Abstract
It has recently become clear that ribosomes are much more heterogeneous than previously thought, with diversity arising from rRNA sequence and modifications, ribosomal protein (RP) content and posttranslational modifications (PTMs), as well as bound nonribosomal proteins. In some cases, the existence of these diverse ribosome populations has been verified by biochemical or structural methods. Furthermore, knockout or knockdown of RPs can diversify ribosome populations, while also affecting the translation of some mRNAs (but not others) with biological consequences. However, the effects on translation arising from depletion of diverse proteins can be highly similar, suggesting that there may be a more general defect in ribosome function or stability, perhaps arising from reduced ribosome numbers. Consistently, overall reduced ribosome numbers can differentially affect subclasses of mRNAs, necessitating controls for specificity. Moreover, in order to study the functional consequences of ribosome diversity, perturbations including affinity tags and knockouts are introduced, which can also affect the outcome of the experiment. Here we review the available literature to carefully evaluate whether the published data support functional diversification, defined as diverse ribosome populations differentially affecting translation of distinct mRNA (classes). Based on these observations and the commonly observed cellular responses to perturbations in the system, we suggest a set of important controls to validate functional diversity, which should include gain-of-function assays and the demonstration of inducibility under physiological conditions.
Keywords: ribosome deficiency, ribosome heterogeneity, ribosomopathy, specialized ribosomes
INTRODUCTION
The literature on functional ribosome heterogeneity is growing rapidly, and each new study inevitably proposes an exciting new role or ability for such ribosomes. Nevertheless, some of these conclusions remain controversial in the field, and merit critical examination. To do this, we need to first define the term “functional ribosome heterogeneity,” starting with a definition of the first term, ribosome heterogeneity.
Ribosomes are massive macromolecular complexes that have, with some early notable exceptions, mostly been thought of as uniform molecular factories with little regulatory capacity (a detailed history of this field can be found in Genuth and Barna 2018b). The conventional depiction was that all ribosomes were constructed according to identical specifications. Recent evidence suggests that ribosomes may be more heterogeneous than previously thought. Heterogeneity in this context means that ribosomes vary in the composition of their rRNA, proteins, or the post-transcriptional/translational modifications to either set of components. For the purpose of this review, we chose to ignore the additional layer of complexity brought by proteins that bind ribosomes (McMullin and Hallberg 1986; Darnell et al. 2011; Fuchs et al. 2011; Matsuo et al. 2014; Simsek et al. 2017; Muto et al. 2018). While such interacting partners can unquestionably modify the functionality of ribosomes and promote specialization, this arises not from the ribosomes per se, and for simplicity is left out of the discussions here, even though many considerations herein will hold for such specialization as well. Similarly, a growing body of literature demonstrates that other parts of the translational machinery can also provide for functional specialization (Richter and Sonenberg 2005; Park et al. 2010; Starck et al. 2012, 2016; Valásek 2012; Gutierrez et al. 2013; Wolfe et al. 2014; Lee et al. 2015a, 2016; Meyer et al. 2015; Truitt et al. 2015; Schuller et al. 2017; Zinshteyn et al. 2017). Again, this is not considered herein, even though it is an exciting area of research.
Studies of heterogeneous ribosomes also suggest that ribosome composition can influence function. For a ribosome to be functionally different, it must still be able to perform its core duties of decoding and peptidyl transfer (a ribosome that cannot make proteins is hardly functional). Thus, it is in the application of the ribosome's core catalytic activities where functional differences are manifest. These differences can include an altered preference for initiating translation of certain mRNAs, changes in fidelity resulting in alternative start- or stop-codon usage, etc. Thus, we define functional ribosome heterogeneity as variations in ribosome composition that influence its activity, thereby changing the output of translation. Importantly, while altered selectivity is the most commonly considered functional difference, other translational activities of the ribosome could be equally affected. An important example for functional differences arising from differences in ribosome composition is in No-go-decay (NGD), a ribosome-mediated mRNA quality control mechanism (Graille and Séraphin 2012; Buskirk and Green 2017; Joazeiro 2017; Simms et al. 2017). NGD depends on the presence of Asc1/RACK1 in the small subunit (Ikeuchi and Inada 2016), thus ribosomes lacking this nonessential protein are defective in NGD.
Importantly, the separation of these terms indicates that it is possible that heterogeneous ribosome populations might exist within the cell, but not have divergent functions. Furthermore, as will be shown below, the evidence for heterogeneity is in most cases much stronger than the evidence for functional specialization, and even more rare is the evidence for physiological roles of such specialization.
WHY WOULD CELLS USE RIBOSOME SPECIALIZATION FOR THE REGULATION OF GENE EXPRESSION?
Gene expression is regulated at multiple levels (transcription, splicing, translation, mRNA and protein stability), but it is not immediately clear why regulation by ribosome customization would be useful. Building ribosomes is extremely resource-intensive for the cell compared to mRNA synthesis and turnover (Warner 1999), so the customization of the translation machinery is expected to serve an important function. We suggest several explanations for why the cell would invest in translational regulation through ribosome modification, which range from accidental to highly beneficial. It is important here to keep in mind that in contrast to transcriptional induction, which is often switch-like, translational control via distinct ribosome pools appears to act more as a modulator, as the preferences exist on a continuum instead of being binary (Thompson et al. 2016; Ferretti et al. 2017, 2018; Shi et al. 2017).
Temporal control of gene expression is vital because cells must rapidly respond to changing external conditions. This is true for both unicellular organisms (which must account for environmental fluctuations in osmolarity, nutrient availability, temperature, etc.) and multicellular life forms (which require rapid temporal regulation of gene expression during cell cycle progression, intense exertion, oxidative stress, etc.). While transcriptional regulation occurs within minutes (Gasch et al. 2000), it still requires the engagement of the transcriptional machinery, mRNA export and translation. On the other hand, a posttranslational change to a ribosomal protein (RP) or the addition/removal of a protein from the ribosome requires only a single step, and so could in principle occur extremely quickly. This level of regulation could also allow cells to link their translational state to local metabolic conditions, as is seen in the case of Rps10-mediated phosphorylation of eIF2α by Gcn2 (Lee et al. 2015b) and translational activator feedback loops in mitochondrial ribosomes (Herrmann et al. 2013).
Complex regulation of cellular activities is necessary for cellular survival. Specialized ribosomes could facilitate such regulation by allowing for a concurrent up-regulation of some mRNAs while others are down-regulated. In contrast, traditional transcriptional induction alone only allows for up-regulation of the induced genes. In this regard, specialized ribosomes could act similarly to alternative transcriptional start sites, which can produce mRNAs with long UTRs that are poorly translated (Cheng et al. 2018b).
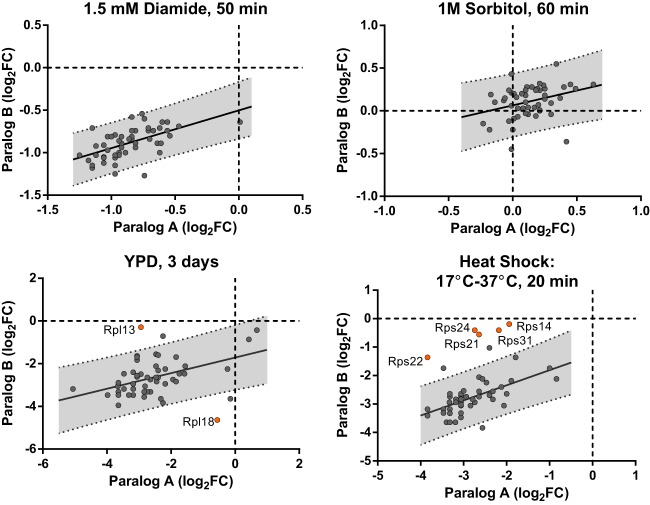
Conversely, if changes in ribosome composition mirror transcriptional changes, then they could also function to sharpen the boundaries of protein gradients, such as those observed during embryogenesis (Fig. 1A; Christian 2012), or yeast mitotic cell division (Long et al. 1997; Takizawa et al. 1997). This would be similar to the function of proteins such as nanos or bicoid (Driever and Nüsslein-Volhard 1988a, 1988b; Irish et al. 1989). Interestingly, early embryonic development in zebrafish is coupled to a change in the rDNA template used for rRNA transcription, thus changing ribosome composition (Locati et al. 2017), perhaps to facilitate such a scenario.
FIGURE 1.
Specialized ribosomes can help regulate gene expression. (A) A comparison of a protein gradient generated by mRNA localization alone (blue), compared to one (purple) created by mRNA localization paired with a gradient of specialized ribosomes (red) that preferentially translate the mRNA. The graph compares the distribution of protein in these two scenarios. (B) A comparison of two theoretical stimuli that activate the same transcriptional signaling pathway, X, but lead to different outputs because stimulus A also activates changes in ribosome composition.
Translational control via ribosome composition might also allow for different cellular outputs, depending on different stimuli, even though overlapping signaling pathways are used. This could arise if a given stimulus, A, induces transcriptional and translational changes (including ribosomal composition changes), via two different signaling pathways. An alternative stimulus, B, could result in the same transcriptional changes via the same signaling cascade, but if it does not induce the same changes in ribosome composition these transcriptional changes could have different outputs (Fig. 1B).
Furthermore, specializing ribosomes might not be as costly if one considers that rapidly growing cells constantly make new ribosomes. In addition, some RPs are replaced during the lifetime of a ribosome (Pulk et al. 2010; Mathis et al. 2017; Lilleorg et al. 2019). In both cases, generating a pool of specialized ribosomes, especially to make longer term shifts in translational control, requires only subtle adjustments to already existing processes. Similarly, mammals undergo turnover of their ribosome pool after the initial developmental stages (Locati et al. 2017), perhaps reflecting the fact that these ribosomes are “old” and possibly damaged as they were deposited into the oocyte prior to the animal's birth. Differentiating cells experience large changes in ribosome numbers, and this also presents an ideal opportunity to shift the ribosome population through specialization (Ramagopal and Ennis 1984; Jacobs et al. 1985).
Finally, the evolution of functional ribosome specialization might simply have taken advantage of accidental occurrences. Ensuring all RPs are correctly incorporated into ribosomes is not trivial: Given that most RPs are small polypeptides and ribosomes are mega-Dalton-sized machines, this is like finding needles in haystacks. Moreover, this task is further complicated by the fact that RP assembly occurs via parallel pathways (Mulder et al. 2010; Davis et al. 2016; Sanghai et al. 2018), producing dozens of correct assembly intermediates and thus rendering quality control for the incorporation of individual proteins challenging. So long as they remain functional (and are therefore not degraded), subpopulations lacking individual proteins might have originally just been unavoidable byproducts. Similarly, given the fact that there are multiple repeats of the rDNA operon in all organisms, it is probably nearly impossible to avoid mutations, as there would be selective pressure only against nonfunctional ribosomes, not differentially functional ribosomes. Cases where small changes in ribosome composition yielded useful functions would be selected for over time, potentially giving rise to a “ribosome code” (Mauro and Edelman 2002, 2007). From this perspective, we note that biological processes that are regulated appear to be predisposed to regulation by specialized ribosomes (Ferretti et al. 2018). This is perhaps because mRNAs encoding important regulatory proteins are often scarce and poorly translated, rendering them sensitive to up-regulation via specialized ribosome populations (Ferretti et al. 2017, 2018). As a result, adaptation via mutations in the Kozak sequence might simply take advantage of preexisting biology and therefore occur frequently (Ferretti et al. 2018; Peter et al. 2018).
RIBOSOMAL DIVERSITY EXISTS UNDER PHYSIOLOGICAL AND PATHOLOGICAL CONDITIONS
Examples of, and evidence for, ribosome heterogeneity have been extensively reviewed elsewhere (Gilbert 2011; Xue and Barna 2012, 2015; Filipovska and Rackham 2013; Sauert et al. 2015; Shi and Barna 2015; Dinman 2016; Genuth and Barna 2018a; Guo 2018). Here we will only briefly review the routes to ribosome diversity, and instead focus on the potential pitfalls that can arise when studying their functional consequences.
Posttranscriptional modification of rRNA
Of the nearly 5,500 rRNA bases in Saccharomyces cerevisiae, over 2% (∼110) can be post-transcriptionally modified (for a more extensive review, see Sloan et al. 2017). Many of these modifications involve 2′-O-methylations of ribose or isomerization of uridine to pseudouridine, and are thought to stabilize the structure of the ribosome, helping to maintain proper orientation of key regions (Liang et al. 2007; Polikanov et al. 2015).
The importance of these modifications is underscored by the consequences of their loss. While the loss of a single modification tends to have no discernible effect under the conditions studied (generally rich medium), multiple deletions often disturb ribosomal function. Loss of modifications to rRNA near the decoding center, effected through deletion of their corresponding small nucleolar RNAs (snoRNAs), impairs cell growth, reduces rates of amino acid incorporation, and disrupts ribosome biogenesis (Liang et al. 2009). In other studies, multiple modifications were removed using the same technique; translational accuracy and antibiotic resistance decreased as more modifications were lost (Baxter-Roshek et al. 2007; Baudin-Baillieu et al. 2009).
Since the majority of rRNA modifications are guided by so-called box C/D or H/ACA snoRNPs (Lestrade 2006), and installed during assembly, they are unlikely to be helpful during rapid adaptation to changing environmental conditions. This is especially true if these changing conditions represent cellular stress, under which ribosome assembly is generally down-regulated (Warner 1999; Gasch et al. 2000; Viladevall et al. 2004; Boulon et al. 2010). Furthermore, rRNA modifications are generally (although not universally) irreversible, and thus removal of a specialized ribosome pool would require ribosome turnover, which is exceedingly slow (LaRiviere et al. 2006). For these reasons, variations in rRNA modification may be a strategy to enact long-term changes in translational preference for a cell.
As the tools to analyze base modifications have become more sensitive, instances of substoichiometric base modifications have been discovered. In these cases, only a subset of ribosomes within a cell are modified, creating populations of ribosomes with a heterogenous set of modifications (Krogh et al. 2016; Taoka et al. 2016; Erales et al. 2017). Furthermore, studies have shown that active regulation of rRNA modification is a conserved behavior. For example, pseudouridylation of rRNA is up-regulated in the bloodstream form of the Trypanosoma brucei parasite (Chikne et al. 2016), and rRNA methylation by NSUN5 regulates life span and stress resistance in flies, worms and yeast (Schosserer et al. 2015). Similarly, a study in X. tropicalis showed that failure to ubiquitylate the ribosome biogenesis factors NOLC1 and TCOF1 destabilizes these proteins, and subsequently disrupts neural crest formation during cell differentiation (Werner et al. 2015). Tcof1 is causally linked to the developmental disorder Treacher Collins syndrome (Dixon et al. 2006) and involved in rDNA transcription and rRNA modification (Lin and Yeh 2009; Larsen et al. 2014; Walker-Kopp et al. 2017). While these changes do not appear to affect overall ribosome number or global translation, they moderately affect the translation of a subset of mRNAs encoding proteins involved in neural crest formation (Werner et al. 2015). These observations may be explained by differences in the rRNA modifications in wild type and TCOF1 mutants, although such different ribosome populations have not yet been shown to exist.
Finally, altered expression of snoRNAs, such as overexpression of SNORA42 in lung cancers and SNORA21 in colorectal cancers, have been associated with oncogenesis (Mei et al. 2012; Yoshida et al. 2017). In addition, multiple transcriptome-wide surveys have identified snoRNAs as commonly differentially expressed in cancers, although for none of these studies has it been shown that alterations actually involve changes in rRNA modifications (Mannoor et al. 2012; Krishnan et al. 2016; Gong et al. 2017). Whether the divergent ribosome populations arising from rRNA modifications have physiological (as opposed to pathological) roles remains to be seen.
Diversity of rRNA sequence
In most organisms, rDNA is encoded in multiple (often hundreds) of repetitive operons throughout the genome. Initial characterization of these regions using restriction digests led to the conclusion that they were relatively homogenous, but this was quickly called into question (Gonzalez et al. 1988). Most bacteria and archaea have between 1 and 15 rDNA operons, which are often not identical (Hillebrand et al. 2005; Sun et al. 2013). Intriguingly, Streptomyces coelicolor transcribes different large-subunit rRNAs during its various stages of morphological development (Kim et al. 2007, 2008). Similarly, the halophilic archaeon Haloarcula marismortui possesses three rDNA operons: A, B and C. While A and C are nearly identical, operon B is highly divergent, and produces a 16S rRNA with a lower predicted free energy, and is likely more stable. At 50°C, rRNA B is expressed at four times the level of the other two operons combined, and strains lacking operon B are deficient at high-temperature growth. These data are consistent with a model wherein the archaeon produces specialized ribosomes with enhanced structural stability in order to grow at high temperatures (López-López et al. 2007), while maintaining the ability to produce a distinct set of ribosomes for “normal” temperature growth, where thermophilic ribosomes might be “frozen” and therefore not functional. Nevertheless, to date no functional data have been reported to support this model.
In Escherichia coli, there are seven rDNA operons with similar, but not identical, sequences (Hillebrand et al. 2005). The promoters of these rDNA operons are differentially responsive to various stimuli, causing the accumulation of distinct ribosome species under various environmental conditions (Condon et al. 1992; Kurylo et al. 2018; see also Maeda et al. 2015). Specifically, in conditions of nutrient limitation, E. coli switches the predominant rDNA template, thereby changing its 16S rRNA makeup. The resulting ribosomes are resistant to tetracycline, and more likely to bind AdhE, a putative RNA helicase, demonstrating functional differences for these ribosomes (Kurylo et al. 2018).
rRNA heterogeneity is also present in eukaryotes. In humans and mice, the many rDNA sequences throughout an individual genome are variable (Parks et al. 2018) and transcription from individual loci can vary across tissues and over the course of a cell's life cycle (Kuo et al. 1996; Tseng et al. 2008; Parks et al. 2018). In zebrafish, a study of rRNA in oocytes and embryos found that maternally derived rRNA dominates during the initial stages of development and is eventually replaced by somatic rRNA, which differs significantly in sequence, although it remains to be seen whether and how these differences impact ribosomal function (Locati et al. 2017). Like rRNA modification, customizing ribosome function with rRNA sequence variation only works over long timescales. Ribosome synthesis occurs on the scale of minutes, so using alternative rRNA transcripts is not helpful for immediate adaptation to acute stress conditions. Instead, this form of ribosome specialization may be more useful for extended tolerance to new environments.
Mitochondrial ribosomes
An extreme case of ribosome modification is found in mitochondria. These organelles produce distinctive mito-ribosomes dedicated to synthesizing the small number of genes found in the mitochondrial genome (Ott et al. 2016). Because mitochondria are descended from an ancient bacterial ancestor, their ribosomes bear similarities to bacterial ribosomes and are sensitive to many of the same antibiotics (De Silva et al. 2015). Nevertheless, mito-ribosomes have diverged significantly, in the content of their rRNA, RPs, and tRNA usage. Note that ribosomes found in chloroplasts appear to be equally divergent (Tiller and Bock 2014; Bieri et al. 2017), but are beyond the scope of this review.
Relative to cytosolic ribosomes, mito-ribosomes possess many structural specializations. The peptide exit channel is lined with hydrophobic protein residues, which may be beneficial as most genes on the mtDNA encode membrane proteins (Ott et al. 2016). In mammals, the mito-ribosome protein mL45 is able to attach to the inner mitochondrial membrane and has been suggested to facilitate protein insertion (Greber et al. 2014; Englmeier et al. 2017). In yeast mito-ribosomes, the mRNA exit channel is significantly remodeled and may act as a binding site for translational activators (Desai et al. 2017). These proteins bind the long 5′-UTRs of these mito-mRNAs to effect translation (Herrmann et al. 2013; Desai et al. 2017).
While some of these structural adaptations could facilitate the production of the mitochondrial proteins, whether this is actually the case has not been probed. In fact, the observation that the proteins encoded by mitochondrial genomes are almost entirely nonoverlapping between yeast and mammals suggests that the mito-ribosome adaptations are not necessary for efficient translation, although it should be noted that mito-ribosomes vary much more significantly between organisms than the cytoplasmic ribosomes. Conversely, a super-folding GFP reporter inserted into a mitochondrial ORF was successfully translated in the organelle (Suhm et al. 2018), again suggesting that specialization, if it exists, is moderate, and does not preclude the translation of a cytosolic protein.
Posttranslational modification of ribosomal proteins
One of the most straightforward paths to ribosome customization is the posttranslational modification (PTM) of RPs (for review, see Sauert et al. 2015; Simsek and Barna 2017; Genuth and Barna 2018a). These changes can be enacted rapidly and do not require the construction of new ribosomes. However, while there are many PTMs of RPs, in most cases it remains unclear whether they are regulated or have functional consequences for ribosome activity.
One example of a PTM that affects ribosome function, is the hydroxylation of Rps23/uS12, which is conserved from yeast to humans, and important for preventing stop-codon readthrough (Loenarz et al. 2014; Singleton et al. 2014). Mutation of the hydroxylated proline residue in Rps23/uS12 in yeast is linked to a syndromic presentation, including microcephaly and hearing loss, and in yeast, this mutation causes defects in ribosome biogenesis and polysome formation that may be due to reduced Rps23 recruitment (Paolini et al. 2017). Likewise, mutation of the methylation site of Rpl3/uL3, or deletion of the cognate methyltransferase, decreases translational accuracy (Al-Hadid et al. 2016). However, at this point, neither Rps23 hydroxylation or Rpl3 methylation are known to be regulated by the cell to exert translational control in certain growth conditions.
Ribosomal protein content
Although the catalytic functions of the ribosome are carried out by the rRNA, most RPs are essential for its proper function. While some RPs promote structural integrity, thereby supporting fidelity, others have roles in the binding of initiation, elongation, and release factors. Some directly contact bound mRNA (Chiu et al. 2010; Elantak et al. 2010; Korostelev 2011; Kouba et al. 2012; Nikolay et al. 2015; Haimov et al. 2017; Hinnebusch 2017). The function of most RPs is, however, unclear.
Multiple studies have shown that ribosomes lacking specific RPs are present in cells. For instance, when ribosomes from mouse embryonic stem cells (ESCs) were fractionated on sucrose gradients and then analyzed by mass spectroscopy, RP content varied in ribosomes isolated from different translational pools (Slavov et al. 2015). Likewise, in another similar study of mouse ESCs, four RPs, Rpl10A/uL1, Rpl38, Rps7, and Rps25, were present at substoichiometric levels when analyzed by selected-reaction monitoring-based mass spectroscopy (Shi et al. 2017). A recent manuscript used cryo-EM and mass spectrometry to identify a fraction of S. cerevisiae ribosomes without Rps1 and Rpl10/uL16 (Sun et al. 2018). Similarly, cryo-EM has identified populations of bacterial ribosomes lacking uS2 (Loveland et al. 2016). Finally, exposing yeast to high salt or high pH causes a fraction of ribosomes to become Rps26-deficient, and accumulation of these ribosomes increases tolerance to those stresses (Ferretti et al. 2017). Together these studies demonstrate that ribosomes lacking the full complement of RPs can be found in cells.
In addition to ribosomes lacking RPs being present in normal tissues, they are also associated with various cancers. The protein RACK1/Asc1 affects ribosomal activity in multiple contexts, and ribosomes lacking RACK1/Asc1 are deficient in the translation of short mRNAs and have an increased association with certain autophagy-related mRNAs (Thompson et al. 2016; Kim et al. 2017). Furthermore, both cap-dependent and IRES-mediated translations are reduced in RACK1-depleted cells (Majzoub et al. 2014; Gallo et al. 2018). Thus, RACK1/Asc1 impinges on translation generally, but can also act on specific mRNAs. Studies from our laboratory have shown that Asc1 (and Rps10)-deficient ribosomes accumulate in some breast cancer cell lines, relative to glioma cell lines (Collins et al. 2018). This is due to insufficient amounts of the assembly factor Ltv1, whose function is to ensure the stoichiometric incorporation of these two proteins, apparently by promoting the proper incorporation of Rps3 (Collins et al. 2018). While it is possible that this heterogeneity is an accidental consequence of the aneuploidy found in cancers, it is noteworthy that while functionally related ribosome assembly factors tend to be amplified in cancer genomes, Ltv1 is frequently deleted (Cerami et al. 2012; Gao et al. 2013), indicating that Ltv1 deletion is not unique to breast cancers and selected for in cancer cells.
In the context of RP-dysregulation and cancer, Asc1 is not unique. Changes in the expression of other RPs, including Rps10 and Rps26, are found in other cancers (Guimaraes and Zavolan 2016; Ajore et al. 2017; Kulkarni et al. 2017), and are associated with a poor prognosis (although in these cases ribosome composition per se has not yet been analyzed). Rps10/Asc1-deficient ribosomes are also more prone to mistakes during translation, and are deficient in ribosome-mediated RNA quality control (Thompson et al. 2016; Sundaramoorthy et al. 2017; Collins et al. 2018; Gallo et al. 2018). How these properties, as well as the mRNA specificity of Rps10/Asc1-deficient ribosomes, promote cancer progression remains to be determined, although the connection to autophagy is intriguing.
Ribosomes can also be specialized through incorporation of different paralogs. In bacteria, RP paralogs are infrequent and, when present, often have significant sequence divergence (Makarova et al. 2001). A recent study using pulse chase labeling of amino acids to analyze ribosome composition changes during different growth phases in E. coli revealed that bL31 and bL36 paralogs changed between exponential and stationary growth phase, and that the A and B paralogs of each appeared to be mutually exclusive on ribosomes (Lilleorg et al. 2019). Finally, both paralogs were validated to be components of the 70S ribosome by X-ray crystallography. The functional consequence of switching between paralogs is unclear, though only the A paralogs of both proteins can bind zinc. It has been proposed that this pattern among bacterial RPs is tied to survival during zinc starvation (Graham et al. 2009; Hensley et al. 2012).
FUNCTIONAL SPECIALIZATION
While the examples presented above (as well as the many not mentioned here but previously reviewed [Gilbert 2011; Xue and Barna 2012, 2015; Filipovska and Rackham 2013; Sauert et al. 2015; Shi and Barna 2015; Dinman 2016; Genuth and Barna 2018a; Guo 2018]) provide compelling evidence for ribosome heterogeneity, demonstrating that these differences in ribosome composition have functional consequences is challenging. This is because most manipulations of ribosomal composition also change ribosome number (Rotenberg et al. 1988; Choesmel et al. 2007; Li et al. 2009; Bhattacharya et al. 2010; McIntosh et al. 2011; Steffen et al. 2012; Ferretti et al. 2017; Palumbo et al. 2017; Cheng et al. 2018a; Segev and Gerst 2018). Furthermore, changing the expression of one paralog often affects the expression of the other. Additionally, biochemical handles used to study functional ribosomes can perturb ribosome function. Finally, as ribosome degradation is initiated on mRNAs (Cole et al. 2009), it might be impossible to entirely rule out that ribosomes lacking individual proteins are not degradation intermediates. Below, we will discuss each of these cases, before presenting a “cheat-sheet” to control for these possible artifacts as much as possible. Furthermore, we suggest the importance of gain-of-function studies to demonstrate that the ribosome populations have physiological roles, an important consideration when ruling out artifacts.
Ribosome numbers matter
Because initiation is the limiting step in translation (Lackner et al. 2007; Shah et al. 2013), and involves association of ribosomal subunits with the mRNA, first principle considerations suggest that initiation rates (for all mRNAs) should be proportional to ribosome concentrations. However, more detailed mathematical modeling of global protein synthesis rates, validated in in vitro translation systems, predicts that an overall reduction in 40S subunit numbers can cause mRNA-specific effects on translation (Lodish 1974). The modeling (for review, see Mills and Green 2017) predicts that 40S ribosome depletion only marginally attenuates the translation of mRNAs with high initiation rates, while translation of poorly initiated mRNAs is greatly attenuated. This is because a term for the probability of finding an empty start codon is introduced into the equation, which is affected by 40S concentration. Importantly, the Lodish model does not make any predictions about 60S subunit concentrations (Lodish 1974).
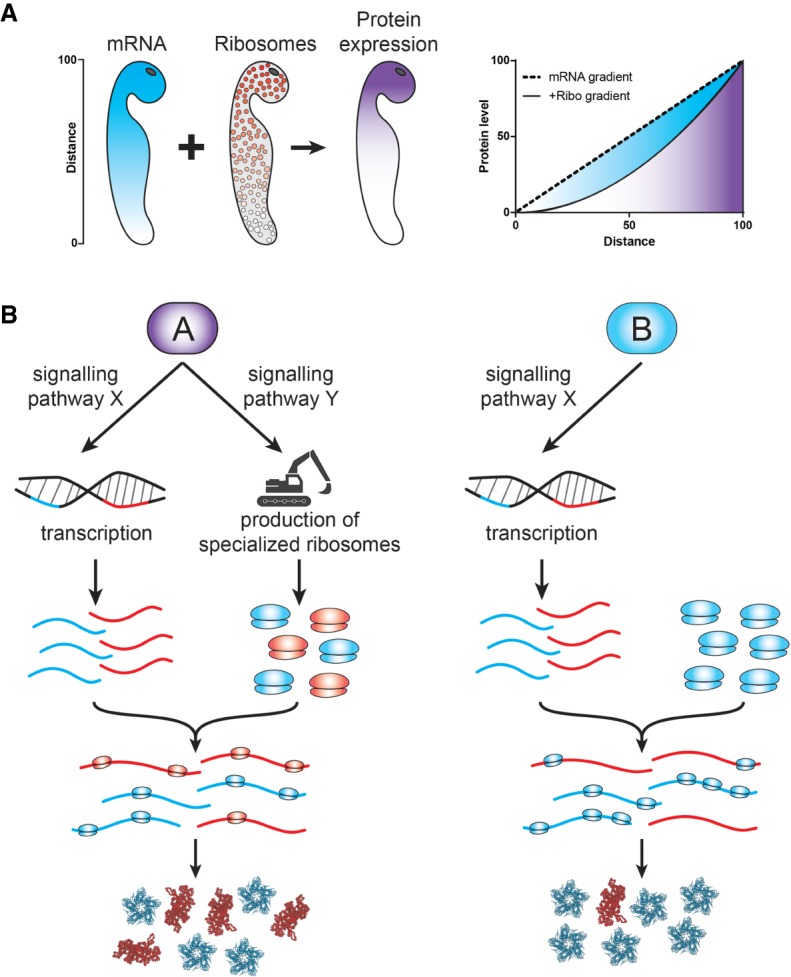
However, our current understanding of translation initiation is that 40S ribosomes are first recruited to the mRNA cap (Fig. 2A). Assuming that cap-40S contacts are broken during scanning in order to recruit another ribosome, one specific consequence of the initial 40S recruitment to the cap is that the probability of finding an empty cap is likely a function of the 5′-UTR length, or more precisely, the distance between the cap and the first start codon (in the case of upstream open reading frames in the 5′-UTR): The caps of mRNAs with short 5′-UTRs might not be available because a ribosome is still bound downstream at the start codon, trying to initiate, and thus blocking ribosome binding to the cap.2 In contrast, cap availability for mRNAs with longer 5′-UTRs is unlikely to be limited by start codon clearance, and one might assume it is always available. This latter point is generally consistent with recent experimental data in yeast (Archer et al. 2016). Thus, the prediction that well-translated mRNAs are insensitive to 40S ribosome concentration, is likely broadly true only for mRNAs with short 5′-UTRs, while it is unlikely to be correct for mRNAs with long 5′-UTRs as their caps seem to be generally available. (Note that Lodish tested his predictions with globin mRNA, which has a short 5′-UTR of 55 nt.)
FIGURE 2.
Model of ribosome translation and its dependence on subunit concentration. (A) Binding of the 40S subunit to the mRNA cap is normally mRNA-agnostic and depends solely on 40S and mRNA concentration. The exception is if a pileup of 40S subunits blocks additional binding. (B) After cap binding, the 40S subunit scans through the 5′-UTR for a start codon. Its probability of success is a function of UTR length and structure. (C) Once a start codon is recognized there are two outcomes modeled by the equation on the right: (D1) The 40S resumes scanning, or (D2) a 60S subunit binds and translation begins. We assume for simplicity that successful binding of 60S always results in translation. The equation describing the chance of initiation is influenced by both the concentration of 60S and the time the 40S spends on the start codon. The latter factor is controlled by the start codon's context and the composition of the 40S. For the equation, the extreme cases are illustrative. When the complex is very stable, such that kjoin [60] ≫ kresume, then the chance of initiation approaches 1, and is insensitive to 60S concentration. In contrast, if the complex is unstable (due to a poor Kozak context), then initiation could become linearly dependent on 60S concentration.
In contrast, mRNAs with a poor Kozak context could be sensitive to 60S subunit concentration if we consider that not all initiation attempts at start sites might be successful (Fig. 2D). This possibility is consistent with stochastic bypass of uORFs, which is dependent on the strength of the Kozak sequence (Calvo et al. 2009; Loughran et al. 2012; Chew et al. 2016) and would introduce an initiation probability function. This can be considered in simplified terms as a partitioning at the start codon between joining of 60S (which is assumed here to render initiation irreversible), and the resumption of scanning (Fig. 2D). According to this model, less stable initiation complexes, such as those with a suboptimal Kozak sequence, should be less successful at initiation (as observed for uORFs), and their success would depend on the concentration of 60S subunits. In contrast, preinitiation complexes on mRNAs with an optimal start codon context might be so stable that the partitioning does not change in the range of physiologically attainable 60S changes. Thus, we predict that translation initiation of mRNAs with poor Kozak context is sensitive to 60S concentrations.
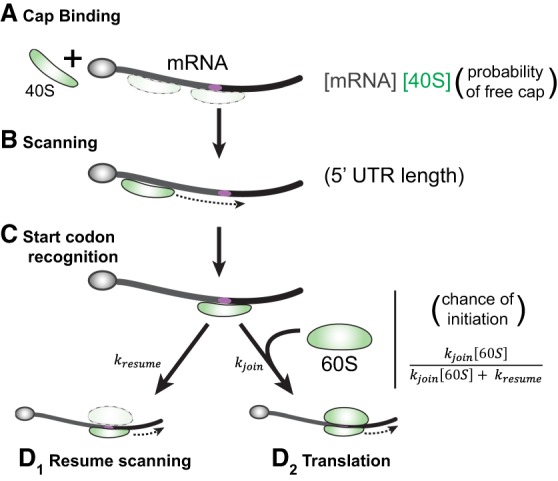
These considerations broadly suggest four classes of mRNAs (Fig. 3), distinguished by their mRNA features (long or short 5′-UTR, strong or weak Kozak context), for which different sensitivities to variances in 40S and 60S concentrations are predicted. Naturally, these predictions are complicated by other features in the 5′-UTR. For instance, uORFs, in particular the 5′-most uORF, will be similarly sensitive to subunit availability, affecting its translatability, and thus, the way in which it regulates the translation of the downstream coding sequence.
FIGURE 3.
Four classes of mRNAs. On the left are mRNAs that are sensitive to 40S concentration because their 5′ cap is always available, and binding is a function of 40S concentration. On the top are mRNAs where preinitiation complexes are stable enough to always lead to subunit joining, while the mRNAs on the bottom are sensitive to 60S concentration. Because well-translated mRNAs tend to have both strong Kozak sequences and short UTRs (Hurowitz and Brown 2003; Ingolia et al. 2009; Liu et al. 2012), while poorly translated mRNAs have poor Kozak contexts and long UTRs, the top right and bottom left corners are biologically relevant. Translation of the top right mRNA is 40S and 60S concentration independent, but the bottom left is dependent on subunit concentration.
The “ribosome concentration hypothesis” (Mills and Green 2017) is important to consider as ribosome numbers are reduced in many experimental systems used to study specialized ribosomes (Rotenberg et al. 1988; Bhattacharya et al. 2010; McIntosh et al. 2011; Vesper et al. 2011; Steffen et al. 2012; Thompson et al. 2016; Ferretti et al. 2017; Palumbo et al. 2017; Cheng et al. 2018a; Segev and Gerst 2018), making these studies particularly vulnerable to this confounding variable, and demonstrating the importance of rigorously controlling for effects arising from ribosome numbers. An important example for such effects from ribosome numbers are recent observations that knockdown of two different RPs had highly overlapping effects on gene-specific translation in hematopoetic progenitor cells (Khajuria et al. 2018). Similarly, as detailed below, several studies on differential effects from knockout of RP paralogs in yeast are most simply consistent with effects arising from reduced ribosome numbers (Rotenberg et al. 1988; Watanabe et al. 1993; Zinn et al. 1994; Strittmatter et al. 2006; Palumbo et al. 2017).
Another example for the likely effects from changes in ribosome numbers comes from the study of mRNAs containing internal ribosome entry sites (IRES). For example, the hepatitis C virus (HCV) IRES has reported sensitivity to the depletion of Rps25, Rpl10A/uL1, Asc1/RACK1, Rps5/uS7, and Rpp1/2 (Landry et al. 2009; Majzoub et al. 2014; Bhat et al. 2015; Campos et al. 2017; Shi et al. 2017), depletion of Rpp1/2 disrupts replication of foot-and-mouth disease virus (Martínez-Azorín et al. 2008), and Rps25 is required by HIV and the Dicistroviridae (Landry et al. 2009; Carvajal et al. 2016), a family of viruses that also requires Asc1/RACK1 (Majzoub et al. 2014). Further, in plants, the replication of turnip mosaic virus is inhibited by depletion of Rps6, Rps2/uS5, Rpl19, Rpl13, Rpl7/uL30, or Rpp1/2 (Yang et al. 2009; Rajamäki et al. 2017). Finally, deletion of Rpl38 prevents polysome recruitment of some Hox mRNAs that contain IRES-like elements (Kondrashov et al. 2011; Xue et al. 2015). Just this enumeration makes it clear that the effects might not be so specific for these individual RPs. Indeed, translation of viral RNAs containing IRES sequences appears to be sensitive to the depletion of nearly any RP (Cherry et al. 2005), strongly suggesting that the effect comes from ribosome numbers and not specific proteins.
Nevertheless, while these considerations suggest that ribosome numbers can matter, and must be considered, it should be noted that they are not incompatible with a model in which ribosome composition also matters. In addition, it is also possible that ribosome numbers matter for some phenotypes (like growth or developmental deficiencies), while ribosome composition might matter for others (like cancer predisposition).
The expression of ribosomal protein paralogs is differentially regulated
In S. cerevisiae, 59 out of 78 RP genes are duplicated into A and B isoforms. These paralogs have highly similar, if not identical, amino acid sequences and have similar patterns of transcriptional regulation (Gasch et al. 2000). Some recent studies have suggested that these paralogs can impart functional specialization (e.g., Haarer et al. 2007; Komili et al. 2007; Samir et al. 2018; Segev and Gerst 2018). Nevertheless, while studying potential instances of such specialization it is important to keep in mind that the RP knockout collection has acquired additional mutations and duplications, which might mask or produce phenotypes (Steffen et al. 2012). Thus, yeast strains need to be carefully evaluated. Furthermore, in general, one of the two RP paralogs is dominant such that RP levels, but not specific paralogs, are the more important factor.
An ideal example is Rps26 (Strittmatter et al. 2006). Deletion of the minor paralog, Rps26B, has minimal effect on cellular growth rate or the ability to engage in filamentous differentiation (pseudohyphal growth for haploid cells, filamentous growth for diploid cells). Deletion of Rps26A, however, leads to slow growth and virtually eliminates filamentous differentiation. When ΔRps26A cells are supplemented with either Rps26A or B on a high expression plasmid, however, the defects are rescued in a paralog-independent manner, demonstrating that the effects arise from insufficiency of Rps26 and not one of the specific isoforms.
A similar case is seen with Rpl7/uL30, where phenotypes that appear paralog-specific are actually responsive to Rpl7 dose (Palumbo et al. 2017). Deletion of Rpl7A leads to specific phenotypes including slow growth, tunicamycin resistance and defects in bud tip localization of the ASH1 mRNA. If the loss of Rpl7A, specifically, is responsible for these defects, a rescue with Rpl7B should fail. Instead, replacement of the Rpl7A coding region and introns (CRI) with the Rpl7B CRI rescues the defects. These data show that the defects which appear to be paralog-specific come instead from differences in paralog expression that originate in the mRNA UTRs, their promoter regions, or both.
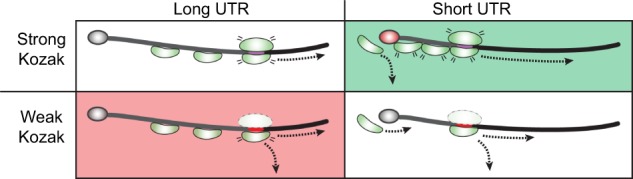
Importantly, paralog mRNA levels do not always react similarly to cellular perturbations (Gasch et al. 2000; Yadav et al. 2016). For example, most RP paralogs are similarly regulated during stress, but the Rpl13 and Rpl18 paralogs diverge during cell quiescence, and the expression of 5 paralogs is decoupled during the response to heat shock (Fig. 4). These differences might arise at the level of transcription or splicing (Parenteau et al. 2011; Petibon et al. 2016), but regardless are often used to support functional relevance. For example, while both Rpl13 paralogs are equally expressed during vegetative growth, in quiescent cells expression of Rpl13A is reduced by nearly eightfold, so nearly all mRNA is from the B paralog. Thus, while in rich media ribosome numbers are not differentially affected by deletion of a specific paralog, in quiescent cells deletion of Rpl13B will have a much larger effect. This underscores the importance of careful controls for expression levels and ribosome numbers under all relevant growth conditions.
FIGURE 4.
Ribosomal protein paralog expression ratio is differentially regulated between environmental conditions. Each graph compares the change in expression of the A and B paralogs of ribosomal proteins. The gray area around the regression line is the 95% prediction band. Orange data points were identified as outliers by ROUT (Motulsky and Brown 2006) with a Q value of 1%. Data are from Gasch et al. (2000).
Some “specialized” ribosomes may be degradation intermediates
Ribosome degradation is an important factor to keep in mind when considering specialized ribosomes. While the degradation of ribosomes has been described in some cases (Kraft et al. 2008; Cole et al. 2009; Lafontaine 2010; Ossareh-Nazari et al. 2010; Cebollero et al. 2012; Niki et al. 2014; An and Harper 2018), the process is not fully understood. More importantly, the steps in ribosome degradation, and which degradation intermediates are stable, are still unknown. This is critical because what appear to be specialized ribosomes could, in some cases, be nonfunctional ribosomes in the process of being destroyed, especially because at least one form of decay occurs on mRNAs (Cole et al. 2009) and would therefore lead to RP-depleted ribosomes in the polysome fraction.
For instance, a recent study describes a fraction of ribosomes lacking Rps1 and Rpl10/uL10, identified by cryo-EM. This fraction increased upon shifting the cells from their preferred carbon source, glucose, to glycerol (Sun et al. 2018). In contrast to other ribosomes identified, these RP-deficient ribosomes did not have any tRNA bound. We can only speculate on the nature of these ribosomes, but they could simply be in the process of being recycled. This possibility is consistent with the increase in their number after the cells are switched to less preferred growth conditions and the absence of tRNAs.
In another case, the existence of mazF-induced specialized ribosomes in bacteria has recently been questioned (Culviner and Laub 2018). Previous work, relying on reporter assays and assaying only the “specialized” ribosome populations, appeared to show that under stress site-specific cleavage by mazF generates a pool of leaderless mRNAs that are preferentially translated by ribosomes cleaved by the same nuclease (Vesper et al. 2011; Sauert et al. 2016; Nikolic et al. 2017). However, a recent genome-wide analysis of this system shows that these ribosomes and mRNAs are instead the product of widespread mRNA and rRNA cleavages, and thus possibly degradation intermediates, and failures of ribosome biogenesis (Culviner and Laub 2018).
These examples demonstrate that caution is warranted when examining claims of specialized ribosomes, as cellular perturbations can potentially generate stable ribosome degradation products or dead-end assembly intermediates that look like specialized ribosomes.
Tagging RPs can create phenotypes
The addition of epitope tags to RPs of interest, while powerful and often necessary, can also suffer from distinct drawbacks. Tagged proteins can have altered function (Swulius and Jensen 2012; Heo et al. 2017; Collins et al. 2018) or stability (De Marco et al. 2004; Kim et al. 2013; Natsume et al. 2016). These problems are especially acute when dealing with RPs, which are often small and highly charged. Thus, when a tagged RP is used to isolate a specialized ribosome, extra care must be taken.
For example, we have recently shown that HA-tagged Rps29 interacts genetically with deletion of the assembly factor Ltv1 (Collins et al. 2018), consistent with either mispositioning or partial loss of this protein upon introduction of the tag. Similarly, Rpl40-HA confers resistance to the elongation inhibitor sordarin (Fernández-Pevida et al. 2012) and C-terminal tags on Rpl15 prevent its incorporation into ribosomes (Simoff et al. 2009). Furthermore, Rps31 (and Rpl40) are translated as ubiquitin fusion pre-proteins, which are processed during or before assembly (Ozkaynak et al. 1987). In the case of Rps31, failure to do so affects ribosome activity (Finley et al. 1989; Lacombe et al. 2009).
A recent study examining ribosomes lacking Rpl10A/uL1 and Rps25 utilized Flag-tagged versions of each protein to pull down ribosomes containing the protein, and compared them to heterogeneous populations of ribosomes with and without the target protein (Shi et al. 2017). These tagged, target RP-containing ribosomes were associated with different mRNAs compared to total (tagged and untagged/target RP-bound and deficient) ribosomes from their host cells. However, the mRNA levels (which could reveal ribosome stress) in these different cell types were not probed, and moreover, the polysome distribution of the Flag-tagged Rpl10A varied slightly from that of untagged Rpl10A, indicating that the Flag-tag might subtly affect function (Shi et al. 2017). As a control, ribosomes with HA-tagged Rpl22 were analyzed and did not exhibit significant mRNA bias. However, a primary component analysis reveals that the samples derived from Rpl22-HA cells diverged significantly compared to samples from Rps25-Flag and Rpl10A-Flag cells (Fig. 5). Whether these differences arise because of subtle defects in the tagged cell lines, or perhaps more likely, because the Rpl22-HA cell line came from mice with a different background is impossible to know, but illustrates the caveats involved with tagging of RPs.
FIGURE 5.
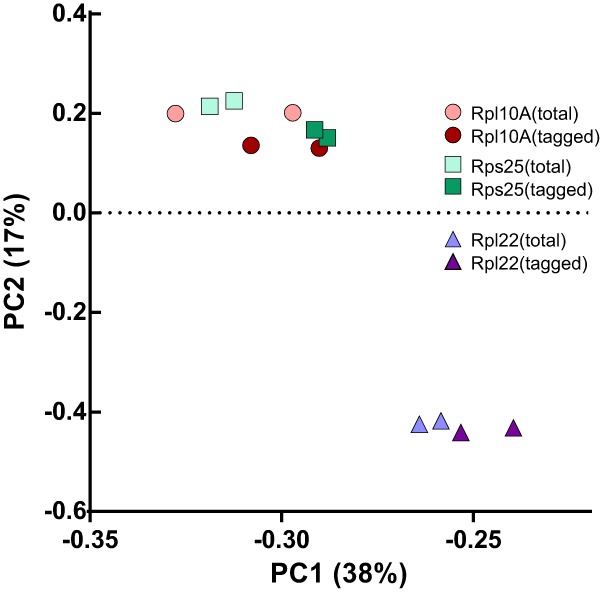
Primary component analysis of footprint data in Shi et al. (2017). RPKMs from each footprinting experiment were downloaded from NCBI and analyzed using the prcomp function in R (R Core Team 2018).
Comparisons across strains do not always unveil ribosome-derived phenotypes
General effects on cellular translation are easy to mistake for ribosome-specific regulation. For example, researchers might suspect that the modification guided by a snoRNA alters mRNA-specific translation, knock out the snoRNA, and then compare ribosome footprints between the knockout and wild type strains. While there may be clear differences between the samples, this comparison alone cannot determine if the effect is caused by altered ribosome activity or some other change in the cells. Two recent careful studies demonstrate vast changes in transcription in response to subtle variations in ribosome composition (Cheng et al. 2018a; Kurylo et al. 2018).
In E. coli, Kurylo et al. demonstrate specific induction of one of the seven rDNA operons by nutrient stress and go on to generate strains differentiated only by the small difference in rDNA sequence between the induced and the otherwise predominant operon (Kurylo et al. 2018). The authors demonstrate how this difference in rDNA sequence triggered changes in mRNA levels for nearly 20% of the annotated genes, leading to a vastly changed gene expression program. Without measuring mRNA levels, the transcriptional changes could easily have been misconstrued as originating from translational regulatory changes by the ribosome. Similarly, a study of multiple yeast strains deleted individually for several proteins of the small or large subunit demonstrates substantial transcriptional changes in these yeast strains (Cheng et al. 2018a). Intriguingly, these changes in mRNA levels are similar for different strains with deletions in each subunit, but differ between the two subunits. In other words, there are subunit-specific responses to gene expression. In these cases, the effects on TE are nearly nonexistent, although other translational effects (e.g., on pausing, translocation or stop codon read through) might be hidden deeper in the data (Cheng et al. 2018a). These examples powerfully demonstrate the importance of controlling for changes in transcription as a result from perturbations to the ribosome, as well as the importance of carrying out controls to demonstrate protein specificity.
BEST PRACTICES FOR DESCRIBING SPECIALIZED RIBOSOMES
As described above, ribosomes and ribosome assembly are connected to nearly every process in the cell, so the study of ribosome specialization is inherently complex. In addition, as described above, there are caveats to consider when interpreting the often-tantalizing findings in our laboratories. Here we lay out some best practices that we hope will help in the evaluation of the literature and direct future studies of specialized ribosomes.
Account for total ribosome numbers
Experimental systems developed to study specialized ribosomes will often drive their accumulation to levels not normally seen in the cell. Examples might include expression of an alternative rRNA, inactivation of a snoRNA, or depletion of a RP. Experiments examining ribosome specialization due to a specific RP paralog are particularly vulnerable to these artifacts. These systems generally require the deletion of one paralog to determine the functional changes caused by the other. As described above, a single RP locus is often unable to compensate for the loss of its partner, and the importance of each paralog can vary based on cellular conditions (Fig. 4). At minimum, expression of each paralog in a common UTR-context should be tested to ensure that differential effects arise from the ORF and not the noncoding regions. Ideally, western blot or mass spectroscopy should be used to validate that an RP is present at the same level as before the deletion of one paralog. Furthermore, when looking for specialization by loss of a specific RP, one should control for effects arising from a reduction in ribosome number, perhaps by ensuring the effects are observed only for the protein under investigation and not another RP.
Control for other perturbations
Tagging RPs is an effective method for isolating specific populations of ribosomes. Adding tags to proteins and RNA can also affect their structure and function; thus, experimentalists utilizing these techniques must exercise caution. Experimental systems utilizing ribosome tags should demonstrate that the tagged ribosomes are functional, potentially by generating a strain in which the tagged RP is the only source of RP expression and showing full complementation and incorporation of the tagged protein into ribosomes (we have observed tags to be proteolytically removed in ribosome-bound molecules). Changes in the overall translational state of the cells should be reported in the form of polysome profiles that can be compared. Evidence of cellular stress, and ribosome stress in particular, should also be measured and can be identified by changes in the transcription of RPs, altered polysome profiles, or both. Finally, pulldowns that compare tagged and untagged ribosomes that are otherwise identical should show that the tag itself is not the cause of any differences.
Furthermore, when two strains (or cell lines) are compared, it is critical to evaluate both transcriptional and translational changes, to ensure that observed differences arise directly from translation and no other steps in gene expression. Furthermore, the analysis of such changes should include a critical evaluation of changes in known translational modifiers. For example, if accumulation of a specific ribosome subpopulation leads to transcriptional up-regulation of a translational regulator, then this regulator could be responsible for subsequent translational changes if those do not involve up-regulated mRNAs.
Test for functionality
Finding ribosomes with altered composition does not mean they are functional. As noted above, degradation or assembly intermediates can be mistaken for specialized ribosomes, as can experimental artifacts, such as ribosomes that have lost RPs during isolation. In neither case can these ribosomes exert translational control.
To avoid this pitfall, ribosomal functionality should be tested. An in vitro translation assay would be the most direct measurement of activity. However, ensuring the purity or sufficient quantity of the ribosomes in this assay can be challenging. Testing for functional consequences of specialized ribosomes is viable but requires careful consideration of potential confounders. As we have described above, cells in which ribosomes have been altered can have unexpected defects that can be mistaken for specialized ribosome activity. For this reason, experiments that use binding or other observed functional differences to predict, and test for, gain-of-function phenotypes are the most powerful in vivo evidence of specialized ribosome functionality.
Look for regulation
Specialized ribosomes can exist without cellular mechanisms that regulate their construction. Nevertheless, evidence that cells control the levels of a particular kind of ribosome helps to establish that such ribosomes are functional. This kind of data would ideally be paired with independent reporter assays to confirm that ribosome composition is responsible for the change and is not simply being co-regulated. The best way to design such experiments is to identify possible mechanisms by which a specialized ribosome is modifying the translational landscape, and then use molecular biology to probe for predicted changes.
IS THERE EVIDENCE FOR PHYSIOLOGICAL ROLES OF SPECIALIZED RIBOSOMES?
The study of ribosome specialization is advancing rapidly, and much is still unknown about its frequency, conservation, and impact on gene expression. The criteria above lay out the considerations that claims of specialized ribosomes should be measured against. And while some potential alternative interpretations might be nearly impossible to entirely rule out (specialized ribosomes are degradation intermediates or perturbations from the tag), there are a now a few examples that suggest that ribosome heterogeneity has functional consequences and biological roles.
These include the production of Rps26-depleted ribosomes in yeast cells exposed to high salt or high pH values to promote the translation of mRNAs supporting the response to these stresses. This is mediated by loss of sequence selectivity within the Kozak sequence normally imparted by Rps26 (Ferretti et al. 2017, 2018). In other cases, mRNA specificity for individual ribosome populations has been demonstrated or suggested (Werner et al. 2015; Thompson et al. 2016; Shi et al. 2017), but biological roles for this specificity in the physiological regulation of gene expression remain unclear, and these might mainly manifest in disease-states. Similarly, whether the dependence of NGD on Asc1/RACK1 is ever exploited to block this pathway under certain conditions (or in certain cell types) remains to be seen. Kurylo et al. have provided evidence for shifting of ribosome populations in E. coli to effect functional differences in tetracycline binding that arise from variations in 16S rRNA (Kurylo et al. 2018). These variations also lead to changes in the association of AdhE, an alcohol dehydrogenase also implicated as an RNA helicase (Kurylo et al. 2018). Whether these are the cause for additional changes in ribosome function, or perhaps a reflection of other functional perturbations, which are compensated for by binding of this putative RNA helicase to the mRNA entry channel, remains to be seen. Nevertheless, this study strongly suggests the need to look beyond effects on mRNA selectivity that could arise from ribosome specialization. Such effects could include mRNA quality control, as already shown for NGD (Ikeuchi and Inada 2016), but could also include effects on termination, pausing, nascent protein modification and folding. Importantly, such effects can be hard to uncover in a conventional analysis of a ribosome profiling data set (Thompson et al. 2016).
ACKNOWLEDGMENTS
We thank Arlen Johnson, Rachel Green, and Wendy Gilbert for helpful discussion, and John Cleveland, Murray Deutscher, and members of the Karbstein laboratory for comments on the manuscript. Work on specialized ribosomes in our laboratory is supported by grants from the National Institutes of Health (R01-GM086451 and R01-GM117093 to K.K. and F31-GM116406 to M.B.F.), the Department of Defense Congressionally Directed Medical Research Program (W81XWH-16-1-0008 to K.K.), and a Howard Hughes Medical Institute Faculty Scholar grant (55108536 to K.K.). K.K. is a Howard Hughes Medical Institute Faculty Scholar.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.069823.118.
While a typical footprint for translating ribosomes is only ∼30 nt, providing a rough size cutoff for this effect, scanning and initiating ribosomes can have footprints of up to 75 nt in size (Archer et al. 2016), possibly due to the bound initiation machinery. Thus, mRNAs with 5′-UTRs of ≈75 nt might also be affected.
REFERENCES
- Ajore R, Raiser D, McConkey M, Jöud M, Boidol B, Mar B, Saksena G, Weinstock DM, Armstrong S, Ellis SR, et al. 2017. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol Med 9: 498–507. 10.15252/emmm.201606660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hadid Q, Roy K, Chanfreau G, Clarke SG. 2016. Methylation of yeast ribosomal protein Rpl3 promotes translational elongation fidelity. RNA 22: 489–498. 10.1261/rna.054569.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Harper JW. 2018. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20: 135–143. 10.1038/s41556-017-0007-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T. 2016. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535: 570–574. 10.1038/nature18647 [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. 2009. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res 37: 7665–7677. 10.1093/nar/gkp816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Roshek JL, Petrov AN, Dinman JD. 2007. Optimization of ribosome structure and function by rRNA base modification. PLoS One 2: e174 10.1371/journal.pone.0000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P, Shwetha S, Sharma DK, Joseph AP, Srinivasan N, Das S. 2015. The β hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function. Nucleic Acids Res 43: 2888–2901. 10.1093/nar/gkv110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR. 2010. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol 30: 5562–5571. 10.1128/MCB.00618-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N. 2017. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 36: 475–486. 10.15252/embj.201695959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40: 216–227. 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk AR, Green R. 2017. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc B Biol Sci 372: 20160183 10.1098/rstb.2016.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512. 10.1073/pnas.0810916106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RK, Wong B, Xie X, Lu YF, Shi PY, Pompon J, Garcia-Blanco MA, Bradrick SS. 2017. RPLP1 and RPLP2 are essential Flavivirus host factors that promote early viral protein accumulation. J Virol 91: e01706-16 10.1128/JVI.01706-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal F, Vallejos M, Walters B, Contreras N, Hertz MI, Olivares E, Cáceres CJ, Pino K, Letelier A, Thompson SR, et al. 2016. Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J 283: 2508–2527. 10.1111/febs.13756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F, Kraft C. 2012. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol 2012: 182834 10.1155/2012/182834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Mugler CF, Keskin A, Hodapp S, Chan LYL, Weis K, Mertins P, Regev A, Jovanovic M, Brar GA. 2018a. Small and large ribosomal subunit deficiencies lead to distinct gene expression signatures that reflect cellular growth rate. Mol Cell 73: 36–47.e10. 10.1016/j.molcel.2018.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Otto GM, Powers EN, Keskin A, Mertins P, Carr SA, Jovanovic M, Brar GA. 2018b. Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell 172: 910–923.e16. 10.1016/j.cell.2018.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19: 445–452. 10.1101/gad.1267905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF. 2016. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun 7: 11663 10.1038/ncomms11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikne V, Doniger T, Rajan KS, Bartok O, Eliaz D, Cohen-Chalamish S, Tschudi C, Unger R, Hashem Y, Kadener S, et al. 2016. A pseudouridylation switch in rRNA is implicated in ribosome function during the life cycle of Trypanosoma brucei. Sci Rep 6: 25296 10.1038/srep25296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG. 2010. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol Cell Biol 30: 4415–4434. 10.1128/MCB.00280-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Crétien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. 2007. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood 109: 1275–1283. 10.1182/blood-2006-07-038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian JL. 2012. Morphogen gradients in development: from form to function. Wiley Interdiscip Rev Dev Biol 1: 3–15. 10.1002/wdev.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. 2009. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 34: 440–450. 10.1016/j.molcel.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JC, Ghalei H, Doherty JR, Huang H, Culver RN, Karbstein K. 2018. Ribosome biogenesis factor Ltv1 chaperones the assembly of the small subunit head. J Cell Biol 217: 4141–4154. 10.1083/jcb.201804163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Philips J, Fu ZY, Squires C, Squires CL. 1992. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J 11: 4175–4185. 10.1002/j.1460-2075.1992.tb05511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culviner PH, Laub MT. 2018. Global analysis of the E. coli toxin MazF reveals widespread cleavage of mRNA and the inhibition of rRNA maturation and ribosome biogenesis. Mol Cell 70: 868–880.e10. 10.1016/j.molcel.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Ying K, Hung S, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Tan YZ, Carragher B, Potter CS, Lyumkis D, Williamson JR. 2016. Modular assembly of the bacterial large ribosomal subunit. Cell 167: 1610–1622.e15. 10.1016/j.cell.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco V, Stier G, Blandin S, de Marco A. 2004. The solubility and stability of recombinant proteins are increased by their fusion to NusA. Biochem Biophys Res Commun 322: 766–771. 10.1016/j.bbrc.2004.07.189 [DOI] [PubMed] [Google Scholar]
- De Silva D, Tu YT, Amunts A, Fontanesi F, Barrientos A. 2015. Mitochondrial ribosome assembly in health and disease. Cell Cycle 14: 2226–2250. 10.1080/15384101.2015.1053672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V. 2017. The structure of the yeast mitochondrial ribosome. Science 355: 528–531. 10.1126/science.aal2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD. 2016. Pathways to specialized ribosomes: the Brussels lecture. J Mol Biol 428: 2186–2194. 10.1016/j.jmb.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. 2006. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci 103: 13403–13408. 10.1073/pnas.0603730103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. 1988a. A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93. 10.1016/0092-8674(88)90182-1 [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. 1988b. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54: 95–104. 10.1016/0092-8674(88)90183-3 [DOI] [PubMed] [Google Scholar]
- Elantak L, Wagner S, Herrmannová A, Karásková M, Rutkai E, Lukavsky PJ, Valášek L. 2010. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eiF1A in stringent AUG selection. J Mol Biol 396: 1097–1116. 10.1016/j.jmb.2009.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier R, Pfeffer S, Förster F. 2017. Structure of the human mitochondrial ribosome studied in situ by cryoelectron tomography. Structure 25: 1574–1581.e2. 10.1016/j.str.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, Garcia M, Laforêts F, Marcel V, Baudin-Baillieu A, et al. 2017. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci 114: 12934–12939. 10.1073/pnas.1707674114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pevida A, Rodríguez-Galán O, Díaz-Quintana A, Kressler D, De La Cruz J. 2012. Yeast ribosomal protein L40 assembles late into precursor 60 S ribosomes and is required for their cytoplasmic maturation. J Biol Chem 287: 38390–38407. 10.1074/jbc.M112.400564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti MB, Ghalei H, Ward EA, Potts EL, Karbstein K. 2017. Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat Struct Mol Biol 24: 700–707. 10.1038/nsmb.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti MB, Barre JL, Karbstein K. 2018. Translational reprogramming provides a blueprint for cellular adaptation. Cell Chem Biol 25: 1372–1379. 10.1016/j.chembiol.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A, Rackham O. 2013. Specialization from synthesis: how ribosome diversity can customize protein function. FEBS Lett 587: 1189–1197. 10.1016/j.febslet.2013.02.032 [DOI] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A. 1989. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401. 10.1038/338394a0 [DOI] [PubMed] [Google Scholar]
- Fuchs G, Diges C, Kohlstaedt LA, Wehner KA, Sarnow P. 2011. Proteomic analysis of ribosomes: translational control of mRNA populations by glycogen synthase GYS1. J Mol Biol 410: 118–130. 10.1016/j.jmb.2011.04.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo S, Ricciardi S, Manfrini N, Pesce E, Oliveto S, Calamita P, Mancino M, Maffioli E, Moro M, Crosti M, et al. 2018. RACK1 specifically regulates translation through its binding to ribosomes. Mol Cell Biol 38 10.1128/MCB.00230-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth NR, Barna M. 2018a. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat Rev Genet 19: 431–452. 10.1038/s41576-018-0008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth NR, Barna M. 2018b. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol Cell 71: 364–374. 10.1016/j.molcel.2018.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. 2011. Functional specialization of ribosomes? Trends Biochem Sci 36: 127–132. 10.1016/j.tibs.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Li Y, Liu CJ, Xiang Y, Li C, Ye Y, Zhang Z, Hawke DH, Park PK, Diao L, et al. 2017. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep 21: 1968–1981. 10.1016/j.celrep.2017.10.070 [DOI] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE, Schmickel RD. 1988. Human 28S ribosomal RNA sequence heterogeneity. Nucleic Acids Res 16: 10213–10224. 10.1093/nar/16.21.10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. 2009. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J Biol Chem 284: 18377–18389. 10.1074/jbc.M109.001503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Séraphin B. 2012. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol 13: 727–735. 10.1038/nrm3457 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. 2014. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505: 515–519. 10.1038/nature12890 [DOI] [PubMed] [Google Scholar]
- Guimaraes JC, Zavolan M. 2016. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol 17: 236 10.1186/s13059-016-1104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. 2018. Specialized ribosomes and the control of translation. Biochem Soc Trans 46: 855–869. 10.1042/BST20160426 [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45. 10.1016/j.molcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC. 2007. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev 21: 148–159. 10.1101/gad.1477507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Martin F, Ulitsky I, Emmanuel R, Tamarkin-Ben-Harush A, Vardy A, Dikstein R. 2017. Efficient and accurate translation initiation directed by TISU involves RPS3 and RPS10e binding and differential eukaryotic initiation factor 1A regulation. Mol Cell Biol 37: e00150-17 10.1128/MCB.00150-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley MP, Gunasekera TS, Easton JA, Sigdel TK, Sugarbaker SA, Klingbeil L, Breece RM, Tierney DL, Crowder MW. 2012. Characterization of Zn(II)-responsive ribosomal proteins YkgM and L31 in E. coli. J Inorg Biochem 111: 164–172. 10.1016/j.jinorgbio.2011.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo M, Nord AL, Chamousset D, van Rijn E, Beaumont HJE, Pedaci F. 2017. Impact of fluorescent protein fusions on the bacterial flagellar motor. Sci Rep 7: 12583 10.1038/s41598-017-11241-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Woellhaf MW, Bonnefoy N. 2013. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta 1833: 286–294. 10.1016/j.bbamcr.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Hillebrand A, Wurm R, Menzel A, Wagner R. 2005. The seven E. coli ribosomal RNA operon upstream regulatory regions differ in structure and transcription factor binding efficiencies. Biol Chem 386: 523–534. 10.1515/BC.2005.062 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42: 589–611. 10.1016/j.tibs.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Hurowitz EH, Brown PO. 2003. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol 5: R2 10.1186/gb-2003-5-1-r2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Inada T. 2016. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci Rep 6: 28234 10.1038/srep28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. 1989. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338: 646–648. 10.1038/338646a0 [DOI] [PubMed] [Google Scholar]
- Jacobs FA, Bird RC, Sells BH. 1985. Differentiation of rat myoblasts: regulation of turnover of ribosomal proteins and their mRNAs. Eur J Biochem 150: 255–263. 10.1111/j.1432-1033.1985.tb09015.x [DOI] [PubMed] [Google Scholar]
- Joazeiro CA. 2017. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu Rev Cell Dev Biol 33: 343–368. 10.1146/annurev-cellbio-111315-125249 [DOI] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, et al. 2018. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173: 90–103.e19. 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HL, Shin EK, Kim HM, Ryou SM, Kim S, Cha CJ, Bae J, Lee K. 2007. Heterogeneous rRNAs are differentially expressed during the morphological development of Streptomyces coelicolor. FEMS Microbiol Lett 275: 146–152. 10.1111/j.1574-6968.2007.00872.x [DOI] [PubMed] [Google Scholar]
- Kim HL, Song WS, Kim K, Lee K. 2008. Characterization of heterogeneous LSU rRNA profiles in Streptomyces coelicolor under different growth stages and conditions. Curr Microbiol 57: 537–541. 10.1007/s00284-008-9238-1 [DOI] [PubMed] [Google Scholar]
- Kim I, Miller CR, Young DL, Fields S. 2013. High-throughput analysis of in vivo protein stability. Mol Cell Proteomics 12: 3370–3378. 10.1074/mcp.O113.031708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Bin KE, Kim YJ, Chang JS, Kim J. 2017. RACK1 depletion in the ribosome induces selective translation for non-canonical autophagy. Cell Death Dis 8: e2800 10.1038/cddis.2017.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. 10.1016/j.cell.2007.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev AA. 2011. Structural aspects of translation termination on the ribosome. RNA 17: 1409–1421. 10.1261/rna.2733411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T, Dányi I, Gunišová S, Munzarová V, Vlčková V, Cuchalová L, Neueder A, Milkereit P, Valášek LS. 2012. Small ribosomal protein RPS0 stimulates translation initiation by mediating 40S-binding of eIF3 via its direct contact with the eIF3a/TIF32 subunit. PLoS One 7: e40464 10.1371/journal.pone.0040464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610. 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- Krishnan P, Ghosh S, Wang B, Heyns M, Graham K, Mackey JR, Kovalchuk O, Damaraju S. 2016. Profiling of small nucleolar RNAs by next generation sequencing: potential new players for breast cancer prognosis. PLoS One 11: e0162622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh N, Jansson MD, Häfner SJ, Tehler D, Birkedal U, Christensen-Dalsgaard M, Lund AH, Nielsen H. 2016. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res 44: 7884–7895. 10.1093/nar/gkw482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Dolezal JM, Wang H, Jackson L, Lu J, Frodey BP, Dosunmu-Ogunbi A, Li Y, Fromherz M, Kang A, et al. 2017. Ribosomopathy-like properties of murine and human cancers. PLoS One 12: e0182705 10.1371/journal.pone.0182705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo BA, Gonzalez IL, Gillespie DA, Sylvester JE. 1996. Human ribosomal RNA variants from a single individual and their expression in different tissues. Nucleic Acids Res 24: 4817–4824. 10.1093/nar/24.23.4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurylo CM, Parks MM, Juette MF, Zinshteyn B, Altman RB, Thibado JK, Vincent CT, Blanchard SC. 2018. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep 25: 236–248.e6. 10.1016/j.celrep.2018.08.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bähler J. 2007. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155. 10.1016/j.molcel.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe T, García-Gómez JJ, De La Cruz J, Roser D, Hurt E, Linder P, Kressler D. 2009. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol 72: 69–84. 10.1111/j.1365-2958.2009.06622.x [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ. 2010. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci 35: 267–277. 10.1016/j.tibs.2009.12.006 [DOI] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. 10.1101/gad.1832209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. 2006. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell 24: 619–626. 10.1016/j.molcel.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask MB, et al. 2014. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol 16: 792–803. 10.1038/ncb3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Cate JHD. 2015a. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522: 111–114. 10.1038/nature14267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Swanson MJ, Sattlegger E. 2015b. Gcn1 contacts the small ribosomal protein Rps10, which is required for full activation of the protein kinase Gcn2. Biochem J 466: 547–559. 10.1042/BJ20140782 [DOI] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Doudna JA, Cate JHD. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536: 96–99. 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162. 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. 2009. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol 7: e1000213 10.1371/journal.pbio.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Liu Q, Fournier MJ. 2007. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 28: 965–977. 10.1016/j.molcel.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. 2009. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15: 1716–1728. 10.1261/rna.1724409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilleorg S, Reier K, Pulk A, Liiv A, Tammsalu T, Peil L, Cate JD, Remme J. 2019. Bacterial ribosome heterogeneity: changes in ribosomal protein composition during transition into stationary growth phase. Biochimie 156: 169–180. 10.1016/j.biochi.2018.10.013 [DOI] [PubMed] [Google Scholar]
- Lin CI, Yeh NH. 2009. Treacle recruits RNA polymerase I complex to the nucleolus that is independent of UBF. Biochem Biophys Res Commun 386: 396–401. 10.1016/j.bbrc.2009.06.050 [DOI] [PubMed] [Google Scholar]
- Liu H, Yin J, Xiao M, Gao C, Mason AS, Zhao Z, Liu Y, Li J, Fu D. 2012. Characterization and evolution of 5′ and 3′ untranslated regions in eukaryotes. Gene 507: 106–111. 10.1016/j.gene.2012.07.034 [DOI] [PubMed] [Google Scholar]
- Locati MD, Pagano JFB, Girard G, Ensink WA, van Olst M, Van Leeuwen S, Nehrdich U, Spaink HP, Rauwerda H, Jonker MJ, et al. 2017. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 23: 1188–1199. 10.1261/rna.061515.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF. 1974. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251: 385–388. 10.1038/251385a0 [DOI] [PubMed] [Google Scholar]
- Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, Mackeen MM, McDonough MA, Cockman ME, Kessler BM, Ratcliffe PJ, et al. 2014. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci 111: 4019–4024. 10.1073/pnas.1311750111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 227: 383–387. 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- López-López A, Benlloch S, Bonfá M, Rodríguez-Valera F, Mira A. 2007. Intragenomic 16S rDNA divergence in Haloarcula marismortui is an adaptation to different temperatures. J Mol Evol 65: 687–696. 10.1007/s00239-007-9047-3 [DOI] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906. 10.1093/nar/gkr1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Bah E, Madireddy R, Zhang Y, Brilot AF, Grigorieff N, Korostelev AA. 2016. Ribosome•RelA structures reveal the mechanism of stringent response activation. eLife 5: e17029 10.7554/eLife.17029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Shimada T, Ishihama A. 2015. Strength and regulation of seven rRNA promoters in Escherichia coli. PLoS One 10: e0144697 10.1371/journal.pone.0144697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, et al. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159: 1086–1095. 10.1016/j.cell.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Ponomarev VA, Koonin EV. 2001. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol 2: research0033.1–research0033.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannoor K, Liao J, Jiang F. 2012. Small nucleolar RNAs in cancer. Biochim Biophys Acta 1826: 121–128. 10.1016/j.bbcan.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Azorín F, Remacha M, Martínez-Salas E, Ballesta JPG. 2008. Internal translation initiation on the foot-and-mouth disease virus IRES is affected by ribosomal stalk conformation. FEBS Lett 582: 3029–3032. 10.1016/j.febslet.2008.07.039 [DOI] [PubMed] [Google Scholar]
- Mathis AD, Naylor BC, Carson RH, Evans E, Harwell J, Knecht J, Hexem E, Peelor FF, Miller BF, Hamilton KL, et al. 2017. Mechanisms of in vivo ribosome maintenance change in response to nutrient signals. Mol Cell Proteomics 16: 243–254. 10.1074/mcp.M116.063255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas RG, Tollervey D, Hurt E. 2014. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505: 112–116. 10.1038/nature12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. 2002. The ribosome filter hypothesis. Proc Natl Acad Sci 99: 12031–12036. 10.1073/pnas.192442499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. 2007. The ribosome filter redux. Cell Cycle 6: 2246–2251. 10.4161/cc.6.18.4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh KB, Bhattacharya A, Willis IM, Warner JR. 2011. Eukaryotic cells producing ribosomes deficient in rpl1 are hypersensitive to defects in the ubiquitin-proteasome system. PLoS One 6: e23579 10.1371/journal.pone.0023579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin TW, Hallberg RL. 1986. Effect of heat shock on ribosome structure: appearance of a new ribosome-associated protein. Mol Cell Biol 6: 2527–2535. 10.1128/MCB.6.7.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR, et al. 2012. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 31: 2794–2804. 10.1038/onc.2011.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R. 2017. Ribosomopathies: there's strength in numbers. Science 358: eaan2755 10.1126/science.aan2755 [DOI] [PubMed] [Google Scholar]
- Motulsky H, Brown R. 2006. Detecting outliers when fitting data with nonlinear regression—a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7: 123 10.1186/1471-2105-7-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. 2010. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science 330: 673–677. 10.1126/science.1193220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Sugihara Y, Shibakawa M, Oshima K, Matsuda T, Nadano D. 2018. The mRNA-binding protein Serbp1 as an auxiliary protein associated with mammalian cytoplasmic ribosomes. Cell Biochem Funct 36: 312–322. 10.1002/cbf.3350 [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT. 2016. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep 15: 210–218. 10.1016/j.celrep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Niki T, Saito S, Gladish DK. 2014. Granular bodies in root primary meristem cells of Zea mays L. var. Cuscoensis K. (Poaceae) that enter young vacuoles by invagination: a novel ribophagy mechanism. Protoplasma 251: 1141–1149. 10.1007/s00709-014-0622-3 [DOI] [PubMed] [Google Scholar]
- Nikolay R, van den Bruck D, Achenbach J, Nierhaus KH. 2015. Ribosomal proteins: role in ribosomal functions In eLS, pp. 1–12. Wiley, Chichester, UK. [Google Scholar]
- Nikolic N, Didara Z, Moll I. 2017. MazF activation promotes translational heterogeneity of the grcA mRNA in Escherichia coli populations. PeerJ 5: e3830 10.7717/peerj.3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C. 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11: 548–554. 10.1038/embor.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Amunts A, Brown A. 2016. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem 85: 77–101. 10.1146/annurev-biochem-060815-014334 [DOI] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. 1987. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J 6: 1429–1439. 10.1002/j.1460-2075.1987.tb02384.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo RJ, Fuchs G, Lutz S, Curcio MJ. 2017. Paralog-specific functions of RPL7A and RPL7B mediated by ribosomal protein or snoRNA dosage in Saccharomyces cerevisiae. G3 7: 591–606. 10.1534/g3.116.035931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini NA, Attwood M, Sondalle SB, Vieira CMDS, van Adrichem AM, di Summa FM, O'Donohue MF, Gleizes PE, Rachuri S, Briggs JW, et al. 2017. A ribosomopathy reveals decoding defective ribosomes driving human dysmorphism. Am J Hum Genet 100: 506–522. 10.1016/j.ajhg.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Ve Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Abou Elela S. 2011. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147: 320–331. 10.1016/j.cell.2011.08.044 [DOI] [PubMed] [Google Scholar]
- Park M, Nishimura K, Zanelli C, Valentini S. 2010. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38: 491–500. 10.1007/s00726-009-0408-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, Blanchard SC. 2018. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv 4: eaao0665 10.1126/sciadv.aao0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A, Yue JX, Pflieger D, Bergström A, Sigwalt A, Barre B, Freel K, Llored A, et al. 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petibon C, Parenteau J, Catala M, Elela SA. 2016. Introns regulate the production of ribosomal proteins by modulating splicing of duplicated ribosomal protein genes. Nucleic Acids Res 44: 3878–3891. 10.1093/nar/gkw140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov SV, Söll D, Steitz TA. 2015. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 22: 342–344. 10.1038/nsmb.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, Liiv A, Peil L, Maiväli Ü, Nierhaus K, Remme J. 2010. Ribosome reactivation by replacement of damaged proteins. Mol Microbiol 75: 801–814. 10.1111/j.1365-2958.2009.07002.x [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Rajamäki ML, Xi D, Sikorskaite-Gudziuniene S, Valkonen JPT, Whitham SA. 2017. Differential requirement of the ribosomal protein S6 and ribosomal protein S6 kinase for plant-virus accumulation and interaction of S6 kinase with potyviral VPg. Mol Plant Microbe Interact 30: 374–384. 10.1094/MPMI-06-16-0122-R [DOI] [PubMed] [Google Scholar]
- Ramagopal S, Ennis HL. 1984. Decay and synthesis of ribosomal proteins during Dictyostelium discoideum development. MGG Mol Gen Genet 194: 466–470. 10.1007/BF00425559 [DOI] [Google Scholar]
- Richter JD, Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480. 10.1038/nature03205 [DOI] [PubMed] [Google Scholar]
- Rotenberg MO, Moritz M, Woolford JL. 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev 2: 160–172. 10.1101/gad.2.2.160 [DOI] [PubMed] [Google Scholar]
- Samir P, Browne CM, Rahul SM, Shen B, Li W, Frank J, Link AJ. 2018. Identification of changing ribosome protein compositions using mass spectrometry. Proteomics 18: e1800217 10.1002/pmic.201800217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai ZA, Miller L, Molloy KR, Barandun J, Hunziker M, Chaker-Margot M, Wang J, Chait BT, Klinge S. 2018. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556: 126–129. 10.1038/nature26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauert M, Temmel H, Moll I. 2015. Heterogeneity of the translational machinery: variations on a common theme. Biochimie 114: 39–47. 10.1016/j.biochi.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauert M, Wolfinger MT, Vesper O, Müller C, Byrgazov K, Moll I. 2016. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res 44: 6660–6675. 10.1093/nar/gkw115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. 2015. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 6: 6158 10.1038/ncomms7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CCC, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205.e5. 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N, Gerst JE. 2018. Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J Cell Biol 217: 117–126. 10.1083/jcb.201706059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. 2013. Rate-limiting steps in yeast protein translation. Cell 153: 1589–1601. 10.1016/j.cell.2013.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Barna M. 2015. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu Rev Cell Dev Biol 31: 31–54. 10.1146/annurev-cellbio-100814-125346 [DOI] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, Barna M. 2017. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83.e7. 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Thomas EN, Zaher HS. 2017. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev RNA 8: e1366 10.1002/wrna.1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoff I, Moradi H, Nygärd O. 2009. Functional characterization of ribosomal protein L15 from Saccharomyces cerevisiae. Curr Genet 55: 111–125. 10.1007/s00294-009-0228-z [DOI] [PubMed] [Google Scholar]
- Simsek D, Barna M. 2017. An emerging role for the ribosome as a nexus for post-translational modifications. Curr Opin Cell Biol 45: 92–101. 10.1016/j.ceb.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, Barna M. 2017. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169: 1051–1065. 10.1016/j.cell.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton RS, Liu-Yi P, Formenti F, Ge W, Sekirnik R, Fischer R, Adam J, Pollard PJ, Wolf A, Thalhammer A, et al. 2014. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci 111: 4031–4036. 10.1073/pnas.1314482111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Semrau S, Airoldi E, Budnik B, van Oudenaarden A. 2015. Differential stoichiometry among core ribosomal proteins. Cell Rep 13: 865–873. 10.1016/j.celrep.2015.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. 2017. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 14: 1138–1152. 10.1080/15476286.2016.1259781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. 10.1126/science.1220270 [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867 10.1126/science.aad3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, McCormick MA, Pham KM, Mackay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. 2012. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191: 107–118. 10.1534/genetics.111.136549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter AW, Fischer C, Kleinschmidt M, Braus GH. 2006. FLO11 mediated filamentous growth of the yeast Saccharomyces cerevisiae depends on the expression of the ribosomal RPS26 genes. Mol Genet Genomics 276: 113–125. 10.1007/s00438-006-0127-7 [DOI] [PubMed] [Google Scholar]
- Suhm T, Habernig L, Rzepka M, Kaimal JM, Andréasson C, Büttner S, Ott M. 2018. A novel system to monitor mitochondrial translation in yeast. Microb cell 5: 158–164. 10.15698/mic2018.03.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun DL, Jiang X, Wu QL, Zhou NY. 2013. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol 79: 5962–5969. 10.1128/AEM.01282-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Samir P, Shen B, Li W, Browne CM, Frank J, Link AJ. 2018. Identification of changing ribosome protein compositions using cryo-EM and mass spectrometry. bioRxiv 10.1101/271833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. 2017. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell 65: 751–760.e4. 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Jensen GJ. 2012. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J Bacteriol 194: 6382–6386. 10.1128/JB.00505-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389: 90–93. 10.1038/38015 [DOI] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T. 2016. The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res 44: 8951–8961. 10.1093/nar/gkw564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, Gilbert WV. 2016. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife 5: e11154 10.7554/eLife.11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R. 2014. The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120. 10.1093/mp/ssu022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. 2015. Differential requirements for eIF4E dose in normal development and cancer. Cell 162: 59–71. 10.1016/j.cell.2015.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Chou W, Wang J, Zhang X, Zhang S, Schultz RM. 2008. Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS One 3: e1843 10.1371/journal.pone.0001843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valásek LS. 2012. ‘Ribozoomin’—translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 13: 305–330. 10.2174/138920312801619385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147: 147–157. 10.1016/j.cell.2011.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L, Serrano R, Ruiz A, Domenech G, Giraldo J, Barceló A, Ariño J. 2004. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J Biol Chem 279: 43614–43624. 10.1074/jbc.M403606200 [DOI] [PubMed] [Google Scholar]
- Walker-Kopp N, Jackobel AJ, Pannafino GN, Morocho PA, Xu X, Knutson BA. 2017. Treacher Collins syndrome mutations in Saccharomyces cerevisiae destabilize RNA polymerase I and III complex integrity. Hum Mol Genet 26: 4290–4300. 10.1093/hmg/ddx317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Zinn AR, Page DC, Nishimoto T. 1993. Functional equivalence of human X- and Y-encoded isoforms of ribosomal protein S4 consistent with a role in Turner syndrome. Nat Genet 4: 268–271. 10.1038/ng0793-268 [DOI] [PubMed] [Google Scholar]
- Werner A, Iwasaki S, McGourty CA, Medina-Ruiz S, Teerikorpi N, Fedrigo I, Ingolia NT, Rape M. 2015. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525: 523–527. 10.1038/nature14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al. 2014. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513: 65–70. 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369. 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. 2015. Cis-regulatory RNA elements that regulate specialized ribosome activity. RNA Biol 12: 1083–1087. 10.1080/15476286.2015.1085149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. 2015. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38. 10.1038/nature14010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Radhakrishnan A, Panda A, Singh A, Sinha H, Bhanot G. 2016. The modular adaptive ribosome. PLoS One 11: e0166021 10.1371/journal.pone.0166021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhang C, Dittman JD, Whitham SA. 2009. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology 390: 163–173. 10.1016/j.virol.2009.05.018 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J, Nagasaka T, Ma Y, Takayama T, Fujiwara T, et al. 2017. SNORA21—an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine 22: 68–77. 10.1016/j.ebiom.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn AR, Alagappan RK, Brown LG, Wool I, Page DC. 1994. Structure and function of ribosomal protein S4 genes on the human and mouse sex chromosomes. Mol Cell Biol 14: 2485–2492. 10.1128/MCB.14.4.2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Rojas-Duran MF, Gilbert WV. 2017. Translation initiation factor eIF4G1 preferentially binds yeast transcript leaders containing conserved oligo-uridine motifs. RNA 23: 1365–1375. 10.1261/rna.062059.117 [DOI] [PMC free article] [PubMed] [Google Scholar]