Abstract
The interferon-inducible protein kinase R (PKR) is a key component of host innate immunity that restricts viral replication and propagation. As one of the four eIF2α kinases that sense diverse stresses and direct the integrated stress response (ISR) crucial for cell survival and proliferation, PKR's versatile roles extend well beyond antiviral defense. Targeted by numerous host and viral regulators made of RNA and proteins, PKR is subject to multiple layers of endogenous control and external manipulation, driving its rapid evolution. These versatile regulators include not only the canonical double-stranded RNA (dsRNA) that activates the kinase activity of PKR, but also highly structured viral, host, and artificial RNAs that exert a full spectrum of effects. In this review, we discuss our deepening understanding of the allosteric mechanism that connects the regulatory and effector domains of PKR, with an emphasis on diverse structured RNA regulators in comparison to their protein counterparts. Through this analysis, we conclude that much of the mechanistic details that underlie this RNA-regulated kinase await structural and functional elucidation, upon which we can then describe a “PKR code,” a set of structural and chemical features of RNA that are both descriptive and predictive for their effects on PKR.
Keywords: innate immunity, noncoding RNA, PKR, allosteric regulation, antiviral defense
INTRODUCTION
The ability for a host organism to quickly and efficiently respond to pathogens is critical for its survival. In humans, immune response is carried out through a sophisticated network of signal-transduction pathways involving numerous effector proteins (Katze et al. 2002; Hoang et al. 2018). This is initiated by the recognition of pathogen-associated molecular patterns (PAMPs) from infectious agents by pattern recognition receptors (PRRs) of the innate immune system. PAMPs recognition by PRRs initiate an immune response by induction of signaling proteins. Protein kinase RNA-activated (PKR) was originally characterized in 1972 as a key player in innate immune response to viral infection in vertebrates (Metz and Esteban 1972). Since its discovery, the physiological importance of PKR in integrative stress response (ISR) has been widely investigated under diverse cellular stresses (Pindel and Sadler 2011; Yim and Williams 2014; Pakos-Zebrucka et al. 2016). Additionally, PKR has been recently implicated in metabolic disorders and Alzheimer's disease, suggesting a role for PKR in inflammation control (Ohno 2014; Nakamura et al. 2015). The exact functions of PKR in many of these stress response pathways remains poorly understood. The best characterized substrate of PKR is the α subunit of eukaryotic translation initiation factor 2 (eIF2α). Following recognition of an adequate stimulus and subsequent activation and autophosphorylation of PKR, S51 of eIF2α is phosphorylated (P-eIF2α) and acts as a competitive inhibitor for its nucleotide exchange factor eIF2B (Dar et al. 2005; Scheuner et al. 2006; Hoang et al. 2018). Due to the limited level of eIF2B protein in the cell, relatively small amounts of P-eIF2α are sufficient to block the recycling of GDP-eIF2 to GTP-eIF2 required for cap-dependent translation initiation (Dar et al. 2005; Scheuner et al. 2006; Hoang et al. 2018). Via this mechanism, activated PKR halts global translation, subsequently inducing pathways for either cellular recovery or apoptosis. PKR thus acts as a critical gatekeeper for cellular stress management and survival.
The biological importance of PKR is further accentuated by the vast array of PKR inhibitors produced by pathogens to counteract its debilitating restriction (Nallagatla et al. 2011; Hull and Bevilacqua 2016). Some viruses encode double-stranded RNAs (dsRNAs) that bind PKR with nanomolar affinity and prevent binding of activating RNAs. Other viruses encode proteins that interact directly with PKR to inhibit it and/or sequester dsRNAs that would otherwise lead to PKR activation. In addition to foreign regulators, various host PKR inducers, activators, and inhibitors serve to maintain tight control of PKR expression and activity. Given the structural diversity among various PKR modulators, a rigorous set of standardized activating and inhibiting qualifications have yet to be established. The challenge in systematically characterizing activating or inhibiting elements lies in the inherent complexity of the allosteric mechanism of PKR activation (Cole 2007). Several excellent reviews have tackled the diverse and complex biological functions of PKR (Pindel and Sadler 2011; Ohno 2014; Yim and Williams 2014; Pakos-Zebrucka et al. 2016). In this review, we discuss the evolving understanding of human PKR activation/inhibition and focus on describing and comparing the molecular mechanisms used by its diverse RNAs and protein regulators.
The modular organization of PKR
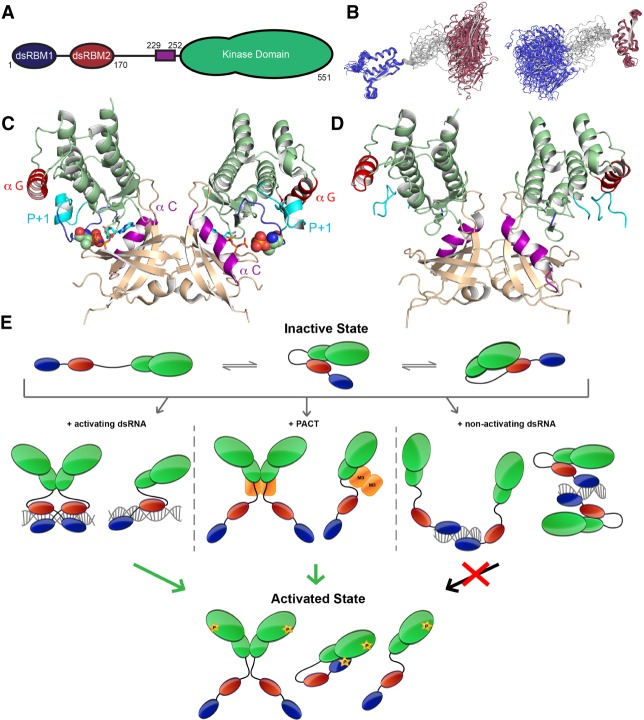
PKR features a modular organization with an amino-terminal regulatory domain, namely two double-stranded RNA-binding motifs (dsRBMs), and a carboxy-terminal effector/catalytic kinase domain (KD) (Fig. 1A; Dar et al., 2005). Despite the absence of high-resolution structural information for PKR in its different signaling states, structural studies of its isolated domains and extensive biochemical analysis have identified important regulatory elements of this kinase and unveiled portions of its allosteric mechanism.
FIGURE 1.
Structural organization of PKR and schematic representation of different activation mechanisms. (A) Different domains of PKR: dsRBM1 is shown in blue, dsRBM2 in red, the basic patch at the amino terminus of the KD in purple, and the Ser/Thr KD in green with a smaller N-lobe than C-lobe. (B) Superposition of the different NMR conformers of PKR dsRBMs (pdb 1qu6) on dsRBM1 as reference (left) or on dsRBM2 (right), showing the dynamic nature of their intervening linker. Colored as in panel A. (C) Activate dimeric KD structure of PKR as seen in complex with eIF2α (pdb 2a1a and 2a19). The nonhydrolyzable ATP analog (AMP-PNP) is represented in sticks and the phosphorylated T446 in spheres. The N-lobe is in wheat and the C-lobe in green. Helix αC (involved in allosteric regulation) is highlighted in magenta and helix αG (involved in eIF2α recognition) is in red, while the activation loop is in dark blue and the P + 1 loop in cyan. (D) Dimeric KD of PKR K296R mutant (pdb 3uiu) colored as in C. Wild-type and K296R PKR share a similar dimeric interface, with the following exception: P-T446, absent from K296R, is required for stabilization/folding of the P + 1 loop for proper substrate recognition. This involves a network of interaction ranging from helix αC recognizing P-T446 further rigidifying the activation loop, allowing for P + 1 loop folding and its interaction with helix αG. (E) PKR is proposed to exist in a multitude of states with distinct structures and activities. PKR phosphorylation following activation is indicated with a yellow star (phosphorylation of T446 or dsRBM1). In its inactive state, PKR is mainly monomeric either extended in solution or locked in an auto-inhibited state with dsRBM2 bound to the KD. Two different dsRBM2-binding interfaces on KD have been proposed; one involving the N-lobe while the other was mapped at the C-lobe next to helix αG. Binding to >30 bp dsRNA induces PKR activation either by relieving the auto-inhibited state and/or by dimerization of PKR on the same RNA, bringing two KDs in close proximity. PACT is thought to bind PKR, especially via its dsRBM3 (M3, see below), and activates KD through a similar mechanism. Shorter dsRNA length induces PKR dimerization but no activation. This could imply that dsRBM2 remains bound to the KD while only dsRBM1 interacts with the RNA and/or assists in forming inactive dimers. Following activation of PKR by different stimuli, T446 is phosphorylated and both dimeric and monomeric activated PKR have been observed, leading to efficient eIF2α phosphorylation. Subsequently, phosphorylation of dsRBM1 could gradually inactivate PKR by interacting with the KD returning to an auto-inhibited state post-activation.
Regulatory domain
The NMR structure of human PKR's tandem dsRBMs revealed a canonical α1-β1-β2-β3-α twofold, separated by a 20 amino acid (a.a.) flexible linker (Fig. 1B; Nanduri et al. 1998, 2000; Patel et al. 2012). Common among Class A dsRBMs, both dsRBMs of PKR contain key structural elements that harbor conserved residues involved in dsRNA binding. These include helix 1 and loop β1–β2 which canonically recognize the wide and shallow minor groove of dsRNA, and helix 2 amino terminus which interacts with the adjacent major groove (Masliah et al. 2013). Interestingly, dsRBM1 has a higher affinity for dsRNA than dsRBM2, but both motifs are required for higher affinity binding (Tian and Mathews 2001; McKenna et al. 2006; Ucci et al. 2007). This synergy between tandem RNA binding domains (RBDs) is a common feature of many modular RNA binding proteins (RBPs), whose functions are subject to control by the lengths and dynamics of their inter-domain linkers (Mackereth and Sattler 2012). In the case of PKR, the linker between dsRBM1 and dsRBM2 was suggested to be involved in RNA recognition based on NMR chemical shift displacements (McKenna et al. 2006; Ucci et al. 2007). Several autophosphorylation sites have been mapped to this linker but their exact function remains unknown. It is tempting to speculate that these phosphorylation events could alter RNA binding by charge repulsion. Indeed, phosphorylated PKR exhibits an 80-fold reduced affinity for RNA, although the exact molecular basis remains unknown (Jammi and Beal 2001).
Effector kinase domain
X-ray structures of the isolated phosphorylated PKR KD in complex with the amino-terminal domain of eIF2α (eIF2αNTD) revealed a bilobal organization common to kinases with a shorter N-lobe comprising a five-stranded antiparallel β sheet and a canonical helix αC, followed by a larger helical C-lobe with the ATP binding site between the two lobes (Fig. 1C; Dar et al. 2005). The active KD assumes a back-to-back dimer configuration induced by inter-N-lobe contacts. Importantly, mutations disrupting this dimerization interface hinder PKR activation both in vivo and in vitro, implying that KD dimerization induces activation (Dey et al. 2005) (see below). Direct interaction of helix αC with the autophosphorylated P-T446 (mandatory for PKR activity) stabilizes the activation loop into an active conformation (Zhang et al. 2001). Additionally, an atypical full turn longer helix αG with a 40° counterclockwise rotation allows for specific recognition of eIF2αNTD's OB fold. Indeed, PKR lacks activity on a peptide fragment encompassing S51 and instead requires the tertiary structure of eIF2α for efficient catalysis (Dey et al. 2011). The X-ray structure of a kinase-dead K296R PKR mutant was essentially identical to the wild-type activated structure, except for the disordered activation and P + 1 loops due to unphosphorylated T446 (Fig. 1D; Li et al. 2012). Since high concentrations of PKR can induce dimerization and activation (Lemaire et al. 2005), this mutant structure likely represents an activated state prior to autophosphorylation instead of an authentic inactive state.
Bridging the regulatory and effector domains
The regulatory and effector domains of PKR described above are separated by a poorly characterized dynamic linker, with limited sequence and length conservation. PKR exists in an ensemble of extended and compact conformations (Anderson and Cole 2008; VanOudenhove et al. 2009). Interestingly, removal of this interdomain linker did not abolish dsRNA-dependent autophosphorylation nor inhibition by viral RNAs in vitro (Sunita et al. 2015; Husain et al. 2016). However, the linker's carboxy-terminal portion, a 23-a.a. “basic patch,” is required for full activation and enhances PKR's basal activity (Husain et al. 2016). These findings hint at a regulatory role of the basic patch which may function through affecting PKR inter- or intra-domain motions.
Multiple facets of the PKR activation mechanism
Despite extensive work over the past four decades, the detailed activation mechanism(s) of PKR is still a work in progress. Two nonmutually exclusive mechanisms have been proposed (Nanduri et al. 2000; Gelev et al. 2006; Li et al. 2006; Cole 2007; McKenna et al. 2007; Lemaire et al. 2008): relief of an inactive autoinhibited state of PKR and activator-induced dimerization and activation. PKR activation is dictated by autophosphorylation of T446 in its activation loop, however, uncertainty remains whether PKR undergoes a cis or trans-inter autophosphorylation (Ortega et al. 1996; Dey et al. 2014).
Inactive PKR exists in a weak reversible monomer-dimer equilibrium. As a result, high concentrations of PKR can induce its activation even in the absence of activators, presumably through increased dimerization (Lemaire et al. 2005). Activation by dsRNA is length-dependent, requires a minimum of >30 base pairs (bp), and follows second-order kinetics and a bell-shaped curve. This supports a model in which long dsRNA bind two or more PKR molecules thus inducing activation, while excess dsRNA dilutes and sequesters monomeric PKR (Fig. 1E, left; Tian and Mathews 2001; Lemaire et al. 2008; Husain et al. 2012). As dsRBMs bind to dsRNA, the KDs of two PKR monomers are brought into close proximity, allowing for efficient dimerization. Indeed, mutation of a strictly conserved inter-N-lobe salt bridge (R262-D266 in PKR) hinders dimerization and downstream activation (Dey et al. 2005, 2007). Mutations in dsRBM1 or dsRBM2 abolishing RNA binding also block PKR dimerization and activation (Bevilacqua et al. 1998; Zhang et al. 2001). In addition, isolated KD258–551 was found to be inactive in vivo (Ung et al. 2001), while its GST fusion (dimerization prone tag) or the substitution of its activation loop with that of phosphorylase kinase 1 Phk1 (which does not require autophosphorylation) induced its activation independently of RNA binding (Dey et al. 2014). This suggests that KD dimerization induces T446 autophosphorylation, which in turn licenses activation of the activation loop for full activity. Importantly, autophosphorylation of PKR stabilizes its dimeric form by ∼500-fold (Lemaire et al. 2005), which in turn amplifies the activation through positive feedback. Nevertheless, PKR dimerization on nonactivating RNAs suggests the existence of active and inactive dimers (Fig. 1E, right; Husain et al. 2015).
Weak interactions between dsRBM2 and KD were revealed by NMR titrations but the interface remains poorly characterized, involving either helix αG or the N-lobe (Fig. 1E; Nanduri et al. 2000; Gelev et al. 2006; Li et al. 2006; Anderson and Cole 2008). This led to an alternative model where autoinhibition locks PKR in an inactive state. Upon dsRNA binding, dsRBM2 engages the activator RNA thus releasing the KD and triggering activation. Indeed, in vivo caspase cleavage at residue 251 of PKR induces its activation presumably by relieving dsRBM's autoinhibition (Kalai et al. 2007). Recently, autophosphorylated dsRBM1, following PKR activation, was shown to interact with the KD inducing a negative-feedback (Fig. 1E), possibly to avoid sustained activation (Wang et al. 2017).
RNA regulators of PKR
RNAs constitute the primary regulators of PKR activity mainly through binding to its tandem dsRBMs and can be either activating, inhibiting, or neutral. In addition to A-form model dsRNAs such as poly I:C, naturally occurring RNA activators and inhibitors of PKR contain helices, loops, bulges, local mismatches, noncanonical pairs, junctions, etc. Such sequence variation and structural complexity likely lead to RNA-specific binding patterns by PKR and thus specific PKR activity (Dauber and Wolff 2009; Nallagatla et al. 2011; Hull and Bevilacqua 2016). As a direct result of an incomplete understanding of the detailed mechanisms that govern PKR enzymatic activity (see preceding section “Multiple facets of the PKR activation mechanism”), we also lack a unified code that would distinguish an RNA activator from an inhibitor. Throughout the years, a cornucopia of cellular and viral RNA regulators of PKR, most of which are highly structured, have been discovered. Understanding the structure-function relationships of these diverse PKR regulators is expected to shed light on the PKR activation and regulatory mechanisms and ultimately lead to a “PKR code” that can predict the effect on PKR by a given RNA sequence and structure.
RNA activators of PKR
Contrary to artificial dsRNAs like poly I:C, which are extensively used to study PKR activation, natural PKR RNA activators generally contain multiple helical defects, and do not always satisfy the 30-bp minimal length requirements. This led to models where RNA dimerization or end-to-end coaxial stacking dictate PKR activation, by presenting a near A-form extended duplex structure long enough to bind two PKR monomers (Heinicke et al. 2009; Nallagatla et al. 2011). Recently, short dsRNA flanked by single-stranded termini, termed ss-dsRNA, have been identified as a new class of PKR activators (Nallagatla et al. 2007; Mayo and Cole 2017). Below we provide specific examples of these divergent RNA activators and discuss their structural features related to PKR activation.
Viral RNA activators (HIV, HDV, HCV, etc.)
The cellular presence of dsRNA is a hallmark of viral infection, as a replicative intermediate or byproduct of bidirectional transcription (Weber et al. 2006). Recognition of sufficiently long viral dsRNAs by PKR dsRBMs leads to its activation, triggering downstream phosphorylation of eIF2α and ultimately arrest of global translation initiation. Through this mechanism, the cell prevents further replication and propagation of viral particles.
The human immunodeficiency virus type 1 (HIV-1) transactivation-response region (TAR) RNA was originally recognized for its interaction with the HIV-1 trans-acting protein Tat, essential for transcriptional elongation and enhanced viral replication (Kao et al. 1987). The TAR RNA consists of a dynamic 23-bp stem–loop containing three bulges (Fig. 2; Ippolito and Steitz 1998; Zhang et al. 2007). Early reports suggested that monomeric TAR RNA activate PKR despite its limited length (Maitra et al. 1994; Kim et al. 2006). Later, dimeric TAR RNA was identified as a stronger in vitro PKR activator despite both RNAs exhibiting similar affinities to PKR and its isolated dsRBMs (Heinicke et al. 2009). This has led to a model where TAR RNA dimerization via a kissing-loop motif drives PKR activation by fulfilling the length requirement for binding two PKR monomers. However, it is not yet clear whether TAR RNA dimerizes in vivo (Andersen et al. 2004; Keane et al. 2016). It is tempting to speculate that upon dimerization of HIV-1 genome, two TAR RNAs are spatially close, bridging two PKR monomers.
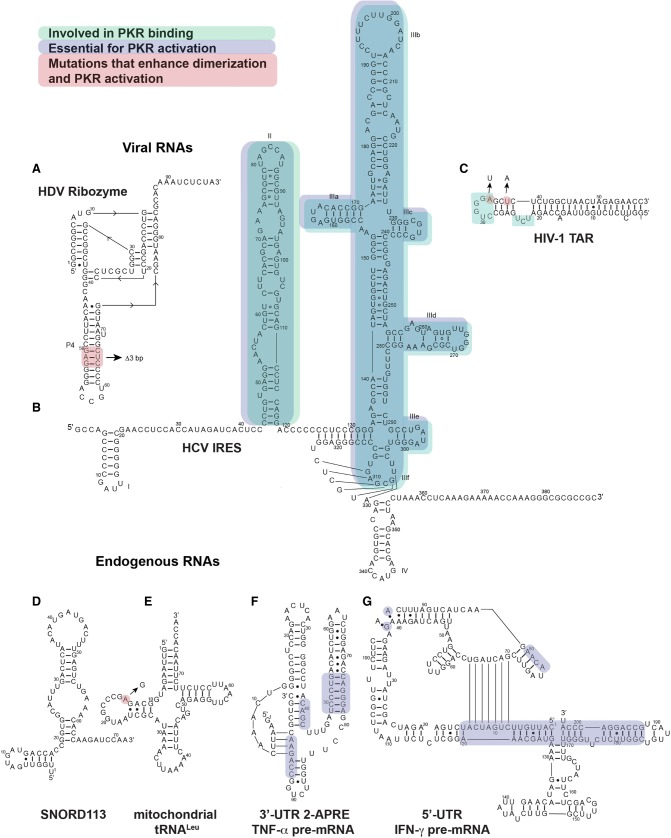
FIGURE 2.
Secondary structures of several naturally occurring RNA activators of PKR. (A) HDV ribozyme, (B) HCV IRES, (C) HIV-1 Tar, (D) SNORD113, (E) human mt-tRNALeu, (F) 3′-UTR 2-APRE of TNF-α pre-Mrna, and (G) 5′-UTR IFN-γ pre-mRNA. Regions relevant to PKR binding are highlighted in cyan, where mutations lead to decreased affinity of PKR. Regions relevant to PKR activation are highlighted in blue, where mutations lead to decreased activation of PKR. Regions that enhance PKR activation by inducing RNA dimerization are highlighted in red.
Purified hepatitis delta virus (HDV) genomic RNA was shown to activate PKR in vitro (Robertson et al. 1996; Circle et al. 1997). The HDV ribozyme domain, responsible for cleaving genomic RNA multimers following viral replication, was found to be the minimal component of the HDV genome capable of activating PKR. The crystal structure of the HDV ribozyme reveals five helical segments forming a nested double pseudoknot, exhibiting extensive coaxial stacking (Ferré-D'Amaré et al. 1998). This helical coaxial stacking could induce dimerization and activation of PKR. Interestingly, deletion of three bases in the P4 loop lead to a misfolded dimeric species that strongly activated PKR (Heinicke and Bevilacqua 2012). Secondary structure probing of this dimeric RNA revealed an extended P4 configuration, resulting in two adjacent 14-bp stem–loops. This extended dsRNA region was proposed to dictate PKR activation by doubling the number of contiguous, stacked pairs, thus satisfying the minimal length requirement for PKR dimerization. Hence, besides multimerization, RNA misfolding appears to be another common trigger for PKR activation (Heinicke and Bevilacqua 2012; Hull and Bevilacqua 2016; Hull et al. 2016).
Translation initiation of Hepatitis C virus (HCV) genome relies on a cap-independent mechanism, involving a highly structured ∼388-nt-long internal ribosome entry site (IRES) located in its 5′-untranslated region (5′-UTR) and is organized into four domains (Fig. 2; Khawaja et al. 2015). Upon IRES binding to the ribosome, eIF3 and eIF2 are recruited, stabilizing the preinitiation complex thus enhancing translation initiation. HCV IRES activates PKR in vitro and enhances eIF2α phosphorylation (Shimoike et al. 2009; Toroney et al. 2010). Interestingly, the isolated domain II, comprised of ∼24 bp and three internal loops, and the fusion construct domain III/IV, comprised of ∼35 bp, bulges and a pseudoknot, were shown to independently activate PKR. Further work is required to establish the synergy, if any, between those domains for maximum PKR activation by full-length HCV IRES. Importantly, HCV IRES-dependent translation was shown to be unaffected by PKR/eIF2α phosphorylation (Shimoike et al. 2009; Karamichali et al. 2014). This hints at a dual pro-viral function of the HCV IRES. In addition to IRES structure-mediated enhancement of translation initiation, the IRES also activates PKR to suppress host cap-dependent translation to gain a competitive advantage.
Endogenous mRNA activators
In addition to playing a key role in the immune response, PKR has been shown to regulate local translation of several specific endogenous mRNAs. This proceeds through the recognition of highly structured regions of their UTRs by the dsRBMs, subsequent activation of the KD, and local suppression of translation initiation through eIF2α phosphorylation. The 3′-UTR of α-tropomyosin mRNA, normally expressed in muscle tissues during differentiation, was shown to inhibit translation in rabbit reticulocyte lysate following PKR activation. In contrast, it did not inhibit translation in wheat germ lysate, a PKR-deficient system. Further analysis showed that purified α-tropomyosin 3′-UTR bound strongly and activated PKR in vitro (Davis and Watson 1996). This implies that PKR can collaborate with 3′-UTR structures to control muscle tissue differentiation and growth. In the course of differentiation, as α-tropomyosin mRNA accumulates, PKR gradually becomes active and curbs the translation of α-tropomyosin mRNA to prevent neoplastic growth.
In addition, PKR regulates its own expression indirectly via its activation by a 5′-UTR. PKR expression is stimulated by the action of interferons (IFNs), including IFN-γ (Katze et al. 2002; Pindel and Sadler 2011). Interestingly, the 5′-terminal 203 nt region of IFN-γ mRNA was shown to specifically activate PKR (Ben-Asouli et al. 2002; Kaempfer 2006; Cohen-Chalamish et al. 2009). Mutations in this region led to enhanced translation of IFN-γ mRNA, with lower levels of phosphorylated PKR. Phylogenetic and mutational analysis of IFN-γ mRNA 5′-UTR identified an H-type pseudoknot and a noncanonical kink-turn, both necessary for PKR autophosphorylation. Those secondary structure features were proposed to align three short helices that together provide a platform long enough for PKR activation (Fig. 2; Cohen-Chalamish et al. 2009). Notably, this region of IFN-γ mRNA also contains the first 26 translated codons, invoking structural rearrangements for its dual function: translation or PKR activation. Since the translation of the CAT mRNA was unaffected when coexpressed with IFN-γ mRNA, it was proposed that PKR activation by this 5′-UTR regulates translation locally. Thus, high levels of IFN-γ protein drive PKR expression while the IFN-γ mRNA increase PKR activation, curbing IFN-γ translation resulting in a negative-feedback loop in which PKR activation attenuates its own expression. This could be an ingenious way by which a robust immune response of limited duration can be triggered upon infection and then gradually tapers off, avoiding harmful effects of excessive or sustained IFN-γ action, i.e., in inflammation.
Tumor necrosis factor-α (TNF-α) is a cytokine regulated by splicing and is essential in protective immunity and inflammatory response. A 2-aminopurine response element (2-APRE), located in the TNF-α3′-UTR, binds and activates PKR in vivo and in vitro (Namer et al. 2017). A conserved pseudoknot that aligns two parallel coaxially stacked helices, individually long enough to accommodate one PKR monomer, was proposed to drive PKR activation (Fig. 2). In this model, binding of one PKR monomer to each parallel helix could bring them into close proximity and allow for on-RNA dimerization and subsequent activation (Namer et al. 2017). Surprisingly, no inhibition of translation, including that of mature TNF-α mRNA, following PKR activation by 2-APRE was observed. Importantly, the splicing of TNF-α pre-mRNA relied on PKR autophosphorylation and downstream phosphorylation of eIF2α (Namer et al. 2017). This novel splicing-based mechanism may provide a fast and efficient way to respond to viral infection or stress. When cellular levels of PKR rise, the splicing and translation of TNF-α pre-mRNA is increased via 2-APRE activation of PKR. This leads to higher levels of TNF-α secretion which induces more PKR expression, forming a positive feedback loop.
Unmodified tRNAs
It has been reported that PKR can be activated both in vivo and in vitro by transfer RNAs (tRNAs) that lack post-transcriptional modifications (Nallagatla et al. 2013). Unmodified tRNAPhe transcripts activate PKR in vitro but not native, extensively modified tRNAPhe. Consistent with this, site-specific incorporation of naturally occurring modifications into the tRNAPhe transcript abolished PKR activation. Interestingly, mitochondrial tRNA (mt-tRNA) extracts from bovine liver activated PKR while cytoplasmic tRNAs did not, despite similar affinities and shared modifications. It has been suggested that dimerization of unmodified tRNAs may be responsible. Indeed, the pathogenic human mt-tRNALeu A14G (Fig. 2) readily dimerizes and was shown to induce PKR autophosphorylation in vitro (Nallagatla et al. 2013). Therefore, post-transcriptional modifications could stabilize tRNA folding thus avoiding misfolded dimers. Alternatively, it could be that the modifications themselves provide local anti-determinants for PKR activation. Since PKR is mainly localized in the cytoplasm with a small fraction in the nucleus (Tian and Mathews 2001), its activation by mt-tRNAs could occur from mitochondrial leakage into the cytoplasm. As such, PKR could be a sentinel for mitochondrial integrity as recently proposed (Kim et al. 2018).
Short double-stranded RNAs with single-stranded tails (ss-dsRNA)
Initially uncovered from in vitro selection experiments that selected for dsRBM/PKR-binding RNAs, RNAs that contain both dsRNA and ssRNA elements have emerged as new, robust PKR activators (Zheng and Bevilacqua 2004; Nallagatla et al. 2007). An imperfect 16-bp or a perfect 15-bp duplex stem flanked by single stranded tails (ss-dsRNA) of 15 nt length on both 5′ and 3′ ends strongly bind and activate PKR autophosphorylation (Nallagatla et al. 2007; Toroney et al. 2012; Mayo et al. 2016; Mayo and Cole 2017). Interestingly, a minimal 10-bp stem is necessary for activation, and the length but not the sequence of the flanking ssRNA is critical for PKR activation (Mayo et al. 2016). Similar to dsRNA activators, a bell-shaped activation curve of PKR autophosphorylation was observed upon titration with increasing ss-dsRNA, implying dimerization driven activation of PKR. The importance of the 5′ phosphorylation state of these ss-dsRNAs has been extensively debated (Nallagatla et al. 2007; Toroney et al. 2012; Mayo et al. 2016). Photochemical cross-linking experiments with 4-thio-U labeled ss-dsRNA identified direct contacts with the dsRBMs as well as the basic patch in the linker that immediately precedes the KD. This led to an intriguing model where a bivalent interaction of ss-dsRNA with both the dsRBMs (via the stem) and the KD (via the flanking ends) drive activation (Mayo and Cole 2017).
Small nucleolar RNAs (snoRNAs), which are typically involved in post-transcriptional modifications of ribosomal RNA and small nuclear RNA (Dupuis-Sandoval et al. 2015), were shown to bind and activate PKR in vivo and in vitro (Youssef et al. 2015). The mechanism by which snoRNAs activate PKR is not well understood. SnoRNAs, such as SNORD113 (Fig. 2), generally do not contain long stretches of dsRNA and are known to harbor regions of ssRNA, reminiscent of the aforementioned ss-dsRNA activators. It is appealing to speculate that snoRNAs may represent natural ss-dsRNA-type activators of PKR. Intriguingly, SNORA3 and SNORD113 required a 5′-triphosphate (5′-ppp) for efficient PKR activation while SNORA71 did not (Youssef et al. 2015). Thus the requirement of a 5′-ppp for PKR activation by ss-dsRNA appears to be variable rather than absolute.
Single-stranded RNAs
Long ssRNAs lacking any discernable secondary structures (such as homopolymeric U30) have also been recently shown to activate PKR in vitro (Toroney et al. 2012; Mayo and Cole 2017). Dimerization of PKR on this ssRNA was observed, corroborated by weak PKR autophosphorylation. Importantly, ssRNAs bound and activated the isolated KD, suggesting that activation is independent of dsRBM binding. Instead, the basic patch neighboring the KD was found to be strictly required for efficient recognition and autophosphorylation of PKR (Mayo and Cole 2017). Similarly to ss-dsRNAs, uncertainty remains whether a 5′-ppp is needed for proper activation.
The notion that PKR can be activated by unstructured ssRNAs via a dsRBM-independent, presumably basic patch-mediated mechanism, opens many new avenues for RNA regulation of PKR in cells. Future experiments will reveal more mechanistic details of how the basic patch and KD interact with RNA activators and coordinate their actions to license enzymatic activation.
RNA inhibitors of PKR
RNA inhibitors of PKR have been identified from both viral and endogenous origins. Down-regulation of PKR through viral RNA presents a major mechanism for viruses to evade host immune response, by inhibiting PKR autophosphorylation and relieving translation inhibition (Dauber and Wolff 2009). In parallel, PKR inhibition by host RNA could set a threshold to avoid inadvertent or premature activation of PKR in noninfected cells. Studies of PKR-inhibitory RNAs have sought to dissect the contributions of RNA length, sequence, secondary and tertiary structures toward PKR inhibition, in order to define a set of distinguishing characteristics for inhibitory versus activating RNAs. However, their sequence and structural diversity, as well as a lack of high-resolution structures, have precluded full mechanistic elucidations, and it remains unknown how these RNA elements confound the immune proteins to escape activation.
Adenovirus virus-associated RNAs (VA)
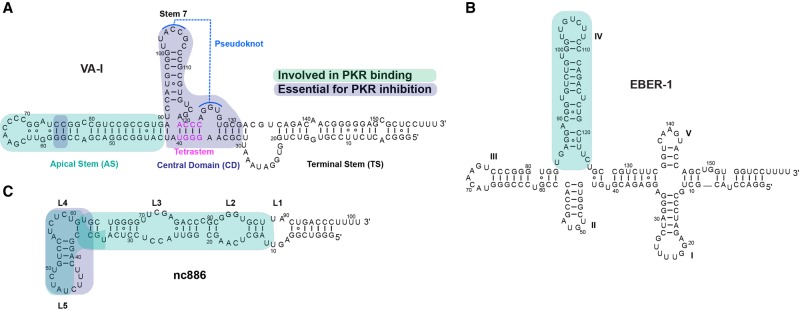
Adenoviruses produce two noncoding RNAs, namely virus-associated RNAs (VA-I and VA-II) involved in PKR inhibition (Vachon and Conn 2016). Both RNAs are highly abundant, GC-rich, and similar in size (∼150–200 nt long) with shared secondary structural features (Mathews and Shenk 1991; Ma and Mathews 1996). The more prevalent and potent species, VA-I, has been extensively characterized. Its secondary structure is comprised of a thermally stable apical stem (AS), a central domain (CD) containing a stem–loop (stem 7) with evidence supporting the formation of a pseudoknot, and a relatively conserved terminal stem (TS) (Clarke and Mathews 1995; Coventry and Conn 2008; Wahid et al. 2008; Launer-Felty and Cole 2014; Wilson et al. 2014; Dzananovic et al. 2017).
VA-I was first recognized as a PKR inhibitor since it is required for efficient translation in infected cells (Thimmappaya et al. 1982; Schneider et al. 1984, 1985; Kitajewski et al. 1986) and it competed with poly I:C for PKR binding, leading to reduced levels of phosphorylated PKR (Mathews and Shenk 1991). VA-I AS is sufficient for binding both full-length PKR and isolated dsRBMs with similar affinities, but does not inhibit PKR (Clarke and Mathews 1995). Importantly, artificial stabilization of a side bulge in the AS stem (C75G:C76G mutant) decreases dsRBM binding and compromises PKR inhibition (Wahid et al. 2009). Hence, dsRBMs binding to VA-I AS is necessary but not sufficient for proper PKR inhibition (McKenna et al. 2006; Wahid et al. 2008; Launer-Felty and Cole 2014; Wilson et al. 2014; Launer-Felty et al. 2015; Dzananovic et al. 2017). Secondary structure probing by SHAPE and DMS support the formation of a pseudoknot at the base of VA-I CD, through the long-range pairing of two trinucleotide sequences ACC (103–105) in loop 8 and the compensatory GGU (124–126) trinucleotide from loop 10. Surprisingly, mutations disrupting the pseudoknot had a limited effect on PKR inhibition (Coventry and Conn 2008; Wilson et al. 2014). Truncations or mismatches in stem 7 in the CD strongly reduced PKR inhibition and converted VA-I to an activator. The function of stem 7 may be to prevent the AS and TS from coaxial stacking to form a long dsRNA activation platform. Alternatively, stem 7 may directly participate in dsRBM binding and PKR inhibition (Dzananovic et al. 2017). In addition, point mutations in the proximal region of the AS (A91U) and in a strictly conserved tetrastem (37–40 GGGU and 119–121 CCCA), in stem 4 of the CD adjacent to stem 7, resulted in weaker binding to PKR and a significant loss of PKR inhibition. A binding stoichiometry of 1:1 PKR:VA-I was identified leading to a model where VA-I strongly binds one PKR monomer thus sequestrating it and blocking its dimerization presumably by steric hindrance (McKenna et al. 2006; Launer-Felty and Cole 2014; Launer-Felty et al. 2015). Intriguingly, point mutations in the loop 2 of the TS were shown to reduce VA-I inhibition while removal of the entire TS enhanced it (Wahid et al. 2008). Though the TS was proposed to be unnecessary for PKR inhibition, conformational changes or misfolding of VA-I termini could impact PKR regulation. Indeed, VA-I was shown to be cleaved by Dicer at its TS downstream from loop 2, suppressing adenoviral replication due to increased phosphorylation levels of eIF2α (Machitani et al. 2016).
Epstein-Barr virus-encoded small RNAs (EBER)
Epstein-Barr virus (EBV) is associated with several human malignancies and its biological function has been studied extensively (Iwakiri 2016). Like adenoviruses, EBV encodes two structured noncoding RNAs, EBER-1 (∼167 nt) and EBER-2 (∼172 nt), that share 54% sequence identity (Rosa et al. 1981). EBER-1 is 10 times more abundant in cells than EBER-2, mirroring the relative abundance of VA-I and VA-II RNAs. EBER RNAs are essential for most EBV-mediated oncogenic activities, although some strains lacking EBER RNAs have also been identified (Clarke et al. 1992; Nanbo et al. 2002). Extensive debate remains regarding EBER function since its cellular localization is controversial (either solely nucleolar or nucleocytoplasmic) (Clarke et al. 1992; Ruf et al. 2005; Fok et al. 2006). Nonetheless, EBER-1 was shown to bind to several host proteins such as La, ribosomal protein L22, and PKR etc., implying its potential involvement in multiple regulatory pathways. Interestingly, EBER-1 inhibits PKR activation by HIV TAR similarly to VA-I in vitro (Sharp et al. 1993; Elia et al. 2004). EBER RNAs can partially compensate for the loss of VA-I in adenoviruses for replication and infectivity (Bhat and Thimmappaya 1983). Nevertheless, further work is required to clearly establish the antagonist function, if any, of EBER on PKR in vivo. The EBER-1 secondary structure comprises five stem–loops that converge at two four-way junctions (Fig. 3). Stem IV (∼13 bp) binds PKR dsRBMs as efficiently as full-length EBER-1, but lacks any inhibitory properties, implying that other elements in EBER-1 structure are required for inhibition (Vuyisich et al. 2002; McKenna et al. 2006). Although this is reminiscent of VA-I AS (∼19 bp), a stoichiometry of 2:1 for dsRBMs:EBER-1 and dsRBMs:Stem IV was reported based on isothermal titration calorimetry (ITC) analyses, suggesting inhibition in a dimeric form. This raises the intriguing possibility that in contrast to VA-I sequestration of PKR in its monomeric form, EBER-1 could induce an inactive dimeric state of PKR to block its trans-phosphorylation. Alternatively, the two PKR monomers trapped on the same EBER-1 RNA may not assume a dimeric configuration. Further investigations are required to distinguish these possibilities.
FIGURE 3.
Secondary structures of PKR-inhibitory RNAs. (A) VA-I, (B) EBER-1, and (C) nc886. Regions involved in PKR binding are highlighted in cyan while those required for inhibition are in blue.
Human noncoding RNA 886 (nc886)
Recently, an endogenous 101 nt long RNA, human noncoding RNA 886 (nc886) was characterized as a PKR inhibitor curbing the basal level of PKR phosphorylation to guard against premature commitment to translation inhibition (Lee et al. 2011, 2016; Fort et al. 2018). nc886 was shown to be repressed in some cancer cell lines leading to higher levels of activated PKR. Strong binding of nc886 to PKR and the isolated dsRBMs was observed by gel-shift assays and surface plasmon resonance (SPR) (Jeon et al. 2012; Calderon and Conn 2017). In addition, nc886 effectively competed with poly I:C, inducing PKR inhibition in vitro. Interestingly, in vitro transcribed nc886 RNA partitions into a slow- and a fast-migrating species, termed conformer 1 and 2, respectively. Separation of the two conformers followed by SHAPE probing identified two distinct structural configurations. While both share a similar single elongated stem structure consisting of ∼25 canonical and noncanonical base pairs and four interior loops (L1–L4; Fig. 3), clear differences were observed in the apical loop, termed L5. L5 is predicted to be involved in PKR binding, as assessed by RNase accessibility. Importantly, conformer 1 exhibited robust binding to PKR and inhibition while conformer 2 only weakly bound PKR and acted as a pseudoactivator. Truncations in the apical stem (L4 and L5) strongly reduced conformer 1 inhibition while a terminal stem truncation had no marked effects. If confirmed, nc886 could represent an endogenous bistable regulatory RNA whose conformational flexibility modulates PKR activity under different cellular states. However, current data do not exclude the possibility that nc886 conformer 1 is in a higher oligomeric state.
Taken together, a coherent theme among known PKR-inhibiting RNAs has yet to emerge and will likely require detailed structural analyses. While many parallels exist between VA and EBER RNAs (length, termini, portions of the secondary structures), their overall structures and mechanisms of action seem to diverge. In addition, understanding the two conformers of nc886 could shed light on the necessary structural features important for PKR inhibition.
Protein regulators of PKR
In addition to RNA-dependent PKR regulation, a number of endogenous and viral proteins have been identified as regulators of PKR in different cellular contexts (Dauber and Wolff 2009). In contrast to the aforementioned RNAs that have been extensively studied in vitro by mutational, functional, and biophysical methods, most of the available work regarding protein-mediated PKR regulation have relied on yeast two-hybrid or coimmunoprecipitation assays. In this section, we review some of the most characterized protein regulators of PKR and describe their proposed binding interfaces (Fig. 4). The first category includes proteins that compete with PKR for binding to RNA activators and/or bind and sequester PKR dsRBMs. The second category relies on binding to PKR's KD through contacts to its N- or C-lobe thus inhibiting its activation and autophosphorylation or competing with its substrate eIF2α in case of preactivated PKR.
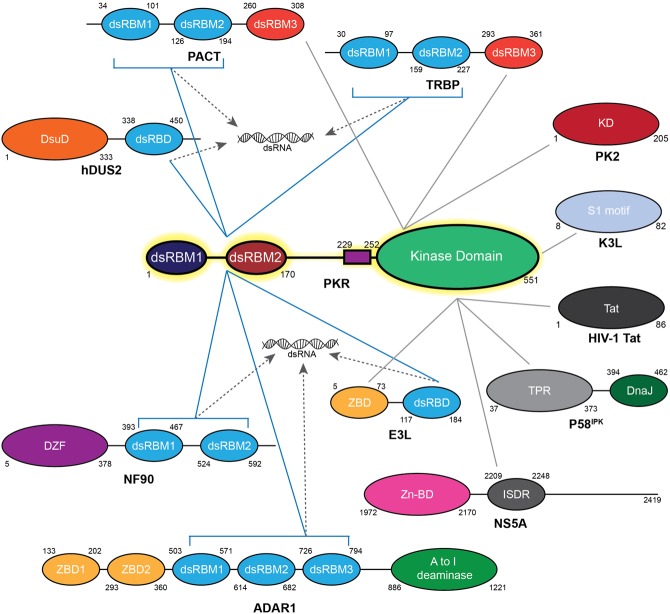
FIGURE 4.
Schematic representation of protein regulators of PKR with their interacting interfaces. Blue lines represent interactions with PKR dsRBMs, while gray lines denote interactions with the Ser/Thr KD. Dashed lines highlight direct competition for dsRNA binding. The domain organization of PKR protein regulators and their boundaries are represented following the color code: Class A's in light blue, Class B's in red, Z-DNA binding domain in yellow, DZF dimerization domain in purple, A to I deaminase catalytic domain in light green, dihydrouridine synthase conserved catalytic domain in orange, Zinc binding domain in pink, DnaJ domain in dark green, ISDR motif in dark gray, and TPR repeat motifs in light gray.
Proteins that target the PKR regulatory domain
Nuclear factor 90
Nuclear factor 90 (NF90) regulates gene expression at transcriptional, post-transcriptional and translation levels by binding miRNAs or modulating the translocation or translation of mRNAs (Jayachandran et al. 2016). More recently, NF90 was shown to exert an antiviral activity since NF90 knockout led to enhanced viral sensitivity (Wen et al. 2014; Li et al. 2016). This has been attributed to NF90's involvement in several host antiviral mechanisms, including relief of PKR inhibition by viral proteins (Wen et al. 2014; Li et al. 2016). Indeed, influenza A virus produces a nonstructural protein 1 (NS1) that inhibits PKR through direct dsRBM binding and/or RNA sequestration. NF90 was shown to bind to NS1 with higher affinity and thus alleviates its inhibition of PKR. Furthermore, direct binding of NF90 to PKR was detected (Parker et al. 2001). NF90 is comprised of a DZF motif at its amino terminus and two dsRBMs at its carboxyl terminus. The latter was identified as the primary PKR binding site, with dsRBM1 of NF90 playing a predominant role while the involvement of its DZF motif is debated. Interestingly, PKR-dependent phosphorylation of NF90 in its dsRBMs was observed, altering intracellular distribution of NF90 from the nucleus to the cytoplasm without changing its RNA affinity (Parker et al. 2001; Parrott et al. 2005). Conversely, NF90 was also shown to inhibit PKR and its overexpression blocks PKR activation during viral infection. This exemplifies the delicate balance and interplay between PKR and NF90 activities, making NF90 an unusual case of dual regulator/substrate of PKR.
tRNA-dihydrouridine synthase 2
Human tRNA-dihydrouridine synthase 2 (hDus2) catalyzes post-transcriptional reduction of uridine into dihydrouridine in tRNAs. Its RNA recognition mechanism was recently shown to rely on a dsRBM while the modification is performed by the conserved Dus domain (Bou-Nader et al. 2015). hDus2 was shown to directly bind to PKR (Mittelstadt et al. 2008). This interaction is mediated by contacts between the hDus2 dsRBM and the tandem dsRBMs of PKR, chiefly involving dsRBM1. Importantly, hDus2 was shown to inhibit PKR autophosphorylation in vitro and in vivo and to block PKR-induced apoptosis upon ER stress, leading to a model where elevated hDus2 expression could down-regulate PKR (Mittelstadt et al. 2008).
ADAR1
Adenosine deaminase acting on RNA (ADAR1) catalyzes the C6 deamination of adenosine (A) to inosine (I) in RNA, termed RNA editing (Pfaller et al. 2011). Since inosine is decoded as guanosine instead of adenosine, RNA editing recodes the message and can also alter the RNA structure. ADAR1 comprises two Z-DNA binding motifs (ZBM) followed by three dsRBMs and the catalytic deaminase domain at its carboxyl terminus. ADAR1 was shown to enhance HIV-I viral replication by inhibiting PKR autophosphorylation (Nie et al. 2007; Clerzius et al. 2009). This was initially proposed to proceed through dsRNA sequestration by ADAR1 as well as its RNA editing activity giving rise to destabilized I:U mismatches in dsRNA. However, constructs lacking the catalytic deaminase domain were still able to inhibit PKR activation in vivo, as did a minimal variant harboring only the three dsRBMs (Wang and Samuel 2009). Coimmunoprecipitation identified direct interaction between ADAR1 and PKR with ADAR1 dsRBM1 being essential for binding and inhibition. It remains unclear whether ADAR1 dsRBM1 is sufficient for PKR inhibition and which region of PKR is targeted. Importantly, the editing function of ADAR was shown to tightly suppress PKR activation by endogenous RNAs such as Alu repeats (Toth et al. 2009; George et al. 2016; Chung et al. 2018). Deamination was proposed to destabilize intra- or intermolecular interactions between Alu repeats thus avoiding the formation of long endogenous dsRNAs, tuning the balance between perceived self versus nonself RNAs (Kim et al. 2014).
Post-translational regulation by ISG15 and SUMO proteins
All proposed models of PKR regulation mentioned so far rely on noncovalent interactions. Recently, covalent modifications targeting both dsRBMs of PKR have been reported. The ubiquitin-like protein ISG15 and its conjugation, termed ISGylation, are strongly induced by type-I interferons upon innate immune response (Haas et al. 1987). Interestingly, PKR was shown to be ISGylated in vivo at K69 in dsRBM1 and K159 in dsRBM2 giving rise to a constitutively autophosphorylated kinase even in the absence of viral infection (Okumura et al. 2013). Similarly, the small ubiquitin-like modifier SUMO1 and SUMO2 were shown to attach to PKR via three SUMOylation sites at K60 in dsRBM1, K150 in dsRBM2, and K440 in KD leading to activation of PKR (de la Cruz-Herrera et al. 2014). SUMOylation did not alter cellular repartition of PKR but SUMOylated PKR was observed mainly in the nucleus. Since all those covalent dsRBMs modifications lie on their predicted RNA binding interface, the effects of alanine substitutions of modification sites need to be interpreted with care. Not only ISGylation and SUMOylation are blocked by the substitutions, RNA binding is also reduced leading to nonactivation of PKR despite viral infection.
Proteins that target the PKR kinase domain
PACT/TRBP
Transactivation response element RNA-binding protein (TRBP) and PKR activating protein (PACT) are intimately involved in RNA interference by tuning both cleavage and strand selection of miRNAs during formation of the RISC complex (Heyam et al. 2015). TRBP and PACT are comprised of three dsRBMs with the first two (M1, M2) being involved in RNA binding while the third motif (M3) was identified as a Class-B dsRBM, solely interacting with proteins. Interestingly, both proteins were shown to interact with PKR but dictate different outcomes (Gupta et al. 2003). Early on, TRBP was identified as a negative regulator of PKR in vivo (Park et al. 1994; Benkirane et al. 1997). In contrast, PACT induces PKR activation and autophosphorylation upon cellular stress (Patel and Sen 1998). In both cases, the first two dsRBMs of TRBP/PACT (M1, M2) interact with the dsRBMs of PKR while their third dsRBM (M3) binds to the KD (Li et al. 2006; Peters et al. 2009; Chukwurah et al. 2018). Removal of M3 does not block PKR binding but abolishes both inhibition and activation. Importantly, swapping M3 between TRBP and PACT in chimeric constructs inverted their functions, proving that each M3 dictates a specific PKR regulatory outcome independently of M1 and M2 (Gupta et al. 2003). In the absence of stress, TRBP and PACT form homo- and hetero-dimers. Under stress conditions, phosphorylation at S287 of PACT M3 was identified as a hallmark for downstream activation of PKR. P-S287 was shown to reduce PACT affinity for TRBP while increasing affinity for PKR in vivo (Singh et al. 2011). Indeed, the S287D substitution in PACT leads to activation of PKR and apoptosis even in the absence of stress. Conversely, phosphorylation of TRBP at S142, S152, S283, and S286 was shown to enhance TRBP-PKR interaction thus allowing cellular recovery (Nakamura et al. 2015; Chukwurah and Patel 2018). This illustrates the delicate balance between TRBP, PACT and PKR activities during stress response and recovery.
P58IPK
P58IPK (58 kDa inhibitor of protein kinase) is a cochaperone regulating the specificity of BiP chaperone (binding immunoglobulin protein), and consists of nine amino-terminal tetratricopeptide repeats (TPRs) and a DnaJ domain at its carboxyl terminus (Svärd et al. 2011). Influenza virus hijacks endogenous P58IPK to inhibit PKR and rescue viral translation (Tang et al. 1996; Goodman et al. 2007, 2011). In the absence of stress, P58IPK is in an inhibited state bound to Hsp40 or P52rIPK. However, viral infection was shown to relieve P58IPK inhibition through an unknown mechanism. Direct interaction between P58IPK and PKR was observed and occurs through the N-lobe of the KD (Tang et al. 1996). Removal of the TPR6 motif of P58IPK alleviates PKR inhibition while mutating the conserved HPD motif in the DnaJ domain had no impact (Yan et al. 2002). Importantly, P58IPK binding was further shown to block dimerization of PKR. This can be rationalized as P58IPK binds at the dimeric interface of PKR N-lobe, thus directly competing with another PKR monomer.
E3L
Poxviruses rely on the multifunctional Vaccinia virus E3L protein to escape host innate immune response and to achieve full pathogenicity (Langland and Jacobs 2002; Kwon and Rich 2005; Koehler et al. 2017). E3L contains an amino-terminal Z-DNA binding domain (ZBD) followed by a carboxy-terminal dsRBM (Ha et al. 2004). E3L was proposed to sequester PKR RNA activators through its dsRBM. Additionally, direct interaction between E3L's dsRBM and PKR-dsRBMs as well as E3L's ZBD and the PKR KD were observed, leading to PKR inhibition (Sharp et al. 1998; Langland and Jacobs 2004; Dueck et al. 2015). Therefore, E3L suppresses PKR activity through either dsRNA sequestration or direct KD association, or both. It was proposed that the latter could be especially important during late stages of infection when accumulated levels of viral dsRNAs surpass that of E3L in the cell, thus triggering an immune response. Curiously, E3L was shown to inhibit both PKR autophosphorylation as well as eIF2α phosphorylation by activated PKR in vitro making E3L a rare example of PKR regulator capable of targeting both its pre and post-activation states.
K3L
In addition to E3L, poxviruses encode a second PKR inhibitor, K3L, to escape host immune response (Langland and Jacobs 2002). K3L exhibits a 28% sequence identity with the NTD of eIF2α, suggesting possible interaction with PKR KD (Kawagishi-Kobayashi et al. 1997). Indeed, K3L was shown to directly bind to the KD C-lobe of PKR in vivo and to inhibit eIF2α phosphorylation in vivo and in vitro (Craig et al. 1996; Dar and Sicheri 2002). In contrast to E3L, K3L could not inhibit PKR autophosphorylation and no interaction was observed with the unphosphorylated kinase. This led to a model where K3L acts as a pseudosubstrate inhibitor of activated PKR by competing with eIF2α binding. The X-ray structure of K3L showed a remarkable mimicry of the eIF2α OB domain with the exception of a rigid helical insert in K3L contrasting with the flexible loop harboring S51 in eIF2α (Dar and Sicheri 2002; Seo et al. 2008). Nonetheless, extensive phylogenetic and functional analyses identified rapid evolution of PKR maintaining eIF2α binding and phosphorylation while evading K3L inhibition (Elde et al. 2009; Rothenburg et al. 2009). This ability of PKR to counteract viral mimicry was attributed to the plasticity of the eIF2α structure.
PK2
Another example of viral mimicry comes from baculovirus protein, PK2, that was shown to inhibit eIF2α phosphorylation in vivo in plants and insects (Dever et al. 1998; Li et al. 2015). Interestingly, PK2 was proposed to share a common fold with the C-lobe of eIF2α kinases, including PKR. Thus, PK2 was described as a truncated eIF2α kinase homolog (Dever et al. 1998). Recently, PK2 was shown to bind to the PKR N-lobe and enhance ATP dissociation from the active site (Li et al. 2015). In addition, PK2 inhibited eIF2α phosphorylation by PKR both in vivo and in vitro despite PK2's inability to bind eIF2α. A domain-swapping model was proposed where PK2 binds to the N-lobe of PKR thus displacing the C-lobe via its mimicry (Li et al. 2015). Nevertheless, baculovirus hosts lack PKR and as such PK2 probably evolved to inhibit another eIF2α kinase. Inhibition of PKR through mimicry of its C-lobe could be a mechanism used by other pathogens that has yet to be identified.
NS5A
Hepatitis C virus (HCV) encodes a single polyprotein that is post-translationally cleaved into individual structural and nonstructural proteins with diverse functions that support virulence, among which is the viral nonstructural 5A protein (NS5A) (He and Katze 2002). NS5A harbors a Zinc-binding motif at its amino terminus followed by the interferon sensitivity-determining region (ISDR) comprised of 39 amino acids and a longer poorly characterized carboxyl terminus (Tellinghuisen et al. 2005). Mutations in NS5A ISDR have been found in many HCV strains that resist interferon response (Gale et al. 1997, 1998; Karamichali et al. 2014). Interestingly, NS5A was shown to directly bind to PKR. The binding interface was mapped to the N-lobe of PKR KD with the ISDR being crucial but not sufficient (Gale et al. 1997, 1998). A minimal construct of 65 amino acids comprising the ISDR and 26 amino acids at its carboxyl terminus can bind PKR as efficiently as the full-length NS5A. Importantly, NS5A inhibited PKR activation and autophosphorylation both in vitro and in vivo rescuing viral translation. Therefore, it was proposed that NS5A inhibits PKR by binding to its N-lobe thus blocking its dimerization and activation.
HIV-I Tat
Human immunodeficiency virus type 1 (HIV-I) produces the trans-activating protein (Tat) crucial for viral replication and propagation (Romani et al. 2010). Tat stimulates HIV-I gene expression by enhancing transcription upon binding to the transactivation response (TAR) element present in the 5′- and 3′-UTRs of the HIV genome. Tat consists of a cysteine-rich region (22–37), a hydrophobic core (38–48), a basic region (49–57), a glutamine-rich region (58–72), and ends with an RGD motif. Both constitutively expressed isoforms of Tat (Tat-86 and Tat-72) were shown to bind to PKR via their basic patch in vivo (McMillan et al. 1995; Vivarini Áde et al. 2015). Interestingly, Tat was shown to inhibit PKR activation by dsRNA in vitro through RNA sequestration (Cai et al. 2000). Conversely, both isoforms were shown to be phosphorylated by activated PKR at S62, T64 and S68, leading to enhanced TAR binding in vitro (Brand et al. 1997; Yoon et al. 2015). Tat binding to PKR competed with eIF2α thus blocking its phosphorylation and promoting cell growth. Recently, phosphorylation of Tat at T23, T40, S46, S62, and S68 by PKR was shown to suppress HIV-I replication by abolishing Tat binding to TAR and cyclin T1 as well as inhibiting Tat nuclear translocation (Yoon et al. 2015). Taken together, Tat binding to and phosphorylation by PKR seems to either enhance or diminish HIV-I transcription depending on cellular contexts.
Conclusions
PKR is a leading molecular sentinel in innate immunity and as such has to recognize a diverse set of stress-inducing stimuli to mount an appropriate defensive response. At the front lines of intracellular antiviral surveillance, PKR encounters and endures a constant onslaught of foreign nucleic acids and proteins and must distinguish self versus nonself versions of these macromolecules. Forced to co-evolve with diverse fast-evolving pathogens, PKR engages in a perpetual tug-of-war with viral and parasitic entities that seek every chance to evade, hoax, or disable this powerful protein defender. Its flexible architecture and rapid evolution have allowed for direct and allosteric modulation by a plethora of RNAs, proteins, and small molecules. Extensive investigations have identified versatile endogenous and exogenous regulators of this crucial kinase. They target essentially all aspects of PKR action from activator (dsRNA) availability, RNA interaction, auto-inhibition relief, dimerization, kinase activation, substrate (eIF2α) interaction, etc. Even more regulators are likely to emerge, especially with the accelerated discovery of novel noncoding RNAs with unusual topologies (e.g., circular RNAs), structure (e.g., riboswitches and ribozymes), and chemical elaborations (e.g., m6A, m1A).
RNA regulators seem to primarily target the dsRBMs and the interdomain linker. It is tantalizing to envisage RNA regulators (especially those with ssRNA tails) that may directly manipulate the KD while being anchored through the dsRBM contacts. How PKR dsRBMs discern and distinguish between activating or inhibitory RNAs remain enigmatic and await high-resolution structural information. RNA activation of PKR usually requires a dsRNA platform of sufficient length, which can also be constructed by RNA oligomerization, misfolding, or coaxial stacking of topologically adjacent helices. The latter is frequently assisted by tertiary structure elements such as K-turns, pseudoknots, etc. and long-range interactions. It is likely that the sequence, helical orientations, symmetry/asymmetry, and flexibility/deformability/dynamics of the RNA structure also modulate how the dsRBMs approach and engage the A-form regions and instruct the kinase accordingly. In contrast, protein regulators appear to use more diverse strategies (at least, known to us) for PKR regulation by targeting either the dsRBMs or the KD and by modulating dsRNA or eIF2α binding. It is likely that different protein and RNA modules interact with PKR dsRBMs in fundamentally similar manners but lead to differential outcomes due to nuances of the mechanisms of activation. Lastly, while known RNA regulators act on PKR in its inactive state, PKR-regulating proteins can act on both the kinase-inactive state (preactivation) and the activated state (post-activation). Identifying one or several coherent, convergent mechanism(s) shared among PKR regulators could shed light on how different environmental and cellular cues trigger a productive or unproductive configuration of the kinase by exploiting the versatility of its modular organization.
ACKNOWLEDGMENTS
We thank K. Suddala and S. Li for insightful discussions. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institutes of Health.
Author contributions: All authors contributed to the preparation of the figures and wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.070169.118.
Freely available online through the RNA Open Access option.
REFERENCES
- Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. 2004. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem 279: 22243–22249. 10.1074/jbc.M314326200 [DOI] [PubMed] [Google Scholar]
- Anderson E, Cole JL. 2008. Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions. Biochemistry 47: 4887–4897. 10.1021/bi702211j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. 2002. Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 108: 221–232. 10.1016/S0092-8674(02)00616-5 [DOI] [PubMed] [Google Scholar]
- Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, Jeang KT. 1997. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J 16: 611–624. 10.1093/emboj/16.3.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, George CX, Samuel CE, Cech TR. 1998. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry 37: 6303–6316. 10.1021/bi980113j [DOI] [PubMed] [Google Scholar]
- Bhat RA, Thimmappaya B. 1983. Two small RNAs encoded by Epstein-Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc Natl Acad Sci 80: 4789–4793. 10.1073/pnas.80.15.4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Nader C, Pecqueur L, Bregeon D, Kamah A, Guérineau V, Golinelli-Pimpaneau B, Guimarães BG, Fontecave M, Hamdane D. 2015. An extended dsRBD is required for post-transcriptional modification in human tRNAs. Nucleic Acids Res 43: 9446–9456. 10.1093/nar/gkv989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand SR, Kobayashi R, Mathews MB. 1997. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem 272: 8388–8395. 10.1074/jbc.272.13.8388 [DOI] [PubMed] [Google Scholar]
- Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. 2000. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys 373: 361–367. 10.1006/abbi.1999.1583 [DOI] [PubMed] [Google Scholar]
- Calderon BM, Conn GL. 2017. Human noncoding RNA 886 (nc886) adopts two structurally distinct conformers that are functionally opposing regulators of PKR. RNA 23: 557–566. 10.1261/rna.060269.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwurah E, Patel RC. 2018. Stress-induced TRBP phosphorylation enhances its interaction with PKR to regulate cellular survival. Sci Rep 8: 1020 10.1038/s41598-018-19360-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwurah E, Willingham V, Singh M, Castillo-Azofeifa D, Patel RC. 2018. Contribution of the two dsRBM motifs to the double-stranded RNA binding and protein interactions of PACT. J Cell Biochem 119: 3598–3607. 10.1002/jcb.26561 [DOI] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, et al. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172: 811–824 e814. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circle DA, Neel OD, Robertson HD, Clarke PA, Mathews MB. 1997. Surprising specificity of PKR binding to δ agent genomic RNA. RNA 3: 438–448. [PMC free article] [PubMed] [Google Scholar]
- Clarke PA, Mathews MB. 1995. Interactions between the double-stranded RNA binding motif and RNA: definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA 1: 7–20. [PMC free article] [PubMed] [Google Scholar]
- Clarke PA, Sharp NA, Clemens MJ. 1992. Expression of genes for the Epstein-Barr virus small RNAs EBER-1 and EBER-2 in Daudi Burkitt's lymphoma cells: effects of interferon treatment. J Gen Virol 73: 3169–3175. 10.1099/0022-1317-73-12-3169 [DOI] [PubMed] [Google Scholar]
- Clerzius G, Gelinas JF, Daher A, Bonnet M, Meurs EF, Gatignol A. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol 83: 10119–10128. 10.1128/JVI.02457-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Chalamish S, Hasson A, Weinberg D, Namer LS, Banai Y, Osman F, Kaempfer R. 2009. Dynamic refolding of IFN-γ mRNA enables it to function as PKR activator and translation template. Nat Chem Biol 5: 896–903. 10.1038/nchembio.234 [DOI] [PubMed] [Google Scholar]
- Cole JL. 2007. Activation of PKR: an open and shut case? Trends Biochem Sci 32: 57–62. 10.1016/j.tibs.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coventry VK, Conn GL. 2008. Analysis of adenovirus VA RNAI structure and stability using compensatory base pair modifications. Nucleic Acids Res 36: 1645–1653. 10.1093/nar/gkn020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AW, Cosentino GP, Donzé O, Sonenberg N. 1996. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem 271: 24526–24533. 10.1074/jbc.271.40.24526 [DOI] [PubMed] [Google Scholar]
- Dar AC, Sicheri F. 2002. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell 10: 295–305. 10.1016/S1097-2765(02)00590-7 [DOI] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F. 2005. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell 122: 887–900. 10.1016/j.cell.2005.06.044 [DOI] [PubMed] [Google Scholar]
- Dauber B, Wolff T. 2009. Activation of the antiviral Kinase PKR and viral countermeasures. Viruses 1: 523–544. 10.3390/v1030523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Watson JC. 1996. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human α-tropomyosin. Proc Natl Acad Sci 93: 508–513. 10.1073/pnas.93.1.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz-Herrera CF, Campagna M, García MA, Marcos-Villar L, Lang V, Baz-Martínez M, Gutiérrez S, Vidal A, Rodríguez MS, Esteban M, et al. 2014. Activation of the double-stranded RNA-dependent protein kinase PKR by small ubiquitin-like modifier (SUMO). J Biol Chem 289: 26357–26367. 10.1074/jbc.M114.560961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Sripriya R, McLachlin JR, Lu J, Fabian JR, Kimball SR, Miller LK. 1998. Disruption of cellular translational control by a viral truncated eukaryotic translation initiation factor 2α kinase homolog. Proc Natl Acad Sci 95: 4164–4169. 10.1073/pnas.95.8.4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. 2005. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2α substrate recognition. Cell 122: 901–913. 10.1016/j.cell.2005.06.041 [DOI] [PubMed] [Google Scholar]
- Dey M, Cao C, Sicheri F, Dever TE. 2007. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J Biol Chem 282: 6653–6660. 10.1074/jbc.M607897200 [DOI] [PubMed] [Google Scholar]
- Dey M, Velyvis A, Li JJ, Chiu E, Chiovitti D, Kay LE, Sicheri F, Dever TE. 2011. Requirement for kinase-induced conformational change in eukaryotic initiation factor 2α (eIF2α) restricts phosphorylation of Ser51. Proc Natl Acad Sci 108: 4316–4321. 10.1073/pnas.1014872108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey M, Mann BR, Anshu A, Mannan MA. 2014. Activation of protein kinase PKR requires dimerization-induced cis-phosphorylation within the activation loop. J Biol Chem 289: 5747–5757. 10.1074/jbc.M113.527796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck KJ, Hu YS, Chen P, Deschambault Y, Lee J, Varga J, Cao J. 2015. Mutational analysis of vaccinia virus E3 protein: the biological functions do not correlate with its biochemical capacity to bind double-stranded RNA. J Virol 89: 5382–5394. 10.1128/JVI.03288-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Sandoval F, Poirier M, Scott MS. 2015. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6: 381–397. 10.1002/wrna.1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzananovic E, Astha, Chojnowski G, Deo S, Booy EP, Padilla-Meier P, McEleney K, Bujnicki JM, Patel TR, McKenna SA. 2017. Impact of the structural integrity of the three-way junction of adenovirus VAI RNA on PKR inhibition. PLoS One 12: e0186849 10.1371/journal.pone.0186849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. 2009. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457: 485–489. 10.1038/nature07529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A, Vyas J, Laing KG, Clemens MJ. 2004. Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein-Barr virus small RNA EBER-1. Eur J Biochem 271: 1895–1905. 10.1111/j.1432-1033.2004.04099.x [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Zhou K, Doudna JA. 1998. Crystal structure of a hepatitis δ virus ribozyme. Nature 395: 567–574. 10.1038/26912 [DOI] [PubMed] [Google Scholar]
- Fok V, Friend K, Steitz JA. 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol 173: 319–325. 10.1083/jcb.200601026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort RS, Mathó C, Geraldo MV, Ottati MC, Yamashita AS, Saito KC, Moreira Leite KR, Méndez M, Maedo N, Méndez L, et al. 2018. Nc886 is epigenetically repressed in prostate cancer and acts as a tumor suppressor through the inhibition of cell growth. BMC Cancer 18: 127 10.1186/s12885-018-4049-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MJ Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230: 217–227. 10.1006/viro.1997.8493 [DOI] [PubMed] [Google Scholar]
- Gale MJ Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol 18: 5208–5218. 10.1128/MCB.18.9.5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelev V, Aktas H, Marintchev A, Ito T, Frueh D, Hemond M, Rovnyak D, Debus M, Hyberts S, Usheva A, et al. 2006. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J Mol Biol 364: 352–363. 10.1016/j.jmb.2006.08.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Ramaswami G, Li JB, Samuel CE. 2016. Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem 291: 6158–6168. 10.1074/jbc.M115.709014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AG, Smith JA, Balachandran S, Perwitasari O, Proll SC, Thomas MJ, Korth MJ, Barber GN, Schiff LA, Katze MG. 2007. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J Virol 81: 2221–2230. 10.1128/JVI.02151-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AG, Tanner BC, Chang ST, Esteban M, Katze MG. 2011. Virus infection rapidly activates the P58(IPK) pathway, delaying peak kinase activation to enhance viral replication. Virology 417: 27–36. 10.1016/j.virol.2011.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Huang X, Patel RC. 2003. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology 315: 283–291. 10.1016/S0042-6822(03)00589-0 [DOI] [PubMed] [Google Scholar]
- Ha SC, Lokanath NK, Van Quyen D, Wu CA, Lowenhaupt K, Rich A, Kim YG, Kim KK. 2004. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zα bound to DNA. Proc Natl Acad Sci 101: 14367–14372. 10.1073/pnas.0405586101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Ahrens P, Bright PM, Ankel H. 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem 262: 11315–11323. [PubMed] [Google Scholar]
- He Y, Katze MG. 2002. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol 15: 95–119. 10.1089/088282402317340260 [DOI] [PubMed] [Google Scholar]
- Heinicke LA, Bevilacqua PC. 2012. Activation of PKR by RNA misfolding: HDV ribozyme dimers activate PKR. RNA 18: 2157–2165. 10.1261/rna.034744.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC. 2009. RNA dimerization promotes PKR dimerization and activation. J Mol Biol 390: 319–338. 10.1016/j.jmb.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyam A, Lagos D, Plevin M. 2015. Dissecting the roles of TRBP and PACT in double-stranded RNA recognition and processing of noncoding RNAs. Wiley Interdiscip Rev RNA 6: 271–289. 10.1002/wrna.1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang HD, Graber TE, Alain T. 2018. Battling for ribosomes: translational control at the forefront of the antiviral response. J Mol Biol 430: 1965–1992. 10.1016/j.jmb.2018.04.040 [DOI] [PubMed] [Google Scholar]
- Hull CM, Bevilacqua PC. 2016. Discriminating self and non-self by RNA: roles for RNA structure, misfolding, and modification in regulating the innate immune sensor PKR. Acc Chem Res 49: 1242–1249. 10.1021/acs.accounts.6b00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Anmangandla A, Bevilacqua PC. 2016. Bacterial riboswitches and ribozymes potently activate the human innate immune sensor PKR. ACS Chem Biol 11: 1118–1127. 10.1021/acschembio.6b00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain B, Mukerji I, Cole JL. 2012. Analysis of high-affinity binding of protein kinase R to double-stranded RNA. Biochemistry 51: 8764–8770. 10.1021/bi301226h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain B, Hesler S, Cole JL. 2015. Regulation of PKR by RNA: formation of active and inactive dimers. Biochemistry 54: 6663–6672. 10.1021/acs.biochem.5b01046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain B, Mayo C, Cole JL. 2016. Role of the interdomain linker in RNA-activated protein kinase activation. Biochemistry 55: 253–261. 10.1021/acs.biochem.5b01171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito JA, Steitz TA. 1998. A 1.3-Å resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc Natl Acad Sci 95: 9819–9824. 10.1073/pnas.95.17.9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D. 2016. Multifunctional non-coding Epstein-Barr virus encoded RNAs (EBERs) contribute to viral pathogenesis. Virus Res 212: 30–38. 10.1016/j.virusres.2015.08.007 [DOI] [PubMed] [Google Scholar]
- Jammi NV, Beal PA. 2001. Phosphorylation of the RNA-dependent protein kinase regulates its RNA-binding activity. Nucleic Acids Res 29: 3020–3029. 10.1093/nar/29.14.3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran U, Grey H, Cook AG. 2016. Nuclear factor 90 uses an ADAR2-like binding mode to recognize specific bases in dsRNA. Nucleic Acids Res 44: 1924–1936. 10.1093/nar/gkv1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SH, Lee K, Lee KS, Kunkeaw N, Johnson BH, Holthauzen LM, Gong B, Leelayuwat C, Lee YS. 2012. Characterization of the direct physical interaction of nc886, a cellular non-coding RNA, and PKR. FEBS Lett 586: 3477–3484. 10.1016/j.febslet.2012.07.076 [DOI] [PubMed] [Google Scholar]
- Kaempfer R. 2006. Interferon-γ mRNA attenuates its own translation by activating PKR: a molecular basis for the therapeutic effect of interferon-β in multiple sclerosis. Cell Res 16: 148–153. 10.1038/sj.cr.7310020 [DOI] [PubMed] [Google Scholar]
- Kalai M, Suin V, Festjens N, Meeus A, Bernis A, Wang XM, Saelens X, Vandenabeele P. 2007. The caspase-generated fragments of PKR cooperate to activate full-length PKR and inhibit translation. Cell Death Differ 14: 1050–1059. 10.1038/sj.cdd.4402110 [DOI] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330: 489–493. 10.1038/330489a0 [DOI] [PubMed] [Google Scholar]
- Karamichali E, Foka P, Tsitoura E, Kalliampakou K, Kazazi D, Karayiannis P, Georgopoulou U, Mavromara P. 2014. HCV NS5A co-operates with PKR in modulating HCV IRES-dependent translation. Infect Genet Evol 26: 113–122. 10.1016/j.meegid.2014.04.015 [DOI] [PubMed] [Google Scholar]
- Katze MG, He Y, Gale M Jr. 2002. Viruses and interferon: a fight for supremacy. Nat Rev Immunol 2: 675–687. 10.1038/nri888 [DOI] [PubMed] [Google Scholar]
- Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. 1997. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol Cell Biol 17: 4146–4158. 10.1128/MCB.17.7.4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane SC, Van V, Frank HM, Sciandra CA, McCowin S, Santos J, Heng X, Summers MF. 2016. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci 113: 13033–13038. 10.1073/pnas.1614785113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaja A, Vopalensky V, Pospisek M. 2015. Understanding the potential of hepatitis C virus internal ribosome entry site domains to modulate translation initiation via their structure and function. Wiley Interdiscip Rev RNA 6: 211–224. 10.1002/wrna.1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Liu CW, Puglisi JD. 2006. Specific recognition of HIV TAR RNA by the dsRNA binding domains (dsRBD1–dsRBD2) of PKR. J Mol Biol 358: 430–442. 10.1016/j.jmb.2006.01.099 [DOI] [PubMed] [Google Scholar]
- Kim Y, Lee JH, Park JE, Cho J, Yi H, Kim VN. 2014. PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes Dev 28: 1310–1322. 10.1101/gad.242644.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park J, Kim S, Kim M, Kang MG, Kwak C, Kang M, Kim B, Rhee HW, Kim VN. 2018. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol Cell 71: 1051–1063 e1056. 10.1016/j.molcel.2018.07.029 [DOI] [PubMed] [Google Scholar]
- Kitajewski J, Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Thimmappaya B, Shenk T. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2α kinase. Cell 45: 195–200. 10.1016/0092-8674(86)90383-1 [DOI] [PubMed] [Google Scholar]
- Koehler H, Cotsmire S, Langland J, Kibler KV, Kalman D, Upton JW, Mocarski ES, Jacobs BL. 2017. Inhibition of DAI-dependent necroptosis by the Z-DNA binding domain of the vaccinia virus innate immune evasion protein, E3. Proc Natl Acad Sci 114: 11506–11511. 10.1073/pnas.1700999114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JA, Rich A. 2005. Biological function of the vaccinia virus Z-DNA-binding protein E3L: gene transactivation and antiapoptotic activity in HeLa cells. Proc Natl Acad Sci 102: 12759–12764. 10.1073/pnas.0506011102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299: 133–141. 10.1006/viro.2002.1479 [DOI] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. 2004. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology 324: 419–429. 10.1016/j.virol.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Launer-Felty K, Cole JL. 2014. Domain interactions in adenovirus VAI RNA mediate high-affinity PKR binding. J Mol Biol 426: 1285–1295. 10.1016/j.jmb.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launer-Felty K, Wong CJ, Cole JL. 2015. Structural analysis of adenovirus VAI RNA defines the mechanism of inhibition of PKR. Biophys J 108: 748–757. 10.1016/j.bpj.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, Kang GY, Bang JY, Park HS, Leelayuwat C, Lee YS. 2011. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA 17: 1076–1089. 10.1261/rna.2701111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Hong SH, Shin S, Lee HS, Lee JS, Park EJ, Choi SS, Min JW, Park D, Hwang JA, et al. 2016. nc886, a non-coding RNA and suppressor of PKR, exerts an oncogenic function in thyroid cancer. Oncotarget 7: 75000–75012. 10.18632/oncotarget.11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire PA, Lary J, Cole JL. 2005. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol 345: 81–90. 10.1016/j.jmb.2004.10.031 [DOI] [PubMed] [Google Scholar]
- Lemaire PA, Anderson E, Lary J, Cole JL. 2008. Mechanism of PKR Activation by dsRNA. J Mol Biol 381: 351–360. 10.1016/j.jmb.2008.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Peters GA, Ding K, Zhang X, Qin J, Sen GC. 2006. Molecular basis for PKR activation by PACT or dsRNA. Proc Natl Acad Sci 103: 10005–10010. 10.1073/pnas.0602317103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li S, Wang Z, Shen Y, Zhang T, Yang X. 2012. Structure of the kinase domain of human RNA-dependent protein kinase with K296R mutation reveals a face-to-face dimer. Chin Sci Bull 58: 998–1002. 10.1007/s11434-012-5461-z [DOI] [Google Scholar]
- Li JJ, Cao C, Fixsen SM, Young JM, Ono C, Bando H, Elde NC, Katsuma S, Dever TE, Sicheri F. 2015. Baculovirus protein PK2 subverts eIF2α kinase function by mimicry of its kinase domain C-lobe. Proc Natl Acad Sci 112: E4364–4373. 10.1073/pnas.1505481112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Li X, Zhu W, Wang H, Mei L, Wu S, Lin X, Han X. 2016. NF90 is a novel influenza A virus NS1-interacting protein that antagonizes the inhibitory role of NS1 on PKR phosphorylation. FEBS Lett 590: 2797–2810. 10.1002/1873-3468.12311 [DOI] [PubMed] [Google Scholar]
- Ma Y, Mathews MB. 1996. Secondary and tertiary structure in the central domain of adenovirus type 2 VA RNAI. RNA 2: 937–951. [PMC free article] [PubMed] [Google Scholar]
- Machitani M, Sakurai F, Wakabayashi K, Tomita K, Tachibana M, Mizuguchi H. 2016. Dicer functions as an antiviral system against human adenoviruses via cleavage of adenovirus-encoded noncoding RNA. Sci Rep 6: 27598 10.1038/srep27598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth CD, Sattler M. 2012. Dynamics in multi-domain protein recognition of RNA. Curr Opin Struct Biol 22: 287–296. 10.1016/j.sbi.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. 1994. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology 204: 823–827. 10.1006/viro.1994.1601 [DOI] [PubMed] [Google Scholar]
- Masliah G, Barraud P, Allain FH. 2013. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol Life Sci 70: 1875–1895. 10.1007/s00018-012-1119-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J Virol 65: 5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo CB, Cole JL. 2017. Interaction of PKR with single-stranded RNA. Sci Rep 7: 3335 10.1038/s41598-017-03047-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo CB, Wong CJ, Lopez PE, Lary JW, Cole JL. 2016. Activation of PKR by short stem-loop RNAs containing single-stranded arms. RNA 22: 1065–1075. 10.1261/rna.053348.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna SA, Kim I, Liu CW, Puglisi JD. 2006. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J Mol Biol 358: 1270–1285. 10.1016/j.jmb.2006.03.003 [DOI] [PubMed] [Google Scholar]
- McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. 2007. Molecular framework for the activation of RNA-dependent protein kinase. J Biol Chem 282: 11474–11486. 10.1074/jbc.M700301200 [DOI] [PubMed] [Google Scholar]
- McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213: 413–424. 10.1006/viro.1995.0014 [DOI] [PubMed] [Google Scholar]
- Metz DH, Esteban M. 1972. Interferon inhibits viral protein synthesis in L cells infected with vaccinia virus. Nature 238: 385–388. 10.1038/238385a0 [DOI] [PubMed] [Google Scholar]
- Mittelstadt M, Frump A, Khuu T, Fowlkes V, Handy I, Patel CV, Patel RC. 2008. Interaction of human tRNA-dihydrouridine synthase-2 with interferon-induced protein kinase PKR. Nucleic Acids Res 36: 998–1008. 10.1093/nar/gkm1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Kunz RC, Zhang C, Kimura T, Yuan CL, Baccaro B, Namiki Y, Gygi SP, Hotamisligil GS. 2015. A critical role for PKR complexes with TRBP in immunometabolic regulation and eIF2α phosphorylation in obesity. Cell Rep 11: 295–307. 10.1016/j.celrep.2015.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 2007. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 318: 1455–1458. 10.1126/science.1147347 [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Toroney R, Bevilacqua PC. 2011. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol 21: 119–127. 10.1016/j.sbi.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Jones CN, Ghosh SK, Sharma SD, Cameron CE, Spremulli LL, Bevilacqua PC. 2013. Native tertiary structure and nucleoside modifications suppress tRNA's intrinsic ability to activate the innate immune sensor PKR. PLoS One 8: e57905 10.1371/journal.pone.0057905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namer LS, Osman F, Banai Y, Masquida B, Jung R, Kaempfer R. 2017. An ancient pseudoknot in TNF-α pre-mRNA activates PKR, inducing eIF2α phosphorylation that potently enhances splicing. Cell Rep 20: 188–200. 10.1016/j.celrep.2017.06.035 [DOI] [PubMed] [Google Scholar]
- Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. 2002. Epstein-Barr virus RNA confers resistance to interferon-α-induced apoptosis in Burkitt's lymphoma. EMBO J 21: 954–965. 10.1093/emboj/21.5.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. 1998. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J 17: 5458–5465. 10.1093/emboj/17.18.5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri S, Rahman F, Williams BR, Qin J. 2000. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J 19: 5567–5574. 10.1093/emboj/19.20.5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Hammond GL, Yang JH. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol 81: 917–923. 10.1128/JVI.01527-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M. 2014. Roles of eIF2α kinases in the pathogenesis of Alzheimer's disease. Front Mol Neurosci 7: 22 10.3389/fnmol.2014.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Okumura AJ, Uematsu K, Hatakeyama S, Zhang DE, Kamura T. 2013. Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J Biol Chem 288: 2839–2847. 10.1074/jbc.M112.401851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega LG, McCotter MD, Henry GL, McCormack SJ, Thomis DC, Samuel CE. 1996. Mechanism of interferon action. Biochemical and genetic evidence for the intermolecular association of the RNA-dependent protein kinase PKR from human cells. Virology 215: 31–39. 10.1006/viro.1996.0004 [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. 2016. The integrated stress response. EMBO Rep 17: 1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, Venkatesan S. 1994. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci 91: 4713–4717. 10.1073/pnas.91.11.4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LM, Fierro-Monti I, Mathews MB. 2001. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J Biol Chem 276: 32522–32530. 10.1074/jbc.M104408200 [DOI] [PubMed] [Google Scholar]
- Parrott AM, Walsh MR, Reichman TW, Mathews MB. 2005. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J Mol Biol 348: 281–293. 10.1016/j.jmb.2005.02.047 [DOI] [PubMed] [Google Scholar]
- Patel RC, Sen GC. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J 17: 4379–4390. 10.1093/emboj/17.15.4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Blose JM, Sokoloski JE, Pollack L, Bevilacqua PC. 2012. Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering. Biochemistry 51: 9312–9322. 10.1021/bi300935p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Dickerman B, Sen GC. 2009. Biochemical analysis of PKR activation by PACT. Biochemistry 48: 7441–7447. 10.1021/bi900433y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol 23: 573–582. 10.1016/j.coi.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindel A, Sadler A. 2011. The role of protein kinase R in the interferon response. J Interferon Cytokine Res 31: 59–70. 10.1089/jir.2010.0099 [DOI] [PubMed] [Google Scholar]
- Robertson HD, Manche L, Mathews MB. 1996. Paradoxical interactions between human δ hepatitis agent RNA and the cellular protein kinase PKR. J Virol 70: 5611–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani B, Engelbrecht S, Glashoff RH. 2010. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J Gen Virol 91: 1–12. 10.1099/vir.0.016303-0 [DOI] [PubMed] [Google Scholar]
- Rosa MD, Gottlieb E, Lerner MR, Steitz JA. 1981. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol 1: 785–796. 10.1128/MCB.1.9.785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. 2009. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol 16: 63–70. 10.1038/nsmb.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf IK, Lackey KA, Warudkar S, Sample JT. 2005. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J Virol 79: 14562–14569. 10.1128/JVI.79.23.14562-14569.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Patel R, Wang F, Lee K, Kumar K, Wu J, Nilsson A, Karin M, Kaufman RJ. 2006. Double-stranded RNA-dependent protein kinase phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 mediates apoptosis. J Biol Chem 281: 21458–21468. 10.1074/jbc.M603784200 [DOI] [PubMed] [Google Scholar]
- Schneider RJ, Weinberger C, Shenk T. 1984. Adenovirus VAI RNA facilitates the initiation of translation in virus-infected cells. Cell 37: 291–298. 10.1016/0092-8674(84)90325-8 [DOI] [PubMed] [Google Scholar]
- Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Shenk T. 1985. Adenovirus VAI RNA prevents phosphorylation of the eukaryotic initiation factor 2α subunit subsequent to infection. Proc Natl Acad Sci 82: 4321–4325. 10.1073/pnas.82.13.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EJ, Liu F, Kawagishi-Kobayashi M, Ung TL, Cao C, Dar AC, Sicheri F, Dever TE. 2008. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci 105: 16894–16899. 10.1073/pnas.0805524105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud CG, Hilse K, Clemens MJ. 1993. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res 21: 4483–4490. 10.1093/nar/21.19.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Moonan F, Romashko A, Joshi B, Barber GN, Jagus R. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250: 302–315. 10.1006/viro.1998.9365 [DOI] [PubMed] [Google Scholar]
- Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. 2009. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res 83: 228–237. 10.1016/j.antiviral.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Singh M, Castillo D, Patel CV, Patel RC. 2011. Stress-induced phosphorylation of PACT reduces its interaction with TRBP and leads to PKR activation. Biochemistry 50: 4550–4560. 10.1021/bi200104h [DOI] [PubMed] [Google Scholar]
- Sunita S, Schwartz SL, Conn GL. 2015. The regulatory and kinase domains but not the interdomain linker determine human double-stranded RNA-activated kinase (PKR) sensitivity to inhibition by viral non-coding RNAs. J Biol Chem 290: 28156–28165. 10.1074/jbc.M115.679738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd M, Biterova EI, Bourhis JM, Guy JE. 2011. The crystal structure of the human co-chaperone P58IPK. PLoS One 6: e22337 10.1371/journal.pone.0022337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NM, Ho CY, Katze MG. 1996. The 58-kDa cellular inhibitor of the double stranded RNA-dependent protein kinase requires the tetratricopeptide repeat 6 and DnaJ motifs to stimulate protein synthesis in vivo. J Biol Chem 271: 28660–28666. 10.1074/jbc.271.45.28660 [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435: 374–379. 10.1038/nature03580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya B, Weinberger C, Schneider RJ, Shenk T. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31: 543–551. 10.1016/0092-8674(82)90310-5 [DOI] [PubMed] [Google Scholar]
- Tian B, Mathews MB. 2001. Functional characterization of and cooperation between the double-stranded RNA-binding motifs of the protein kinase PKR. J Biol Chem 276: 9936–9944. 10.1074/jbc.M007328200 [DOI] [PubMed] [Google Scholar]
- Toroney R, Nallagatla SR, Boyer JA, Cameron CE, Bevilacqua PC. 2010. Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J Mol Biol 400: 393–412. 10.1016/j.jmb.2010.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroney R, Hull CM, Sokoloski JE, Bevilacqua PC. 2012. Mechanistic characterization of the 5′-triphosphate-dependent activation of PKR: lack of 5′-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD. RNA 18: 1862–1874. 10.1261/rna.034520.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM, Li Z, Cattaneo R, Samuel CE. 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem 284: 29350–29356. 10.1074/jbc.M109.045146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. 2007. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry 46: 55–65. 10.1021/bi061531o [DOI] [PubMed] [Google Scholar]
- Ung TL, Cao C, Lu J, Ozato K, Dever TE. 2001. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J 20: 3728–3737. 10.1093/emboj/20.14.3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon VK, Conn GL. 2016. Adenovirus VA RNA: an essential pro-viral non-coding RNA. Virus Res 212: 39–52. 10.1016/j.virusres.2015.06.018 [DOI] [PubMed] [Google Scholar]
- VanOudenhove J, Anderson E, Krueger S, Cole JL. 2009. Analysis of PKR structure by small-angle scattering. J Mol Biol 387: 910–920. 10.1016/j.jmb.2009.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivarini Áde C, Pereira Rde M, Barreto-de-Souza V, Temerozo JR, Soares DC, Saraiva EM, Saliba AM, Bou-Habib DC, Lopes UG. 2015. HIV-1 Tat protein enhances the intracellular growth of Leishmania amazonensis via the ds-RNA induced protein PKR. Sci Rep 5: 16777 10.1038/srep16777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuyisich M, Spanggord RJ, Beal PA. 2002. The binding site of the RNA-dependent protein kinase (PKR) on EBER1 RNA from Epstein-Barr virus. EMBO Rep 3: 622–627. 10.1093/embo-reports/kvf137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid AM, Coventry VK, Conn GL. 2008. Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J Biol Chem 283: 17485–17493. 10.1074/jbc.M802300200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid AM, Coventry VK, Conn GL. 2009. The PKR-binding domain of adenovirus VA RNAI exists as a mixture of two functionally non-equivalent structures. Nucleic Acids Res 37: 5830–5837. 10.1093/nar/gkp595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Samuel CE. 2009. Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2α phosphorylation. J Mol Biol 393: 777–787. 10.1016/j.jmb.2009.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, de Weerd NA, Willard B, Polekhina G, Williams BR, Sadler AJ. 2017. Auto-phosphorylation represses protein kinase R activity. Sci Rep 7: 44340 10.1038/srep44340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80: 5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Huang X, Mok BW, Chen Y, Zheng M, Lau SY, Wang P, Song W, Jin DY, Yuen KY, et al. 2014. NF90 exerts antiviral activity through regulation of PKR phosphorylation and stress granules in infected cells. J Immunol 192: 3753–3764. 10.4049/jimmunol.1302813 [DOI] [PubMed] [Google Scholar]
- Wilson JL, Vachon VK, Sunita S, Schwartz SL, Conn GL. 2014. Dissection of the adenoviral VA RNAI central domain structure reveals minimum requirements for RNA-mediated inhibition of PKR. J Biol Chem 289: 23233–23245. 10.1074/jbc.M114.550046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Gale MJ Jr, Tan SL, Katze MG. 2002. Inactivation of the PKR protein kinase and stimulation of mRNA translation by the cellular co-chaperone P58IPK does not require J domain function. Biochemistry 41: 4938–4945. 10.1021/bi0121499 [DOI] [PubMed] [Google Scholar]
- Yim HC, Williams BR. 2014. Protein kinase R and the inflammasome. J Interferon Cytokine Res 34: 447–454. 10.1089/jir.2014.0008 [DOI] [PubMed] [Google Scholar]
- Yoon CH, Kim SY, Byeon SE, Jeong Y, Lee J, Kim KP, Park J, Bae YS. 2015. p53-derived host restriction of HIV-1 replication by protein kinase R-mediated Tat phosphorylation and inactivation. J Virol 89: 4262–4280. 10.1128/JVI.03087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef OA, Safran SA, Nakamura T, Nix DA, Hotamisligil GS, Bass BL. 2015. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci 112: 5023–5028. 10.1073/pnas.1424044112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, Ozato K, Hinnebusch AG. 2001. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem 276: 24946–24958. 10.1074/jbc.M102108200 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. 2007. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450: 1263–1267. 10.1038/nature06389 [DOI] [PubMed] [Google Scholar]
- Zheng X, Bevilacqua PC. 2004. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10: 1934–1945. 10.1261/rna.7150804 [DOI] [PMC free article] [PubMed] [Google Scholar]