MOLSCRIPT and rendered in RASTER3D, and secondary structure elements are assigned as for caspase 1 (17). (B) Stereoview of the activation loop. The region surrounding the activation loop in the active domain is superimposed on the equivalent region of the inactive domain. In the inactive domain (gray), displacement of the essential specificity determinant Arg341 disrupts the S1 and the S3 subsites; displacement of Trp-340 and Val-338 disrupts the S2 subsite; the backbone of Ser-339 and Arg-341 cannot form the short antiparallel β-strand with the substrate required for its proper alignment. Changes to the catalytic apparatus are equally substantial. Residues that provide hydrogen bonds to stabilize the oxyanion hole are perturbed, and the region at the terminus of the large subunit, which follows the catalytic Cys, are disordered and not defined by electron density. Transition to the active form (red) requires outward movement in the “priming bulge” to compensate the inward movement of Trp-340 and Arg-341. In the active domain residues after the catalytic Cys are well ordered up to Val-296. (C) Stereoview of the electron-density map defining the activation loop in the inactive domain. The loop is well defined by electron density, with an identical conformation in each copy of the inactive domain in the asymmetric unit. Hydrogen bonds are indicated by dotted lines.
The Active Catalytic Domain.
The fold of the active domain is very similar to other caspases. By using a distance cutoff of 1.5 Å, 201 identical Cα positions were identified with an rms deviation of 0.70 Å between caspase 8 (PDB ID code 1QTN; refs. 14 and 18) and capase 9 . The active site itself is almost identical to that of other caspases. Thus, an rms deviation of only 0.17 Å is observed when the active site residues Cys-285, His-237, Trp-340, Arg-341, and Arg-179 are compared between caspase 8 and caspase 9. The S1 pocket is well formed to accept the substrate Asp side chain, making interactions with basic and polar residues characteristic of other known caspase/inhibitor complexes (14–20). Surface loops around the active site of caspases define their specificity for substrates and inhibitors. In caspase 9, these loops are the shortest among all known caspases, revealing a wider substrate-binding cleft, consistent with its broader substrate specificity (31).
The Inactive Catalytic Domain.
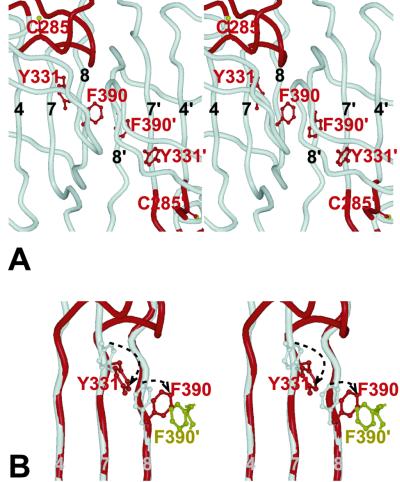
The overall caspase fold is preserved (Fig. 2), but large structural differences are evident in the region of the catalytic site, rendering it inactive. The rearrangements in the small subunit are the most severe, with a displacement of 17 residues that would normally line the substrate binding cleft (the “activation loop”) by as much as 10 Å (Fig. 2B). As a consequence of these rearrangements, substrate recognition is impossible, the active-site Cys is dislocated (Sγ shifts by 7 Å), and the oxyanion hole is annihilated. The activation loop is well structured (Fig. 2C), with internal hydrogen bonds that help to stabilize its conformation. Why are the active and inactive domains not equivalent? We modeled a dimer of two active catalytic domains by superimposition, but found it to create a severe steric clash (Fig. 3). Phe-390 would collide with the symmetry-related residue Phe-390′, and reorientation is necessary to reduce the conflict. Retaining an inactive catalytic domain in the dimeric unit satisfies the conflict.
Figure 3.
The dimer interface. This view focuses on interactions that influence dimer formation. (A) Stereoview as in Fig. 2A showing the side chains of Phe-390 and Tyr-331 and the catalytic Cys-285 in the active (Left) and inactive (Right) catalytic domains, within the dimeric structure. (B) Stereoview of a superposition of the inactive monomer (gray) onto the active one (red). Rotation of Phe-390 about Cα–Cβ occurs in the transition from the inactive to the active conformation, allowing a compensatory rotation of Tyr-331 around its Cα–Cβ. This in turn may help to promote the catalytic conformation of Cys-285. The yellow side-chain represents the position of Phe-390′ in a hypothetical dimer made from two active catalytic domains. Note that it would clash with the active conformation of Phe-390, eliminating the possibility of having two active monomers in this dimer.
The Dimer Interface.
As with other caspases, the dimer is mainly formed by interactions between residues of strand 8 in the small subunits (Fig. 2A). However, there are two distinct features of the interface unique to caspase 9. First, a small β-sheet observed in other caspases between C-terminal residues of the large subunit and the N terminus of the small subunit are absent because of disorder at the C terminus of the inactive domain large subunit. Second, the large subunit of caspase 9 possesses a seven-residue loop inserted at position 240 between strands 3 and 3A that interacts across the dimer interface (Figs. 2A and 3A). It is difficult to discern whether these properties could explain the weak dimer equilibrium. Caspases 1 and 3 are dimers at concentrations below 30 nM (ref. 32; and M.R., unpublished data), whereas caspase 9 remains a monomer above 1 μM. Nevertheless, because inhibitor binding stabilizes the dimer allostericaly (Fig. 1), caspase 9 may be more flexible than other caspase structures and hence a less stable dimer in the inhibitor-free form.
The asymmetry of the dimer thus appears to be an intrinsic property of caspase 9, and is consistent with the half-of-sites reactivity of the inhibitor described above. Furthermore, the details of the conformation of the activation loop in the inactive domain are not affected either by crystal contacts or by the presence of inhibitor in the active domain, because the conformation is identical in the two copies in the asymmetric units of both the uninhibited and inhibited forms. Importantly, the loop probably has a similar conformation in monomeric caspase 9. Limited proteolysis identifies this loop to be unusually protease-sensitive (not shown), which would be favored if the activation loop was in an exposed, rather than in a buried, conformation in solution.
The Mechanism of Caspase 9 Activation.
In contrast to other caspases, proteolysis is not sufficient for the activation of caspase 9 (4, 5). Not only is proteolysis insufficient for caspase 9 activation, it is also unnecessary. The kcat and Km values for cleavage of Ac-LEHD-AFC by the enzymatically active portion of single-chain ΔCARD (4.7 s−1, 185 μM) and two-chain ΔCARD caspase 9 (4.8 s−1, 248 μM) (data not shown) indicate that the structure around the catalytic centers is similar. Moreover, we have demonstrated that these species are predominantly monomeric, and the proteolytic activity is associated with a dimer (Fig. 1). It does not seem to matter whether caspase 9 is cleaved because uncleavable zymogen is able to replace the wild type (5, 21). There is, however, at least one structural difference between single- and two-chain caspase 9, because cleavage is required for interactions with endogenous inhibitors subsequent to the activation event (21, 33).
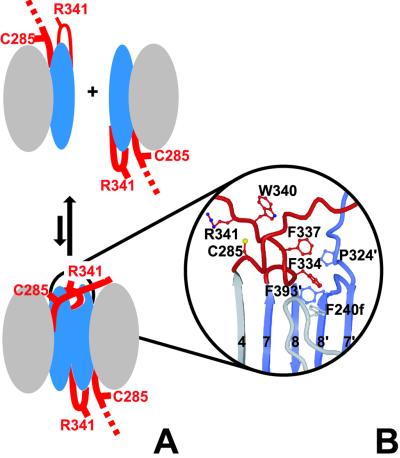
Because proteolysis is not sufficient for activation of caspase 9, there must exist another mechanism to achieve the catalytically competent form. Based on our findings, we propose that caspase 9 is activated on dimerization by a “self priming” mechanism outlined in Fig. 4. In this model, the catalytic machinery of the caspase 9 monomer is in an inactive conformation, presumably like that of the inactive catalytic unit in the dimer (Fig. 2A). Two inactive monomers associate, and a hydrophobic pocket at the dimer interface accepts the side chains of the priming bulge, allowing the activation loop to insert. This motion transmits the activation signal to residues surrounding the catalytic Cys-285, and the catalytic machinery is aligned. Interestingly, it appears that caspase 9 has half of the catalytic sites of other caspases that contain two active sites per catalytic dimer. Although our structural and biochemical data are quite clear about this, the importance of the half-of-site reactivity in vivo requires investigation.
Figure 4.
The mechanism of zymogen activation. Caspase 9 exists as a monomer at physiologic concentrations, with an exposed activation loop that renders the enzyme latent. (A) During dimerization, the activation loop (red) of the left domain is drawn into a pocket on the right domain. (B) The hydrophobic pocket, bordered by Pro-324, Phe-240f, and Phe-393 in the right domain, accepts Phe-334 and Phe-337 from the left domain. This locks into place the priming bulge (Ser-330–Ser-339) of the activation loop, enabling Trp-340 and Arg-341 to sink into their substrate-binding conformation, and simultaneously allowing hydrogen bonding to the segment following Cys-285. This transition generates catalytic potential in the left domain.
Although we could not crystallize full-length caspase 9, the structures of the individual CARD (34, 35) and catalytic domain are now solved. There are no structural data for 40 amino acids between these domains. Results from limited proteolysis studies of full-length caspase 9 (data not shown), and the fact that the CARD domain was removed by an E. coli protease, indicate that this linker region is flexible. How these individual folding domains exist in relation to each other in full-length caspase 9 remains unknown. Regardless, we expect a comparable process for the activation of full-length caspase 9. Single-chain full-length caspase 9, like single-chain ΔCARD caspase 9, exists predominantly as an inactive monomer with the proteolytic activity being associated with a higher molecular weight species (Fig. 1). This indicates that self-priming by means of dimerization is also required for activation of full-length caspase 9.
The activation of caspase 9 by self-priming appears to be a novel strategy for protease activation, and furnishes a satisfying molecular explanation to the mechanism of apoptotic initiation. In its natural setting in human cells, the concentration of procaspase 9 is in the 20 nM range (5), well below the dimerization threshold. This barrier to dimerization is overcome by homophilic interactions between the CARD domains of caspase 9 and Apaf-1 (34, 35), driven by cytochrome c and ATP, to generate the fully functional apoptosome (1). The apoptosome is an oligomeric assembly that recruits several units of caspase 9 and Apaf-1 (30, 36). Most likely, oligomerization of the apoptosome results in caspase 9 dimerization by increasing the local concentration above the Kd. Alternatively, Apaf-1 generates the active conformation in monomeric caspase 9, by providing a complementary surface that simulates the dimerization interface.
In some ways the activation of caspase 9 resembles the activation of members of the trypsin family of serine proteases. Trypsinogen, for example, contains an activation domain with dislocated catalytic machinery and disordered substrate binding sites. Trypsin attains its active conformation after limited proteolysis at Arg-15, and relatively minor shifts in the N terminus lead to huge enhancements in catalysis (6). Interestingly, tight binding trypsin inhibitors can cause a transition of trypsinogen to a trypsin-like state (11), which would be analogous to the inhibitor-driven dimerization of caspase 9 seen in Fig. 1. Perhaps a stronger analogy between caspase 9 and the trypsin family would be coagulation factor VII, which in its processed form has little activity, and which is activated several hundredfold by its cofactor (tissue factor) (13). Notwithstanding these superficial similarities, caspase 9 appears to be the first example of a protease zymogen activation by dimerization, and the first example of a protease with half site occupancy, at least in vitro.
Because caspase 9 is the apical protease of the intrinsic apoptosis pathway its mechanism of activation in vivo cannot be cleavage by an upstream protease. Instead, we propose that activation of caspase 9 in cells is cofactor driven by the mechanism described above. Once the active form has been generated, the zymogens downstream of caspase 9 can be activated by proteolysis. Interestingly, adapter-driven dimerization leading to activation of caspase 9 offers a plausible explanation for the induced proximity hypothesis postulated to explain the activation of apical caspases, including caspase 8 and the worm homolog CED-3 (37). It is likely that cofactor-mediated dimerization represents the primordial method designed to engage the proteolytic pathways of apoptosis that exist in animals from worms and flies to humans (38).
Acknowledgments
We thank John Reed and Erkki Ruoslahti for helpful suggestions. This work was supported by National Institutes of Health Grant NS37878.
Abbreviations
- Z
benzoxycarbonyl
- AFC
7-amino-4-trifluoromethyl coumarin
- FMK
fluoromethyl ketone
- Dcbmk
dichlorobenzyl methylketone
Footnotes
Data deposition: The caspase 9 coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1JXQ).
References
- 1.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 2.Hakem R, Hakem A, Duncan G S, Henderson J T, Woo M, Soengas M S, Elia A, de la Pompa J L, Kagi D, Khoo W, et al. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- 3.Kuida K, Haydar T F, Kuan C Y, Gu Y, Taya C, Karasuyama H, Su M S, Rakic P, Flavell R A. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez J, Lazebnik Y. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stennicke H R, Deveraux Q L, Humke E W, Reed J C, Dixit V M, Salvesen G S. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 6.Khan A R, James M N. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parry M A, Zhang X C, Bode I. Trends Biochem Sci. 2000;25:53–59. doi: 10.1016/s0968-0004(99)01521-2. [DOI] [PubMed] [Google Scholar]
- 8.Stubbs M T, Renatus M, Bode W. Biol Chem. 1998;379:95–103. doi: 10.1515/bchm.1998.379.2.95. [DOI] [PubMed] [Google Scholar]
- 9.Turk B, Turk D, Turk V. Biochim Biophys Acta. 2000;1477:98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 10.Nagase H, Woessner J F J. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 11.Bode W, Schwager P, Huber R. J Mol Biol. 1978;118:99–112. doi: 10.1016/0022-2836(78)90246-2. [DOI] [PubMed] [Google Scholar]
- 12.Stubbs M T, Bode W. Trends Biochem Sci. 1995;20:23–28. doi: 10.1016/s0968-0004(00)88945-8. [DOI] [PubMed] [Google Scholar]
- 13.Ruf W, Dickinson C D. Trends Cardiovasc Med. 1998;8:350–356. doi: 10.1016/s1050-1738(98)00031-0. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard H, Kodandapani L, Mittl P R E, Di Marco S, Krebs J F, Wu J C, Tomaselli K J, Grütter M G. Structure (London) 1999;27:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 15.Mittl P R, Di Marco S, Krebs J F, Bai X, Karanewsky D S, Priestle J P, Tomaselli K J, Grutter M G. J Biol Chem. 1997;272:6539–6547. doi: 10.1074/jbc.272.10.6539. [DOI] [PubMed] [Google Scholar]
- 16.Rotonda J, Nicholson D W, Fazil K M, Gallant M, Gareau Y, Labelle M, Peterson E P, Rasper D M, Tuel R, Vaillancourt J P, Thornberry N A, Becher J W. Nat Struct Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 17.Walker N P C, Talanian R V, Brady K D, Dang L C, Bump N J, Ferenz C R, Franklin S, Ghayur T, Hackett M C, Hammill L D, et al. Cell. 1994;78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 18.Watt W, Koeplinger K A, Mildner A M, Heinrikson R L, Tomasselli G, Watenpaugh K D. Structure (London) 1999;27:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- 19.Wei Y, Fox T, Chambers S P, Sintchak J, Coll J T, Golec J M, Swenson L, Wilson K P, Charifson P S. Chem Biol. 2000;7:423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 20.Wilson K P, Black J A, Thomson J A, Kim E E, Griffith J P, Navia M A, Murcko M A, Chambers S P, Aldape R A, Raybuck S A, Livingston D J. Nature (London) 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasula S M, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee R A, Robbins P D, Fernandes-Alnemri T, Shi Y, Alnemri E S. Nature (London) 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 22.Stennicke H R, Salvesen G S. Methods Enzymol. 2000;322:91–100. doi: 10.1016/s0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- 23.Bury A. J Chromatog. 1981;213:491–500. [Google Scholar]
- 24.Matsudaira P. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 25.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 26.Turk D. Ph.D. thesis. Munich: Technische Universität; 1992. [Google Scholar]
- 27.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Brunger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges N, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 30.Cain K, Brown D G, Langlais C, Cohen G M. J Biol Chem. 1999;274:22686–22692. doi: 10.1074/jbc.274.32.22686. [DOI] [PubMed] [Google Scholar]
- 31.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 32.Talanian R V, Dang L C, Ferenz C R, Hackett M C, Mankovich J A, Welch J P, Wong W W, Brady K D. J Biol Chem. 1996;271:21853–21858. doi: 10.1074/jbc.271.36.21853. [DOI] [PubMed] [Google Scholar]
- 33.Ekert P, Silke J, Hawkins C, Verhagen A, Vaux D. J Cell Biol. 2001;152:483–490. doi: 10.1083/jcb.152.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin H, Srinivasula S M, Wu G, Fernandes-Alnemri T, Alnemri E S, Shi Y. Nature (London) 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 35.Zhou P, Chou J, Olea R S, Yuan J, Wagner G. Proc Natl Acad Sci USA. 1999;96:11265–11270. doi: 10.1073/pnas.96.20.11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou H, Li Y, Liu X, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 37.Salvesen G S, Dixit V M. Proc Natl Acad Sci USA. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier P, Finch A, Evan G. Nature (London) 2000;407:796–801. doi: 10.1038/35037734. [DOI] [PubMed] [Google Scholar]