Abstract
The development of PPARα/γ dual or PPARα/γ/δ pan-agonists could represent an efficacious approach for a simultaneous pharmacological intervention on carbohydrate and lipid metabolism. Two series of new phenyldiazenyl fibrate derivatives of GL479, a previously reported PPARα/γ dual agonist, were synthesized and tested. Compound 12a was identified as a PPAR pan-agonist with moderate and balanced activity on the three PPAR isoforms (α, γ, δ). Moreover, docking experiments showed that 12a adopts a different binding mode in PPARγ compared to PPARα or PPARδ, providing a structural basis for further structure-guided design of PPAR pan-agonists. The beneficial effects of 12a were evaluated both in vitro, on the expression of PPAR target key metabolic genes, and ex vivo in two rat tissue inflammatory models. The obtained results allow considering this compound as an interesting lead for the development of a new class of PPAR pan-agonists endowed with an activation profile exploitable for therapy of metabolic syndrome.
Keywords: PPARs, pan-agonist, docking, metabolic syndrome, type 2 diabetes mellitus, gene expression, PGE2
Peroxisome proliferator-activated receptors (PPARs) are members of the nuclear receptor superfamily and play a decisive role in the regulation of lipid metabolism and glucose homeostasis. PPARs exist as three subtypes with different localizations and physiological functions, designated as PPARα, PPARγ, and PPARδ.1 These receptors are targets for the therapy of metabolic syndrome, a group of risk factors for cardiovascular disease and type 2 diabetes mellitus (T2DM) including insulin resistance, elevated fasting blood glucose, hypertension, obesity, and atherogenic dyslipidemia.1 Furthermore, metabolic syndrome is accompanied by a low-grade inflammatory state in white adipose tissue and liver, which may exacerbate insulin resistance and diabetes.2 Fibrates (e.g., fenofibrate and gemfibrozil), a class of lipid-lowering drugs, are PPARα ligands.3 Thiazolidinediones (TZDs), such as rosiglitazone and pioglitazone, are insulin-sensitizing agents that activate PPARγ.4 PPARδ agonists have not yet reached the market, but some of them, for example, GW501516, GW0741, L165,041, GFT505, and MBX8025, are being investigated for a possible therapeutic usefulness in the treatment of metabolic disorders, inflammation, and angiogenesis (Figure S1).5 However, due to the adverse effects associated with these drug treatments,3,4 new research strategies have been undertaken with the aim to obtain new PPAR ligands with reduced side effects and improved beneficial effects. One of these strategies consists in the synthesis of PPARα/γ dual or PPARα/γ/δ pan-agonists, which beneficially alter carbohydrate and lipid metabolism in a coordinated manner.6−8 In particular, pan-agonists would merge the agonist activities with regard to PPARα, -γ, and -δ in a unique ligand but with a balanced activation profile, which could be useful for treatment of metabolic syndrome and T2DM. In the search for new PPAR ligands with these properties, we have recently discovered a series of derivatives of the antilipidemic drugs gemfibrozil or clofibric acid, in which the clofibric acid skeleton was combined with the lipophilic scaffolds derived from natural products, such as α-asarone, stilbene, chalcone, and their bioisosteres.9−12 In particular, the combination of the clofibric acid with a phenyldiazenyl function resulted in GL479 (R = H, X = O, Y = CH2; Figure 1), a PPARα/γ dual agonist.13 Cocrystal structures of GL479 with PPARα and -γ revealed that the ligand adopts different binding modes at these receptor subtypes, reflecting the distinct activation profile observed for each receptor.14
Figure 1.
General features of synthetic PPAR agonists and structural development of the presented compounds.
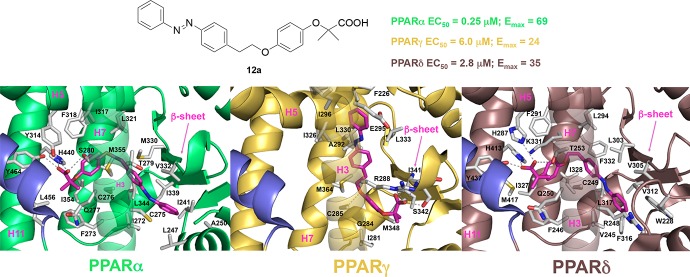
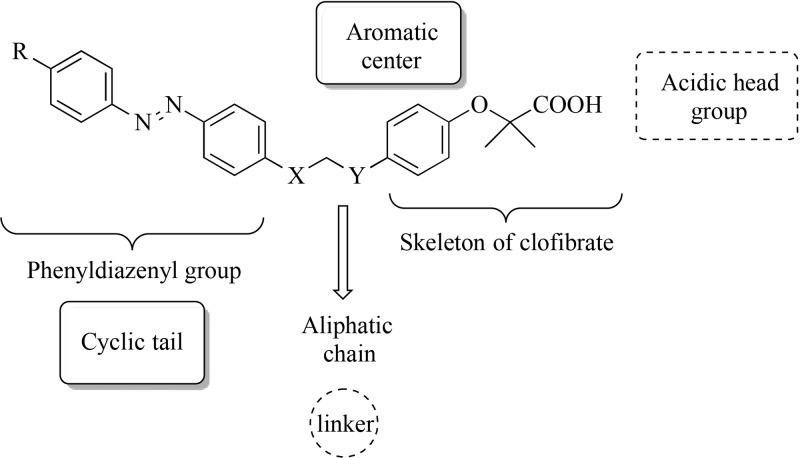
Herein we wish to report the synthesis and biological evaluation of a series of derivatives of GL479 (compound 11a, Table 1) in which (i) small substituents with different stereochemical properties were inserted at the para position of the distal aromatic ring of phenyldiazenyl moiety (compounds 11b–g) and (ii) the oxygen atom of the linker was moved to the para position of the phenoxy-propanoic acid (compounds 12a–f). Such design strategy was planned taking into account the crystallographic pose of GL479 into both PPARα and -γ: the hydrophobic tail binds upward into arm II of PPARα, where it explores a relatively large accessible volume. Instead, this group bends downward into arm III of PPARγ, surrounded by hydrophobic residues such as I388, I325, M329, and F266. Therefore, with the aim of probing further binding interactions in this region of the protein, we further explored the introduction of small substituents at the para position of the phenyldiazenyl group. Furthermore, C285 on H3, which assumes two possible conformations reflecting a certain degree of flexibility, and the oxygen in the linker of GL479 are at a favorable distance for H-bonding. This cysteine residue is also conserved in PPARα. Thus, in the attempt to promote possible interactions with the cysteine residues on H3, we varied the position of the oxygen atom on the linker.
Table 1. In Vitro Transactivation Activity of Compounds with Various Lipophilic Tails and Linkers.
| hPPARα | hPPARγ | hPPARδ | ||||
|---|---|---|---|---|---|---|
| compd | R | X | Y | EC50 (μM)a (Emax)b | EC50 (μM)a (Emax)b | EC50 (μM)a (Emax)b |
| 11a (GL479) | H | O | CH2 | 2.1 ± 0.6 (56±9) | n.d. (13 ± 1) | 6.4 ± 1.2 (39 ± 4) |
| 11b | Cl | O | CH2 | 0.38 ± 0.05 (60 ± 6) | n.d. (12 ± 1) | 3.1 ± 0.4 (23 ± 2) |
| 11c | Br | O | CH2 | 0.81 ± 0.06 (71 ± 10) | n.d. (12 ± 3) | 3.9 ± 0.4 (29 ± 2) |
| 11d | CF3 | O | CH2 | 2.3 ± 1.3 (35 ± 8) | n.d. (8 ± 1) | 1.8 ± 0.4 (24 ± 3) |
| 11e | CN | O | CH2 | 2.6 ± 0.8 (25 ± 7) | n.d. (12 ± 1) | 6.5 ± 1.2 (20 ± 4) |
| 11f | NO2 | O | CH2 | n.d. (11 ± 7) | n.d. (14 ± 1) | 4.1 ± 1.0 (21 ± 3) |
| 11g | OCH3 | O | CH2 | 2.4 ± 0.8 (32 ± 2) | n.d. (10 ± 1) | 1.4 ± 0.2 (23 ± 2) |
| 12a | H | CH2 | O | 0.25 ± 0.07 (69 ± 12) | 6.0 ± 1.5 (24 ± 3) | 2.8 ± 0.3 (35 ± 2) |
| 12b | Cl | CH2 | O | 0.51 ± 0.12 (52 ± 9) | n.d. (10 ± 1) | 1.2 ± 0.3 (41 ± 6) |
| 12c | Br | CH2 | O | 1.9 ± 0.2 (35 ± 4) | n.d. (8 ± 1) | 1.1 ± 0.2 (29 ± 3) |
| 12d | CF3 | CH2 | O | 2.2 ± 1.2 (37 ± 7) | n.d. (9 ± 1) | n.d. (12 ± 4) |
| 12e | CN | CH2 | O | 1.4 ± 0.1 (61 ± 11) | n.d. (12 ± 2) | 2.8 ± 0.3 (50 ± 3) |
| 12f | NO2 | CH2 | O | n.d. (19 ± 8) | n.d. (10 ± 1) | 2.3 ± 0.4 (28 ± 3) |
| clofibric acid | 50 ± 6 (100 ± 10) | - | - | |||
| rosiglitazone | - | 0.04 ± 0.02 (100 ± 9) | - | |||
| L165,041 | - | - | 0.21 ± 0.04 (100 ± 4) |
EC50 values were determined by testing compounds in at least three separate experiments at five concentrations ranging from 1 to 50 or 100 μM. The results are expressed as the mean ± SEM.
Efficacy values were calculated as percentage of the maximum fold induction obtained with clofibric acid, rosiglitazone, and L165,041 as reference compounds for PPARα, PPARγ, and PPARδ, respectively.
n.d.: EC50 not determined.
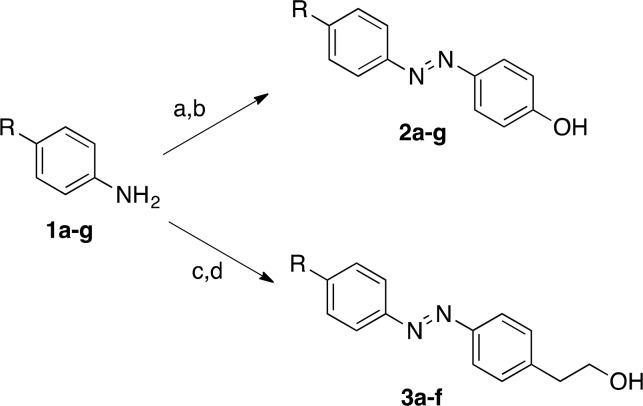
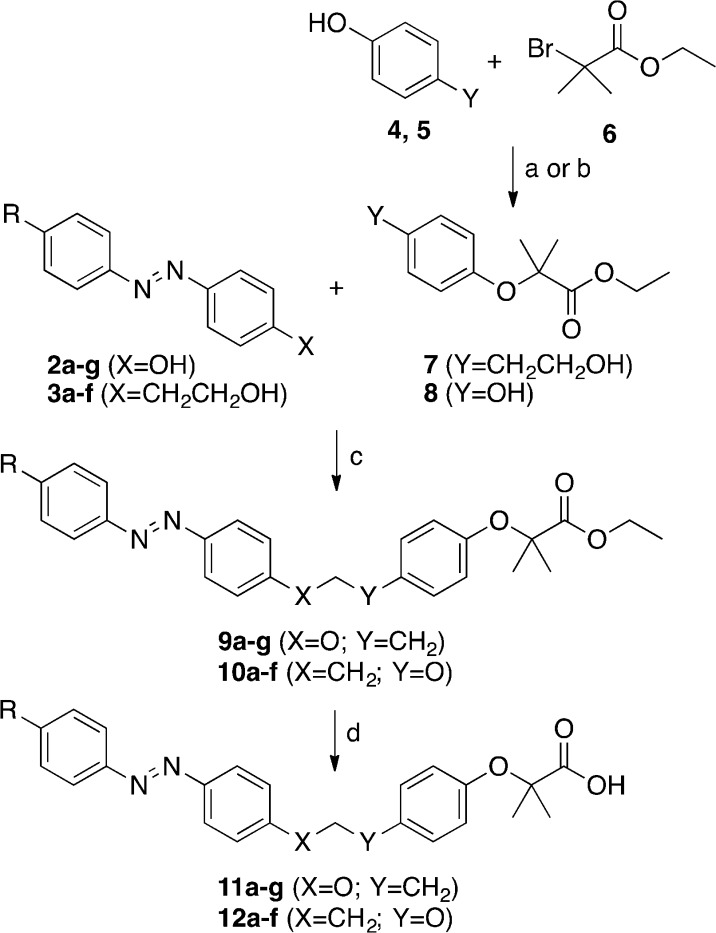
Phenyldiazenyl derivatives 11a–g and 12a–f (Table 1) were easily obtained in good yields using simple synthetic procedures (see Supporting Information for the detailed procedures). The intermediates 2a–g and 3a–f were synthesized according to Scheme 1. Phenols 2a–g were synthesized by diazotization of para-substituted anilines with NaNO2 in 6 N HCl at 0 °C and copulation of the obtained diazonium salts with phenol in 4 N NaOH at 0 °C. The alcohols 3a–f were synthesized by oxidation of para-substituted anilines by oxone in dichloromethane/water at r.t. and subsequent reaction with 2-(4-aminophenyl)ethanol in CH3COOH at r.t. (Scheme 1).
Scheme 1. Synthetic Pathways of Compounds 2a–g and 3a–f.
Reagents and conditions: (a) NaNO2, 6 N HCl, H2O, 0–5 °C, 1 h; (b) phenol, 4 N NaOH, 0–5 °C, 1 h; (c) oxone, CH2Cl2, H2O, r.t., 4 h; (d) 2-(4-aminophenyl)ethanol, CH3COOH, r.t., 24–48 h.
Esters 7 and 8 were obtained by SN2 reaction of phenols 4 or 5 with ethyl 2-bromo-2-methylpropanoate (6), in the presence of dry K2CO3 in DMF at reflux (7) or in the presence of KOH in EtOH at reflux (8) as reported in Scheme 2.
Scheme 2. Synthetic Pathways of Compounds 4–6, 11a–g, and 12a–f.
Reagents and conditions: (a) dry K2CO3, DMF, reflux, 4 h, (Y = CH2CH2OH) (4 → 7); (b) KOH, EtOH, reflux, N2, 72 h, (Y = OH) (5 → 8); (c) PPh3, DIAD, N2, THF dry, r.t., 10–12 h; (d) 1 N NaOH, EtOH, r.t., 15–24 h.
Esters 9a–g and 10a–f were synthesized by Mitsunobu reaction between 2a–g or 3a–f and the appropriate alcohol 7 or phenol 8. These esters were hydrolyzed in basic condition with 1 N NaOH in EtOH at room temperature to obtain the target acids 11a–g and 12a–f (Scheme 2).
All synthesized compounds were evaluated for their agonist activity toward the three subtypes of human PPARs (hPPARs) by a cell-based transactivation assay, according to a previously reported procedure.15 The results were compared with those obtained with clofibric acid (100 μM), rosiglitazone (2 μM), and L-165,041 (2 μM), used as reference compounds for PPARα, -γ, and -δ, respectively (Table 1). Maximum fold induction obtained with the reference agonists was set at 100%. The activity of all compounds was evaluated at 25 μM, and only those showing efficacy percentage (Emax) higher than 20% were selected for determination of EC50. Because of the different cell line used in this assay, GL479 turned out to be a partial agonist instead of a full agonist, as reported earlier;13 moreover, this ligand displayed a significant activation of PPARδ subtype that had not been previously investigated. Derivatives 11b–g displayed partial agonist activity toward PPARα. The introduction of a chlorine or bromine resulted in about 3- and 5-fold increase of potency compared to lead compound GL479 (11b, EC50 = 0.38 ± 0.05 μM; 11c, EC50 = 0.81 ± 0.06 μM; GL479, EC50 = 2.1 ± 0.6 μM) and a slight increase of efficacy, suggesting a favorable influence of the weak electron-withdrawing properties of these substituents on the activity. In contrast, the introduction of stronger electron-withdrawing substituents, such as trifluoromethyl and cyanide, afforded compounds with potency comparable to GL479 (11d, EC50 = 2.3 ± 1.3 μM; 11e, EC50 = 2.6 ± 0.8 μM), but a concomitant loss of efficacy from 56% to about 30%. The presence of the highly hydrophilic nitro group caused an almost complete loss of PPARα activation (11f). Compound 11g, bearing the electron-donating group methoxy, showed a similar activity to the derivative 11d. Therefore, we argued that the electronic properties of the substituent in the para position of the distal aromatic ring of phenydiazenyl moiety do not seem to exert a crucial role on the activity. Surprisingly, compounds 11b–g showed poor PPARγ activity as well as a weak activation of PPARδ. In particular, the substitution with trifluoromethyl or methoxy increased the potency toward this receptor subtype, with 11d and 11g being the most potent.
Derivatives 12a–f displayed partial agonism on PPARα. Compound 12a, a close analogue of GL479, showed about 8-fold potency increase and a concomitant slight increase of Emax (about 70%). Substitutions at the para position of the phenyldiazenyl moiety afforded compounds displaying lower potency and efficacy compared to 12a. The chloro (12b) and cyano (12e) derivatives were the most active. Again, the presence of the hydrophilic and strong electron-withdrawing nitro group caused an almost complete loss of PPARα activation (12f). As the activity is not strikingly affected from the electronic properties of the para-substituent of the distal aromatic ring of phenyldiazenyl moiety and given that the preparation of the derivative bearing the methoxy was somewhat troublesome, this last compound was not synthesized. Derivatives 12b–f displayed very weak activation of PPARγ; only the unsubstituted derivative 12a showed higher potency and efficacy than the lead compound GL479.
Regarding PPARδ, all derivatives 12a–f displayed a partial agonism profile toward this receptor subtype. Introduction of chlorine or bromine increased potency with respect to the unsubstituted derivative 12a, whereas the introduction of trifluoromethyl group was detrimental. The cyano and nitro derivatives displayed a similar potency to 12a. Overall, this second series of derivatives (12a–f) displayed higher potency and a slightly higher efficacy than the corresponding derivatives of the first series (11a–f) toward PPARδ. This suggests that the shift of the oxygen of the linker to the para position of the phenoxy group is more favorable to the interaction and resulting activation of PPARδ. Interestingly, 12a resulted more potent than GL479 but with basically the same significant and well-balanced activity on all three PPAR subtypes. In fact, 12a exhibits a weak/moderate efficacy, which is desirable given that moderately effective PPAR agonists demonstrate good tolerability and safety in a large population of patients.16−18 In addition, the partial agonist activity of 12a allows hypothesizing that this compound behaves like a selective PPAR modulator (SPPARM), which is a ligand that, compared with a full agonist, differentially induces specific receptor effects, ideally uncoupling the benefits of PPAR activation from the adverse side effects.19 Until a few years ago, SPPARM activity was pursued only for PPARγ agonist, but nowadays, it is considered desirable also for the other two subtypes.20 As far as we know, 12a is one of the few pan-agonists showing moderate activity toward all three PPAR subtypes. This is noteworthy if one considers that most pan-agonists introduced and then discontinued in clinical trials (e.g., indeglitazar, etc.)21 were full agonists at least toward one of the three subtypes.
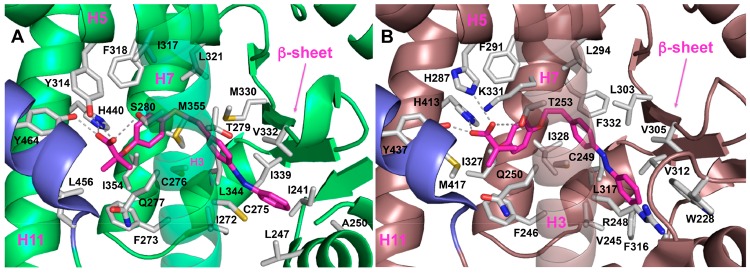
To help interpretation of SAR data and to increase our understanding of the molecular basis for the observed partial agonism of 12a toward PPARα, -γ, and -δ, we undertook docking simulations and cluster analysis (see details in the Supporting Information). Compound 12a occupies arms I and II in both PPARα and -δ (Figure 2), whereas it interacts mainly with arms II and III in PPARγ (Figure S7). As illustrated in Figure 2A,B, the clofibric acid moiety of 12a is located in the arm I, making conventional interactions with the polar side chains (S280, Y314, H440, and Y464 for PPARα and T253, H287, H413, and Y437 for PPARδ), the central phenoxy ring is placed in the center, and the hydrophobic tail part is buried in the arm II. In the PPARα LBD complex, the gem-dimethyl substituents are directed into the lipophilic “benzophenone pocket”, lined by F273, Q277, V444, and L456.22 However, in PPARδ V444 is substituted with a methionine, which has a bulkier side chain and is less prone to accommodate the fibrate headgroup.23 This might account for the potency increase of 12a toward PPARα and for the decrease of PPARδ potency. Noteworthy, the phenyldiazenyl moiety in PPARα forms sulfur–aromatic interactions24 with C275, C276, M355, and M330. This remarkable environment might rationalize the augmented potency of 12a in the PPARα pocket, which is more lipophilic and less solvent-exposed than the corresponding pocket of PPARδ.25
Figure 2.
Binding mode of compound 12a (magenta sticks) into the PPARα (A) and -δ (B) LBDs, represented as green and dirty violet ribbon models, respectively. Only amino acids located within 4.5 Å of the bound ligand are displayed (white sticks) and labeled. H-bonds discussed in the text are depicted as dashed gray lines. H12 is shown in slate.
In fact, with the exception of C276, sulfur-containing residues are not conserved in PPARδ. The 10-fold potency decrease of 11a compared to 12a in PPARα can be explained based on the valence-bond (or resonance) theory (see details in the Supporting Information).
Compound 12a binds to PPARγ LBD in a different binding mode as compared to that of either PPARα LBD or PPARδ LBD because it does not interact directly with H12 (Supporting Information, Figure S7). Therefore, the low affinity and the attenuated transcriptional response of 12a and its derivatives toward PPARγ might be ascribed to the peculiar binding mode, which is not able to replicate the spectrum of contacts of full agonists.26
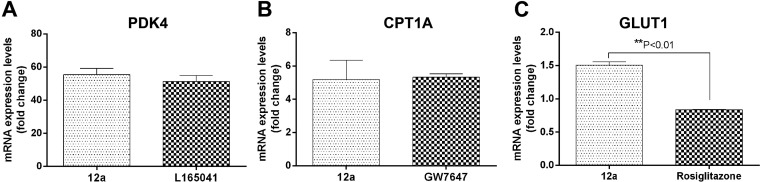
Next, we evaluated whether 12a could affect the expression of three PPAR target genes involved in the control of lipid and carbohydrate metabolism, namely, carnitine palmitoyltransferase 1A (CPT1A), pyruvate dehydrogenase kinase 4 (PDK4), and glucose transporter 1 (GLUT1).27CPT1A is a molecular component of the carnitine shuttle system that catalyzes the entry of fatty acids into the mitochondrial matrix and their subsequent enzymatic oxidation. Importantly, CPT1A gene expression is enhanced by PPARα agonists.27PDK4 is a well-established PPARδ target gene and contributes to the regulation of glucose and fatty acid metabolism and homeostasis.27GLUT1 is a constitutive glucose transporter whose expression is increased by PPARγ ligands, resulting in enhanced glucose uptake and utilization into adipocytes.28 Therefore, a screening of gene activation by real-time quantitative PCR (RTqPCR) analysis was performed for compound 12a in HepG2 human hepatocellular carcinoma cell line, using the PPARδ agonist L165,041, the PPARα agonist GW7647, and the PPARγ agonist rosiglitazone as reference positive controls. As illustrated in Figure 3, treatment with 12a (100 μM) showed enhanced activity on the expression of PDK4 and CPT1A similar to the agonists L165,041 (2 μM) and GW7647 (2 μM, Figure 3A,B). Moreover, 12a significantly increased the mRNA expression level of GLUT1 as compared to rosiglitazone (10 μM, P < 0.01; Figure 3C). The choice of performing the experiment at the fixed concentration of 100 μM was based on the lower potency and/or efficacy of 12a in the transactivation assay compared to the reference compounds as reported in Table 1. However, these results indicate that 12a, even though less potent than positive controls, through its simultaneous and balanced activation of the three PPAR subtypes, may have a beneficial outcome on carbohydrate and lipid metabolism. In light of these results, for a deeper investigation of the biological properties of 12a, we decided to evaluate its anti-inflammatory activity. Numerous studies indicate that the role of PPARs in inflammation is particularly significant in the case of metabolic syndrome and atherosclerosis, which present an inflammatory component.2 This allows hypothesizing that the beneficial effects of PPAR activation on insulin sensitivity are mediated, at least in part, by its anti-inflammatory activities. For this reason, we conducted ex vivo experiments for evaluating the ability of 12a to reduce the LPS-induced PGE2 production in liver and cortex of male adult Sprague–Dawley rats. LPS is often used as a stimulus for the induction of inflammation-related disorders simulating metabolic diseases.29
Figure 3.
Expression of PDK4 (A), CPT1A (B), and GLUT1 (C) after treatment with PPAR agonist 12a (100 μM). The enhanced activity on the gene expression is compared with the commercial compounds L165,041 (2 μM, A), GW7647 (2 μM, B), or rosiglitazone (10 μM, C) as reference positive controls.
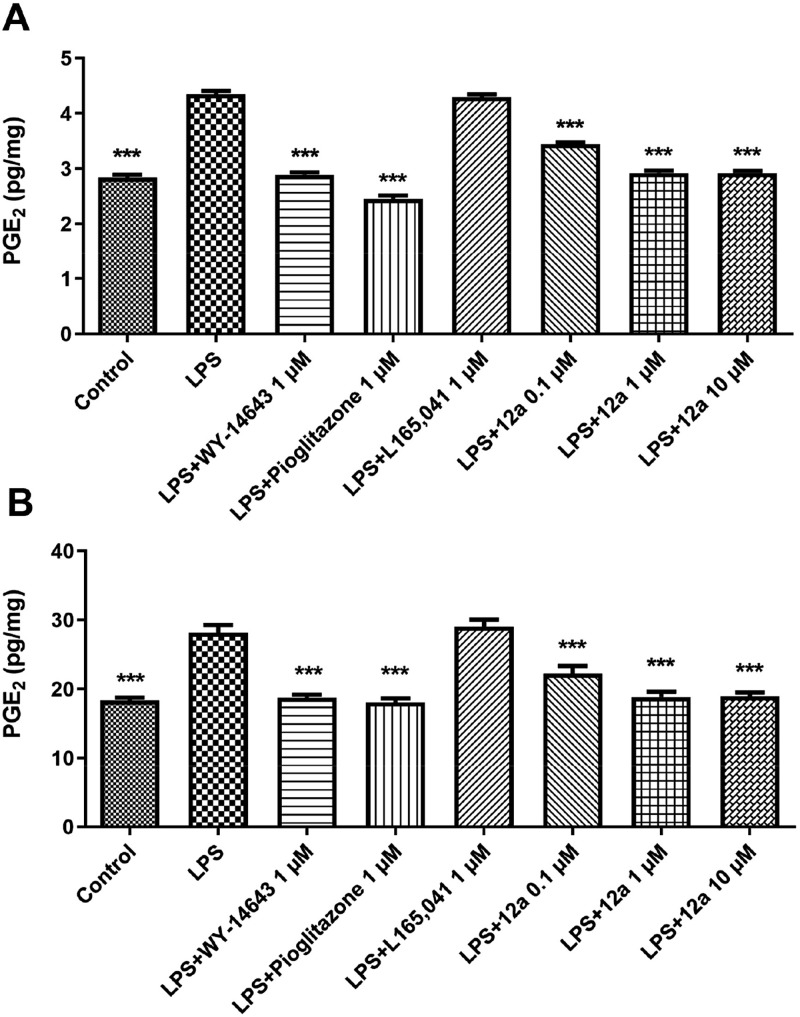
To assess the degree of inflammation, we measured the levels of prostaglandin PGE2, which can contribute to diabetes by inhibiting insulin secretion, leading to impaired glucose tolerance and insulin sensitivity.30 Rats were sacrificed and both liver and cortex specimens were immediately collected and maintained in a humidified incubator in RPMI (Roswell Park Memorial Institute) buffer with added bacterial LPS (10 μg/mL). During the incubation period, tissues were treated alternatively with 1 μM WY14643, pioglitazone, and L165,041 as selective PPARα, -γ, and -δ agonists, respectively, or with scalar concentrations of 12a (0.1–10 μM). Compound 12a was able to significantly inhibit LPS-induced PGE2 production in both rat liver (Figure 4A) and cortex (Figure 4B), two tissues that play a central role in the regulation of glucose homeostasis and metabolism, in a nondose-dependent manner. The efficacy of 12a was comparable with that of WY14643 and pioglitazone, whereas the PPARδ agonist L165,041 did not cause any decrease of the LPS-induced PGE2 production in both tissues.
Figure 4.
Effects of 12a (0.1–10 μM) on LPS-induced production of PGE2 (pg/mg) in rat liver (A) and cortex (B), ex vivo. Data were reported as means ± SEM. Results were analyzed by analysis of variance (ANOVA) followed by Newman–Keuls posthoc test. ANOVA, p < 0.0001; posthoc ***p < 0.001 vs LPS-treated group.
In conclusion, this study led to the identification of 12a endowed with moderate activity on the three PPAR subtypes (α, γ, δ), which might represent a potential lead for the development of a new class of PPAR pan-agonists for treating metabolic syndrome and T2DM.
Acknowledgments
This study was supported by the University of Chieti “G. d’Annunzio” local grants, as well as by the grant "Combattere la resistenza tumorale: piattaforma integrata multidisciplinare per un approccio tecnologico innovativo alle oncoterapie - CAMPANIA ONCOTERAPIE" of Regione Campania (Italy).
Glossary
ABBREVIATIONS
- PPARs
peroxisome proliferator-activated receptors
- T2DM
type 2 diabetes mellitus
- TZDs
thiazolidinediones
- SAR
structure–activity relationship
- CPT1A
carnitine palmitoyltransferase 1A
- PDK4
pyruvate dehydrogenase kinase 4
- GLUT1
glucose transporter 1
- LPS
lipopolysaccharide
- LBD
ligand binding domain
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00574.
Complete experimental procedures, characterization data for all compounds, biological procedures, docking and cluster analysis, as well as spectra (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
This paper was published ASAP on February 28, 2019 with errors in Table 1. The corrected paper was reposted on March 4, 2019.
Supplementary Material
References
- Kota B. P.; Huang T. H.-W.; Roufogalis B. D. An overview on biological mechanisms of PPARs. Pharmacol. Res. 2005, 51, 85–94. 10.1016/j.phrs.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Rehman K.; Akash M. S. Mechanisms of inflammatory responses and development of insulin resistance: how are they interlinked?. J. Biomed. Sci. 2016, 23, 87. 10.1186/s12929-016-0303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L.; Shen W.-J.; Bittner S.; Kraemer F. B.; Azhar S. PPARs: regulators of metabolism and as therapeutic targets in cardiovascular disease. Part I: PPAR-α. Future Cardiol. 2017, 13, 259–278. 10.2217/fca-2016-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjan M. J.; Mohammed M.; Kumar B. R. P.; Chandrasekar M. J. N. Thiazolidinediones as antidiabetic agents: A critical review. Bioorg. Chem. 2018, 77, 548. 10.1016/j.bioorg.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Neels J. G.; Grimaldi P. A. Physiological functions of peroxisome proliferator-activated receptor β. Physiol. Rev. 2014, 94, 795–858. 10.1152/physrev.00027.2013. [DOI] [PubMed] [Google Scholar]
- Boubia B.; Poupardin O.; Barth M.; Binet J.; Peralba P.; Mounier L.; Jacquier E.; Gauthier E.; Lepais V. r.; Chatar M. Design, Synthesis, and Evaluation of a Novel Series of Indole Sulfonamide Peroxisome Proliferator Activated Receptor (PPAR) α/γ/δ Triple Activators: Discovery of Lanifibranor, a New Antifibrotic Clinical Candidate. J. Med. Chem. 2018, 61, 2246–2265. 10.1021/acs.jmedchem.7b01285. [DOI] [PubMed] [Google Scholar]
- Piemontese L.; Cerchia C.; Laghezza A.; Ziccardi P.; Sblano S.; Tortorella P.; Iacobazzi V.; Infantino V.; Convertini P.; Dal Piaz F. New diphenylmethane derivatives as peroxisome proliferator-activated receptor alpha/gamma dual agonists endowed with anti-proliferative effects and mitochondrial activity. Eur. J. Med. Chem. 2017, 127, 379–397. 10.1016/j.ejmech.2016.12.047. [DOI] [PubMed] [Google Scholar]
- Piemontese L.; Fracchiolla G.; Carrieri A.; Parente M.; Laghezza A.; Carbonara G.; Sblano S.; Tauro M.; Gilardi F.; Tortorella P. Design, synthesis and biological evaluation of a class of bioisosteric oximes of the novel dual peroxisome proliferator-activated receptor α/γ ligand LT175. Eur. J. Med. Chem. 2015, 90, 583–594. 10.1016/j.ejmech.2014.11.044. [DOI] [PubMed] [Google Scholar]
- Giampietro L.; Ammazzalorso A.; Bruno I.; Carradori S.; De Filippis B.; Fantacuzzi M.; Giancristofaro A.; Maccallini C.; Amoroso R. Synthesis of Naphthyl-, Quinolin-and Anthracenyl Analogues of Clofibric Acid as PPAR α Agonists. Chem. Biol. Drug Des. 2016, 87, 467–471. 10.1111/cbdd.12677. [DOI] [PubMed] [Google Scholar]
- De Filippis B.; Agamennone M.; Ammazzalorso A.; Bruno I.; D’Angelo A.; Di Matteo M.; Fantacuzzi M.; Giampietro L.; Giancristofaro A.; Maccallini C.; Amoroso R. PPARα agonists based on stilbene and its bioisosteres: biological evaluation and docking studies. MedChemComm 2015, 6, 1513–1517. 10.1039/C5MD00151J. [DOI] [Google Scholar]
- Giampietro L.; D’Angelo A.; Giancristofaro A.; Ammazzalorso A.; De Filippis B.; DiMatteo M.; Fantacuzzi M.; Linciano P.; Maccallini C.; Amoroso R. Effect of stilbene and chalcone scaffolds incorporation in clofibric acid on PPARα agonistic activity. Med. Chem. 2013, 10, 59–65. 10.2174/157340641001131226123613. [DOI] [PubMed] [Google Scholar]
- Amoroso R.; Leporini L.; Cacciatore I.; Marinelli L.; Ammazzalorso A.; Bruno I.; De Filippis B.; Fantacuzzi M.; Maccallini C.; Menghini L. Synthesis, Characterization and Evaluation of Gemfibrozil-stilbene Hybrid as Antioxidant Agent. Lett. Drug Des. Discovery 2018, 15, 1230–1238. 10.2174/1570180815666180321163246. [DOI] [Google Scholar]
- Giampietro L.; D’Angelo A.; Giancristofaro A.; Ammazzalorso A.; De Filippis B.; Fantacuzzi M.; Linciano P.; Maccallini C.; Amoroso R. Synthesis and structure–activity relationships of fibrate-based analogues inside PPARs. Bioorg. Med. Chem. Lett. 2012, 22, 7662–7666. 10.1016/j.bmcl.2012.09.111. [DOI] [PubMed] [Google Scholar]
- dos Santos J. C.; Bernardes A.; Giampietro L.; Ammazzalorso A.; De Filippis B.; Amoroso R.; Polikarpov I. Different binding and recognition modes of GL479, a dual agonist of Peroxisome Proliferator-Activated Receptor α/γ. J. Struct. Biol. 2015, 191, 332–340. 10.1016/j.jsb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Laghezza A.; Pochetti G.; Lavecchia A.; Fracchiolla G.; Faliti S.; Piemontese L.; Di Giovanni C.; Iacobazzi V.; Infantino V.; Montanari R. New 2-(aryloxy)-3-phenylpropanoic acids as peroxisome proliferator-activated receptor α/γ dual agonists able to upregulate mitochondrial carnitine shuttle system gene expression. J. Med. Chem. 2013, 56, 60–72. 10.1021/jm301018z. [DOI] [PubMed] [Google Scholar]
- Fernández-Miranda C.; Pérez-Carreras M.; Colina F.; López-Alonso G.; Vargas C.; Solís-Herruzo J. A. A pilot trial of fenofibrate for the treatment of non-alcoholic fatty liver disease. Dig. Liver Dis. 2008, 40, 200–205. 10.1016/j.dld.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Tenenbaum A.; Motro M.; Fisman E. Z.; Schwammenthal E.; Adler Y.; Goldenberg I.; Leor J.; Boyko V.; Mandelzweig L.; Behar S. Peroxisome proliferator-activated receptor ligand bezafibrate for prevention of type 2 diabetes mellitus in patients with coronary artery disease. Circulation 2004, 109, 2197–2202. 10.1161/01.CIR.0000126824.12785.B6. [DOI] [PubMed] [Google Scholar]
- Cusi K.; Orsak B.; Bril F.; Lomonaco R.; Hecht J.; Ortiz-Lopez C.; Tio F.; Hardies J.; Darland C.; Musi N.; Webb A.; Portillo-Sanchez P. Long-Term pioglitazone treatment for patients with nonalcoholic steatohepatitis and prediabetes or type 2 diabetes mellitus: a randomized trial. Ann. Intern. Med. 2016, 165, 305–315. 10.7326/M15-1774. [DOI] [PubMed] [Google Scholar]
- Higgins S.; DePaoli A. M. Selective peroxisome proliferator-activated receptor γ (PPARγ) modulation as a strategy for a safer therapeutic PPARγ activation. Am. J. Clin. Nutr. 2010, 91, 267S–272S. 10.3945/ajcn.2009.28449E. [DOI] [PubMed] [Google Scholar]
- Fruchart J.-C. Selective peroxisome proliferator-activated receptor α modulators (SPPARMα): the next generation of peroxisome proliferator-activated receptor α-agonists. Cardiovasc. Diabetol. 2013, 12, 82. 10.1186/1475-2840-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheang W. S.; Tian X. Y.; Wong W. T.; Huang Y. The peroxisome proliferator-activated receptors in cardiovascular diseases: experimental benefits and clinical challenges. Br. J. Pharmacol. 2015, 172, 5512–5522. 10.1111/bph.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampe R. T. Jr; Montana V. G.; Lambert M. H.; Miller A. B.; Bledsoe R. K.; Milburn M. V.; Kliewer S. A.; Willson T. M.; Xu H. E. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 2000, 5, 545–555. 10.1016/S1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- Takada I.; Yu R. T.; Xu H. E.; Lambert M. H.; Montana V. G.; Kliewer S. A.; Evans R. M.; Umesono K. Alteration of a single amino acid in peroxisome proliferator-activated receptor-α (PPARα) generates a PPARδ phenotype. Mol. Endocrinol. 2000, 14, 733–740. 10.1210/mend.14.5.0456. [DOI] [PubMed] [Google Scholar]
- Forbes C. R.; Sinha S. K.; Ganguly H. K.; Bai S.; Yap G. P. A.; Patel S.; Zondlo N. J. Insights into thiol–aromatic interactions: A stereoelectronic basis for S–H/π interactions. J. Am. Chem. Soc. 2017, 139, 1842–1855. 10.1021/jacs.6b08415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. E.; Lambert M. H.; Montana V. G.; Plunket K. D.; Moore L. B.; Collins J. L.; Oplinger J. A.; Kliewer S. A.; Gampe R. T.; McKee D. D. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 13919–13924. 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochetti G.; Godio C.; Mitro N.; Caruso D.; Galmozzi A.; Scurati S.; Loiodice F.; Fracchiolla G.; Tortorella P.; Laghezza A. Insights into the mechanism of partial agonism: crystal structures of the peroxisome proliferator-activated receptor γ ligand-binding domain in the complex with two enantiomeric ligands. J. Biol. Chem. 2007, 282, 17314–17324. 10.1074/jbc.M702316200. [DOI] [PubMed] [Google Scholar]
- Sugden M. C.; Zariwala M. G.; Holness M. J. PPARs and the orchestration of metabolic fuel selection. Pharmacol. Res. 2009, 60, 141–150. 10.1016/j.phrs.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Tafuri S. R. Troglitazone enhances differentiation, basal glucose uptake, and Glut1 protein levels in 3T3-L1 adipocytes. Endocrinology 1996, 137, 4706–4712. 10.1210/endo.137.11.8895337. [DOI] [PubMed] [Google Scholar]
- Guo J.; Liu Z.; Sun H.; Huang Y.; Albrecht E.; Zhao R.; Yang X. Lipopolysaccharide challenge significantly influences lipid metabolism and proteome of white adipose tissue in growing pigs. Lipids Health Dis. 2015, 14, 68. 10.1186/s12944-015-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah R.; Hassouneh R.; Hebert R. L. PGE2, Kidney Disease, and Cardiovascular Risk: Beyond Hypertension and Diabetes. J. Am. Soc. Nephrol. 2016, 27, 666–76. 10.1681/ASN.2015050528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.