Abstract
Human DNA polymerase ι (hPolι) promotes translesion synthesis by inserting nucleotides opposite highly distorting or noninstructional DNA lesions. Here, we provide evidence for the physical interaction of hPolι with proliferating cell nuclear antigen (PCNA), and show that PCNA, together with replication factor C (RFC) and replication protein A (RPA), stimulates the DNA synthetic activity of hPolι. In the presence of these protein factors, on undamaged DNA, the efficiency (Vmax/Km) of correct nucleotide incorporation by hPolι is increased ≈80–150-fold, and this increase in efficiency results from a reduction in the apparent Km for the nucleotide. PCNA, RFC, and RPA also stimulate nucleotide incorporation opposite the 3′-T of the (6) thymine–thymine (T-T) photoproduct and opposite an abasic site. The interaction of hPolι with PCNA implies that the targeting of this polymerase to the replication machinery stalled at a lesion site is achieved via this association.
In both prokaryotes and eukaryotes, DNA polymerases belonging to the UmuC/Rad30/DinB family promote synthesis through DNA lesions. Humans contain three DNA polymerases, Polη, Polι, and Polκ, that are members of this family (1, 2). Of these proteins, Polη is present in both the yeast Saccharomyces cerevisiae and humans, whereas Polι and Polκ are absent from this yeast (2). Extensive studies of both yeast and human Polη have indicated a role for this polymerase in the bypass of a variety of distorting DNA lesions. Both yeast and human Polη replicate through a cis-syn thymine–thymine (T-T) dimer with the same efficiency and accuracy as they replicate through undamaged Ts (3–5), and genetic studies in yeast have indicated a role of Polη in the error-free bypass of 5′-TC-3′ and 5′-CC-3′ cyclobutane dimers (6). Mutational inactivation of Polη in humans results in the cancer prone syndrome, the variant form of xeroderma pigmentosum (7, 8). Polη efficiently replicates through DNA lesions such as 8-oxoguanine and O6-methylguanine as well (9, 10).
By contrast to Polη, human Polι (hPolι) is unable to replicate through a cis-syn T-T dimer, and it does not even insert a nucleotide opposite the 3′-T of the dimer (11). Polι, however, incorporates nucleotides opposite the 3′-T of the (6–4) T-T photoproduct, and opposite an abasic site (11). DNA polymerase ζ, which is an efficient extender of mismatched base pairs, readily extends from the nucleotides inserted by Polι opposite these lesion sites (11). Thus, replication through such highly distorting or noninstructional lesions could be achieved by the coordinated action of Polι and Polζ.
For hPolι to function in lesion bypass, it has to be targeted to the replication machinery stalled at the lesion site. Recently, we have provided evidence for the physical and functional interaction of yeast and human Polη with the proliferating cell nuclear antigen (PCNA), and have shown that this interaction is essential for Polη to function in damage bypass (12, 13). Now, we examine whether PCNA also acts as an intermediary for the targeting of hPolι to the replication machinery. Here, we show that hPolι interacts physically with PCNA, and that PCNA, together with replication factor C (RFC) and replication protein A (RPA), stimulates the DNA synthetic activity of hPolι on both undamaged and damaged DNAs. The efficiency (Vmax/Km) for nucleotide incorporation is enhanced in the presence of these protein factors, and this increase in efficiency derives from a reduction in the apparent Km for the nucleotide.
Materials and Methods
Proteins.
Human PCNA, RFC, and RPA were purified as described (14–16). Human Polι in fusion with glutathione S-transferase (GST) was expressed in the yeast strain BJ5464 and bound to a glutathione-Sepharose 4B column as described (4). GST-Polι protein (≈200 μg) bound to 100 μl of glutathione-Sepharose 4B was incubated overnight at 4°C with 4 units of PreScission protease (Amersham Pharmacia), which cleaves the GST-Polι fusion protein seven amino acids amino-terminal from the first methionine of Polι, in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, and 10% glycerol. The cleaved protein was concentrated by using a Microcon 30 (Amicon) and purified hPolι was aliquoted and frozen at −70°C.
Physical Interaction of hPolι with PCNA.
Gel filtration of hPolι, PCNA, and their complexes was performed at 4°C, using a Superdex 200 PC 3.2/30 column (Amersham Pharmacia) equilibrated with buffer I, containing 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.01% Nonidet P-40, and 10% glycerol. To constitute complexes, hPolι (4 μg), PCNA (5 μg), or the mixture of these proteins were incubated in 25 μl buffer I for 60 min at 4°C followed by incubation for 10 min at 25°C. The protein mixture was then gel-filtered by using 20 μl/min flow-rate at 4°C. Fractions were collected and analyzed on 10% sodium-dodecylsulfate-polyacrylamide gels stained with Coomassie blue R-250.
DNA Substrates.
The circular DNA substrate used in Fig. 2 was a circular single-stranded M13 derivative (M13mp7L2) DNA primed with a 5′ 32P-labeled oligomer primer (LP-273, 5′-GGG TTT TCC CAG TCA CGA CGT TGT AAA ACG ACG GCC AG-3′). Linear, running start DNA substrates were generated by annealing a 75-nt oligomer template (5′-biotin-AGC AAG TCA CCA ATG TCT AAG AGT TCG TAXXAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-biotin-3′) or a 75-nt oligomer template (75AP-2biotin, 5′-biotin-AAA AAA AAA AAA AAA AAA AAA AAA AAA GGG 0AT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-biotin-3′), which contained one biotin molecule attached at each end and two undamaged T residues, or a cis-syn T-T dimer, or a (6–4) T-T photoproduct at position 30–31 (XX), or an abasic site (a tetrahydrofuran moiety; Midland Company, Midland, TX) at position 31 (0), respectively, to the 5′ 32P-labeled oligomer primers [(N4577, 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TA-3′) (Fig. 3A) or (N4267, 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA GGC A-3′) (Fig. 3B)]. For steady-state kinetic analyses of nucleotide insertion opposite DNA lesions and opposite undamaged template T, the same 75-nt oligonucleotide templates were annealed to a 31-nt oligomer primer (LP-159, 5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA T-3′). For steady-state kinetic analyses of nucleotide insertion opposite undamaged C, G, and A template residues, the 75AP-2biotin oligomer template was annealed to the 5′ 32P-labeled oligomer primers (LP-158, 5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG-3′), (N4577, 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TA-3′), and N4267, respectively. To bind streptavidin to the biotin present at the ends of the linear DNA substrates, these primer/template DNAs (2.5 pmol) were preincubated with streptavidin (5 μg) in 25 μl of DNA polymerase buffer that contained no MgCl2, for 10 min at 30°C, before their addition to the DNA polymerase reactions.
Figure 2.
Effect of PCNA, RFC, and RPA on hPolι activity. (A) DNA synthesis by hPolι in the presence of different combinations of PCNA, RFC, and RPA. The complete standard reaction mixture contained hPolι (10 nM), PCNA (100 ng), RFC (50 ng), and RPA (250 ng), and all four deoxynucleotides (100 μM each) along with a ssM13 DNA (2 nM) primed with a 5′ 32P-labeled 38-nt oligonucleotide. Various combinations of PCNA, RFC, and RPA were added to the reaction mixture as indicated. (B) Processivity of hPolι in the presence of PCNA, RFC, and RPA. hPolι (3 nM or 10 nM) alone (lanes 1 and 2, and 5 and 6), or in the presence of PCNA (100 ng), RFC (50 ng), and RPA (250 ng; lanes 3 and 4, and 7 and 8), was preincubated with circular ssM13 template DNA (10 nM) singly primed with a 5′ 32P-labeled oligonucleotide for 5 min at 37°C. Primer extension reactions were initiated by adding all four deoxynucleotides (500 μM each; lanes 1–4) or all four deoxynucleotides and excess sonicated herring sperm DNA (0.5 mg/ml) as a trap (lanes 5–8). After incubation for 10 min at 37°C, samples were quenched and run on a 10% polyacrylamide gel. To demonstrate the effectiveness of the trap, hPolι, along with PCNA, RFC, and RPA, was preincubated with the trap DNA together with the DNA substrate before addition of dNTPs (lane 9).
Figure 3.
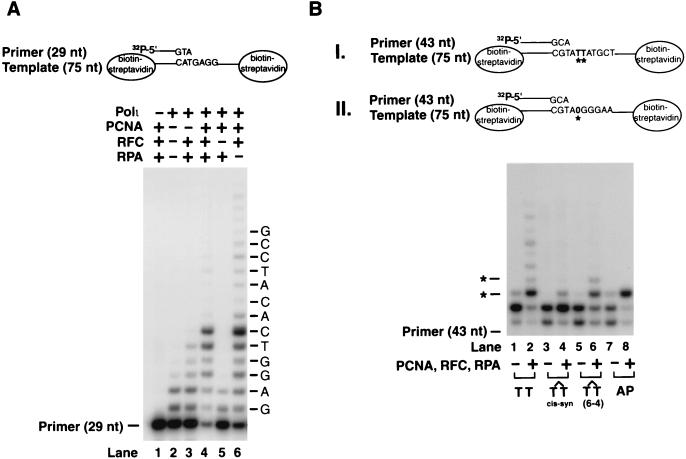
Stimulation of nucleotide incorporation by hPolι in the presence of PCNA, RFC, and RPA on lesion containing DNA substrates. (A) Effect of PCNA, RFC, and RPA on DNA synthesis on an undamaged linear primer:template substrate bearing biotin–streptavidin complex at the ends. The complete reaction mixture contained hPolι (3 nM), PCNA (100 ng), RFC (50 ng), RPA (50 ng), and all four deoxynucleotides (100 μM each) along with the DNA substrate (20 nM). Various combinations of PCNA, RFC, and RPA were added to the reaction mixture as indicated. (B) DNA synthesis by hPolι on lesion containing DNA substrates in the presence or absence of PCNA, RFC, and RPA. A portion of the running start DNA substrate containing the undamaged T-T residues, a cis-syn T-T dimer, or a (6–4) T-T photoproduct is shown in I. The position of the undamaged or damaged TT residues is indicated by asterisks. In the abasic site containing DNA substrate shown in II, the abasic site in the template DNA is indicated by 0 and marked by an asterisk. Polι (3 nM) was incubated with the DNA substrate (20 nM) in the presence of the four dNTPs (100 μM each) under standard reaction conditions.
DNA Polymerase Assays.
The standard DNA polymerase reaction (10 μl) contained 40 mM Tris⋅HCl (pH 7.5), 8 mM MgCl2, 150 mM NaCl, 1 mM DTT, 100 μg/ml BSA, 500 μM ATP, 10% glycerol, and 100 μM of each dGTP, dATP, dTTP, and dCTP. As indicated in the figure legends, hPolι (1–10 nM), PCNA (100 ng), RFC (50 ng), and RPA (50–250 ng) were incubated with primed circular ssM13 DNA (2 or 10 nM) or linear primer/template DNA substrate (20 nM). Assays were assembled on ice, incubated at 37°C for 10 min, and stopped by the addition of loading buffer (40 μl) containing EDTA (20 mM), 95% formamide, 0.3% bromophenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. Quantitation of results was done by using a Molecular Dynamics STORM PhosphoImager and IMAGEQUANT software.
Processivity Assays.
hPolι (3 or 10 nM) in the presence or absence of PCNA, RFC, and RPA was preincubated with circular M13 primer-template DNA substrate (10 nM) in standard reaction buffer that contained no deoxynucleotides for 5 min at 37°C. Reactions were initiated by the addition of all four deoxynucleotides (500 μM each), or all four deoxynucleotides plus excess sonicated herring sperm DNA (0.5 mg/ml) as a trap followed by incubation for 10 min at 37°C. To demonstrate the effectiveness of the trap, hPolι was preincubated with the DNA trap and the primer-template substrate before the addition of dNTPs.
Steady-State Kinetic Analyses.
Steady-state kinetic analyses for deoxynucleotide incorporation opposite undamaged bases, a cis-syn T-T dimer, a (6–4) T-T photoproduct, or an abasic site were performed as described (17, 18). Briefly, hPolι (1 nM) was incubated alone or in the presence of PCNA (100 ng), RFC (50 ng), and RPA (50 ng), with linear primer:template DNA (20 nM) bearing biotin–streptavidin complexes, and with increasing concentration of a single deoxynucleotide for 10 min under standard reaction conditions. Gel band intensities of the substrates and products were quantitated by PhosphorImager and the percentage of primer extensions was plotted as a function of dNTP concentration. The data were fit by nonlinear regression using SIGMAPLOT 5.0 to the Michaelis–Menten equation describing a hyperbola, v = (Vmax × [dNTP]/(Km + [dNTP]). Apparent Km and Vmax steady-state parameters were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (Vmax/Km).
Results
Interaction of hPolι with PCNA.
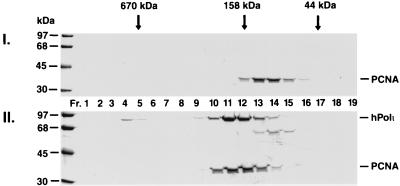
To determine whether hPolι physically interacts with PCNA, we examined whether a complex can be formed between these proteins in vitro. Human Polι was mixed with the human PCNA homotrimer in a 1:1 molar ratio, and after incubation their interaction was examined by gel-filtration, a nonequilibrium technique wherein only stable protein complexes survive. As shown in Fig. 1I, PCNA alone elutes mainly in fractions 13–14, and has a relative molecular mass of ≈110 kDa, which is consistent with its being a homotrimer. However, when PCNA was preincubated with hPolι, PCNA eluted earlier, together with hPolι, primarily in fractions 11–12 (Fig. 1II), which corresponds to a relative molecular mass of ≈200 kDa. This shift in the elution position of PCNA indicates the formation of a complex between hPolι and PCNA, and we estimate that in this complex one hPolι molecule is bound to one PCNA homotrimer.
Figure 1.
Interaction of hPolι with PCNA. hPolι was incubated with human PCNA and gel-filtered (Superdex 200). (I) PCNA is alone (5 μg); (II) the mixture of hPolι (4 μg) and PCNA (5 μg) were gel filtered. Column fractions (Fr.) are indicated, and the positions of molecular weight standards are shown on the left. The elution positions of molecular weight markers for the gel filtration column are indicated on top. PCNA and hPolι are identified on the right.
Stimulation of DNA Synthesis Activity of hPolι by PCNA, RFC, and RPA.
To assess the functional significance of hPolι interaction with PCNA, we examined whether PCNA in combination with RFC, which loads the PCNA ring onto DNA in the presence of ATP, and RPA, a single-stranded DNA-binding protein, stimulates the DNA synthetic activity of hPolι. For monitoring DNA synthesis, we used a single-stranded M13 template DNA primed at a unique site with a 38-nt 5′ 32P-labeled oligomer primer (Fig. 2A). The DNA synthetic activity of hPolι was strongly enhanced upon the addition of PCNA, RFC, and RPA (Fig. 2A, compare lanes 1 and 3); the addition of only PCNA with RFC, PCNA with RPA, or RFC with RPA, however, had no significant stimulatory effect (Fig. 2A, lanes 2, 4, and 5). This result indicates that PCNA cooperates with RFC and RPA to stimulate the DNA synthetic activity of hPolι.
PCNA, along with RFC and RPA, greatly stimulates the processivity of Polδ (19, 20). Next, we examined whether these accessory proteins also enhance hPolι processivity. To test for this, we used a circular ssM13 template DNA primed singly with a 5′ 32P-labeled oligonucleotide primer, and to ensure that we were observing deoxynucleotide incorporation resulting from a single DNA binding event, we monitored DNA synthesis in the presence of an excess of nonradiolabeled, sonicated herring sperm DNA as a trap (Fig. 2B). The reactions were performed by first preincubating hPolι with the DNA substrate without deoxynucleotide in the absence (Fig. 2B, lanes 1 and 2, and 5 and 6) or the presence of PCNA, RFC, and RPA (Fig. 2B, lanes 3 and 4, and 7 and 8). DNA synthesis was initiated by the addition of all four deoxynucleotides (Fig. 2B, lanes 1–4), or a mixture of excess herring sperm DNA and all four deoxynucleotides (Fig. 2B, lanes 5–8) to the reaction. Under these conditions, any hPolι molecules that dissociate from the labeled DNA substrate will be trapped by the excess of nonradiolabeled herring sperm DNA. Even without the DNA trap, and when the enzyme and the DNA substrate molar ratio was 1:1, hPolι synthesized only a short track (≈1–8 nt) of DNA (Fig. 2B, lane 4). In the reactions, wherein single hit conditions were provided by the excess herring sperm DNA, PCNA, RFC, and RPA did not significantly improve the processivity, because hPolι incorporated predominantly only one deoxynucleotide before its dissociation from the DNA substrate (Fig. 2B, lane 8). Thus, hPolι has an extremely low processivity and is able to incorporate only one deoxynucleotide per DNA binding event.
PCNA Stimulates Nucleotide Incorporation Opposite DNA Lesions by hPolι.
We have shown previously that hPolι is unable to bypass a cis-syn T-T dimer, a (6–4) T-T photoproduct, or an abasic site (11). However, hPolι can incorporate a nucleotide opposite a (6–4) photoproduct and an abasic site, whereas the subsequent extension step is carried out by another DNA polymerase, Polζ (11). Next, we examined whether PCNA, RFC, and RPA enable hPolι to replicate through these DNA lesions. However, we found that for these studies, a short linear damage containing primer/template DNA substrate was not satisfactory, probably because the PCNA ring does not remain stably bound on such DNA and may slide off at the ends. Therefore, we constructed a DNA substrate in which the 75-nt template was bound to biotin at both ends, and following its annealing to the 5′ 32P-labeled oligonucleotide primer, both of these biotins were coupled to streptavidin, an ≈50-kDa protein, that would prevent the sliding of PCNA from the ends after it has been loaded onto DNA by RFC. As shown in Fig. 3A, on this linear DNA substrate, PCNA, RFC, and RPA strongly stimulated DNA synthesis by hPolι (Fig. 3A, compare lanes 2 and 4). However, on this DNA substrate, RPA was not required for enhancing the hPolι activity (Fig. 3A, compare lanes 4 and 6). The requirement of RPA when using the ssM13 template DNA might arise from its binding to ssDNA and thereby preventing the nonspecific DNA binding of hPolι, PCNA, or RFC.
Next, we examined synthesis by hPolι on DNA substrates where, after inserting one nucleotide opposite the undamaged template residue, hPolι encounters either an undamaged T residue, a cis-syn T-T dimer, a (6–4) T-T photoproduct, or an abasic site (Fig. 3B). The ability of hPolι to insert a nucleotide opposite the 3′ T of a (6–4) T-T photoproduct (Fig. 3B, lane 6) and opposite an abasic site (Fig. 3B, lane 8) was strongly stimulated by PCNA, RFC, and RPA. However, these accessory proteins were unable to significantly enhance nucleotide insertion opposite the cis-syn T-T dimer by hPolι, and the presence of a strong stall site just before the 3′ T of the dimer indicates that this lesion continues to be a strong block to this polymerase (Fig. 3B, lane 4). In contrast to the strong stimulatory effect of PCNA, RFC, and RPA on nucleotide incorporation opposite a (6–4) T-T photoproduct and an abasic site, hPolι remained inefficient in extending from the nucleotide inserted opposite these lesion sites.
Efficiency of Nucleotide Incorporation by hPolι in the Presence of PCNA, RFC, and RPA.
To determine the extent by which PCNA, RFC, and RPA stimulate nucleotide incorporation by hPolι opposite undamaged template bases, we examined the changes in steady-state kinetic parameters, Km and Vmax for nucleotide insertion by hPolι in the presence of these protein factors. DNA substrates bearing biotin–streptavidin complex on both ends were generated by annealing 32P-labeled oligonucleotide primers to a linear 75-nt template oligomer for standing start assays. The kinetics of insertion of a single deoxynucleotide opposite an undamaged, a cis-syn T-T dimer, a (6–4) T-T photoproduct, and an abasic site were determined as a function of the deoxynucleotide concentration under steady-state conditions (Fig. 4).
Figure 4.
Deoxynucleotide incorporation opposite undamaged G or T residues, and opposite a cis-syn T-T dimer, a (6–4) T-T photoproduct, and an abasic (AP) site by hPolι. The linear DNA substrates contained the biotin–streptavidin complex on both ends. hPolι (1 nM) was incubated with primer:template DNA substrate (20 nM) and increasing concentrations of a single deoxynucleotide in the absence or presence of PCNA (100 ng), RFC (50 ng), and RPA (50 ng).
As judged from the Vmax/Km values, PCNA together with RFC and RPA increases the efficiency of nucleotide incorporation by hPolι opposite undamaged (Table 1), as well as opposite damaged (Table 2) bases. However, the rate of stimulation differs for the various template residues and DNA lesions. In the presence of PCNA, RFC, and RPA, hPolι incorporates the correct nucleotide opposite the undamaged template residues G, A, and C, ≈80–150-fold better, whereas opposite the template T residue, it incorporates an A or a G ≈20–30-fold more efficiently than in the absence of these accessory proteins (Table 1). The increased efficiencies for nucleotide incorporation arise primarily from a marked reduction in the Km for the dNTP substrate, whereas the Vmax stays about the same (Table 1). PCNA, RFC, and RPA did not change significantly the overall fidelity of hPolι, and these protein factors did not alter the preferential incorporation of a G opposite template T (Table 1).
Table 1.
Kinetic parameters of nucleotide insertion reactions catalyzed by hPolι opposite undamaged template residues
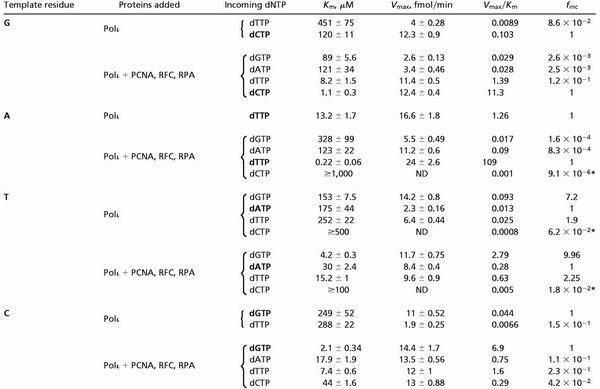 |
ND, not determined.
Because the nucleotide incorporation rate remained linear throughout the nucleotide concentration range used, the Vmax/Km value was obtained from the slope of the line.
Table 2.
Kinetic parameters of nucleotide insertion reactions catalyzed by hPolι opposite damaged bases
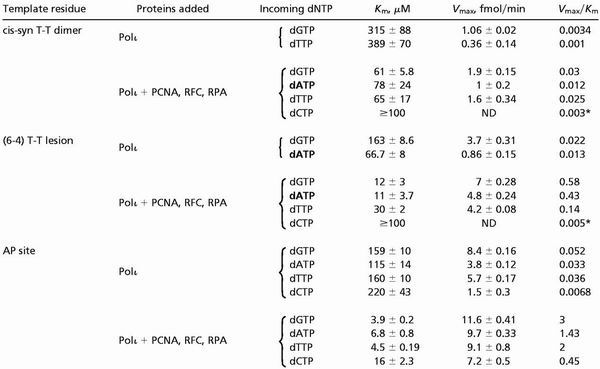 |
AP, abasic; ND, not determined.
Because the nucleotide incorporation rate remained linear throughout the nucleotide concentration range used, the Vmax/Km value was obtained from the slope of the line.
As is shown in Table 2, opposite the 3′ T of a cis-syn T-T dimer, PCNA, RFC, and RPA stimulated G incorporation by hPolι only ≈9-fold, and this nucleotide insertion still occurs ≈100-fold less efficiently than the incorporation of G opposite the undamaged T. Opposite the 3′ T of a (6–4) T-T photoproduct, hPolι incorporates the G nucleotide ≈25-fold more efficiently in the presence of these accessory proteins, which is ≈5-fold less efficient than the incorporation of G opposite the undamaged T (Tables 1 and 2). Opposite the 3′ T of the (6–4) T-T photoproduct, the nucleotide A was inserted ≈30-fold more efficiently in the presence of accessory factors than in their absence. The most efficient nucleotide incorporation occurred opposite a template abasic site, where PCNA, RFC, and RPA stimulated the incorporation of G by hPolι ≈60-fold (Table 2), and in the presence of these proteins, hPolι incorporates a G opposite the abasic site about 50% as efficiently as opposite an undamaged C (Tables 1 and 2). Opposite an abasic site, the A and T nucleotides were incorporated ≈40–50-fold more efficiently in the presence of PCNA, RFC, and RPA, than in their absence (Table 2). Thus, in the presence of PCNA, RFC, and RPA, hPolι efficiently incorporates deoxynucleotides opposite an abasic site and the efficiency of nucleotide insertion opposite the 3′ T of a (6–4) T-T photoproduct is also enhanced. However, even in the presence of these protein factors, hPolι remains highly inefficient at inserting nucleotides opposite the 3′ T of a T-T dimer.
Discussion
Here, we show that hPolι physically interacts with PCNA, and PCNA, together with RFC and RPA, stimulates the DNA synthetic activity of hPolι. Processivity, however, is not affected. Even in the presence of these protein factors, hPolι predominantly incorporates only one nucleotide per DNA binding event. A PCNA binding motif has been identified in a variety of proteins (21, 22), and also in yeast and human Polη (12, 13). A similar conserved motif SRGVLSFF (in which the conserved residues are indicated in bold) is also present in residues 540–547 of hPolι, and may contribute to PCNA binding.
hPolι misincorporates nucleotides at a high rate, and the fidelity of nucleotide incorporation remains low in the presence of PCNA, RFC, and RPA. Although it misincorporates nucleotides opposite templates G, A, and C with frequencies ranging from ≈10−1 to ≈10−4, opposite template T, it preferentially incorporates a G (11, 23, 24). PCNA, RFC, and RPA increase the efficiency of correct nucleotide incorporation opposite templates G, A, and C by ≈80–150-fold, but they also stimulate the incorporation of G opposite template T by ≈30-fold. The increase in the efficiency of nucleotide incorporation results from a marked reduction in the apparent Km for the dNTP.
Based on the ability of hPolι to incorporate nucleotides opposite the 3′ T of a (6–4) T-T photoproduct and opposite an abasic site, we previously inferred a role for this polymerase in lesion bypass (11). However, hPolι did not insert a nucleotide opposite the 3′ T of a cis-syn T-T dimer and was unable to bypass this lesion (11). Here, we show that PCNA, RFC, and RPA stimulate the G nucleotide incorporation opposite the 3′ T of this lesion only ≈9-fold, whereas the incorporation of a G opposite an undamaged T is enhanced ≈30-fold. Thus, a cis-syn T-T dimer continues to be a strong block to hPolι in the presence of these accessory protein factors.
Although a (6–4) T-T photoproduct induces a much larger structural distortion in DNA than a cis-syn T-T dimer, and the 3′ T in the (6–4) lesion is held perpendicular to the 5′ T (25), hPolι displays a greater tendency for nucleotide insertion opposite the 3′ T of a (6–4) T-T lesion than opposite this site in a cis-syn T-T dimer. In the presence of PCNA, RFC, and RPA, hPolι inserts a G or an A nucleotide opposite the 3′ T of the (6–4) T-T lesion ≈30-fold better than in the absence of these protein factors, and the magnitude of this increment is about the same as that seen for the insertion of a G or an A opposite the undamaged T. Moreover, both in the presence or absence of PCNA, RFC, and RPA, hPolι incorporates a G opposite the 3′ T of the (6–4) T-T lesion only about 5-fold less efficiently than the incorporation of this nucleotide opposite an undamaged T. Thus, we expect hPolι to contribute to the mutagenic bypass of a (6–4) T-T lesion, and in this role, it would compete with Polη, which can also insert a G opposite the 3′ T of this lesion (26). PCNA, RFC, and RPA also stimulate the ability of hPolι to insert nucleotides opposite an abasic site ≈50–60-fold, and G, the most frequently inserted nucleotide, is incorporated about as well as its incorporation opposite an undamaged C. Thus, hPolι would also contribute to the mutagenic bypass of an abasic site, and in this role it would compete with Polη and Rev1 (12, 27).
Although PCNA, RFC, and RPA stimulate the ability of hPolι for nucleotide insertion opposite undamaged and damaged bases, they do not alter any of the basic properties of this enzyme. Thus, even in the presence of these protein factors, hPolι remains a low processivity and low fidelity enzyme, and its ability to preferentially insert a G opposite an undamaged T remains unchanged. The major role of PCNA would be to target hPolι to the replication machinery stalled at a lesion site, and PCNA thus would be the primary intermediary governing the access of the replicative and translesion synthesis DNA polymerases to the 3′-primer end. Further, the presence of three monomers in the PCNA ring raises the possibility that as many as three different DNA polymerases could simultaneously bind PCNA. Thus, at a lesion site, on dissociation from the 3′-primer terminus, Polδ may still remain bound to PCNA. A translesion synthesis polymerase such as hPolι may then bind the other PCNA monomer, and DNA polymerase ζ, needed for elongating from the nucleotide inserted opposite the lesion site by hPolι, may bind the third PCNA monomer. The concomitant binding of these polymerases to PCNA may provide for a high degree of coordination in their action in lesion bypass.
Acknowledgments
This work was supported by National Institutes of Health Grants GM19261 and GM38559.
Abbreviations
- hPol
human DNA polymerase
- PCNA
proliferating cell nuclear antigen
- RFC
replication factor C
- RPA
replication protein A
- T-T
thymine–thymine
References
- 1.Goodman M F, Tippin B. Curr Opin Genet Dev. 2000;10:162–168. doi: 10.1016/s0959-437x(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 2.Johnson R E, Washington M T, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 4.Johnson R E, Washington M T, Prakash S, Prakash L. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 5.Washington M T, Johnson R E, Prakash L, Prakash S. Proc Natl Acad Sci USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu S-L, Johnson R E, Prakash S, Prakash L. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 8.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 9.Haracska L, Yu S-L, Johnson R E, Prakash L, Prakash S. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 10.Haracska L, Prakash S, Prakash L. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R E, Washington M T, Haracska L, Prakash S, Prakash L. Nature (London) 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 12.Haracska L, Kondratick C M, Unk I, Prakash S, Prakash L. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 13.Haracska L, Johnson R E, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai J, Gibbs E, Uhlmann F, Phillips B, Yano N, O'Donnell M, Hurwitz J. J Biol Chem. 1997;272:18974–18981. doi: 10.1074/jbc.272.30.18974. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs E, Kelman Z, Gulbis J M, O'Donnell M, Kuriyan J, Burgers P M, Hurwitz J. J Biol Chem. 1997;272:2373–2381. doi: 10.1074/jbc.272.4.2373. [DOI] [PubMed] [Google Scholar]
- 16.Lee S H, Eki T, Hurwitz J. Proc Natl Acad Sci USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creighton S, Bloom L B, Goodman M F. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 18.Goodman M F, Creighton S, Bloom L B, Petruska J. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 19.Prelich G, Kostura M, Marshak D R, Matthews M B, Stillman B. Nature (London) 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 20.Tan C K, Castillo C, So A G, Downey K M. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 21.Kelman Z, Hurwitz J. Trends Biol Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- 22.Warbrick E. BioEssays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 23.Tissier A, McDonald J P, Frank E G, Woodgate R. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Yuan F, Wu X, Wang Z. Mol Cell Biol. 2000;20:7009–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J-K, Patel D, Choi B-S. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 26.Johnson R E, Haracska L, Prakash S, Prakash L. Mol Cell Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haracska L, Unk I, Johnson R E, Johansson E, Burgers P M J, Prakash S, Prakash L. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]