Abstract
The SidA ornithine N5-monooxygenase from A. fumigatus is a flavin monooxygenase that catalyzes the NADPH-dependent hydroxylation of ornithine. Herein we report a mutagenesis study targeting four residues that contact ornithine in crystal structures of SidA: Lys107, Asn293, Asn323, and Ser469. Mutation of Lys107 to Ala abolishes activity as measured in steady-state oxygen consumption and ornithine hydroxylation assays, indicating that the ionic interaction of Lys107 with the carboxylate of ornithine is essential for catalysis. Mutation of Asn293, Asn323, or Ser469 individually to Ala results in >14-fold increases in Km values for ornithine. Asn323 to Ala also increases the rate constant for flavin reduction by NADPH by 18-fold. Asn323 is unique among the four ornithine binding residues in that it also interacts with NADPH by forming a hydrogen bond with the nicotinamide ribose. The crystal structure of N323A complexed with NADP+ and ornithine shows that the nicontinamide riboside group of NADP+ is disordered. This result suggests that the increase in flavin reduction rate results from an increase in conformational space available to the enzyme-bound NADP(H). Asn323 thus facilitates ornithine binding at the expense of hindering flavin reduction, which demonstrates the delicate balance that exists within protein-ligand interaction networks in enzyme active sites.
Keywords: flavin-dependent monooxygneases, siderophore, ornithine hydroxylase, hydroperoxyflavin
Introduction
Pathogenic human fungi of the Aspergillus genus are responsible for severe human diseases ranging from allergic reactions and lung infections to sepsis and death. There are hundreds of members in the Aspergillus genus, but only a few have been identified as pathogenic to humans, with A. fumigatus and A. niger being the most common. The threat of fungal infections to human health has only recently been recognized as a global problem and it is estimated that ~200,000 new cases of invasive aspergillosis occur every year. Even if treated with current antifungal drugs, these infections have a 50% or greater fatality rate [1–3].
A potential avenue for therapeutic intervention in Aspergillus infections is to interfere with the acquisition of iron. Iron is an essential nutrient for fungal growth but is unavailable to invading pathogens in humans, as it is sequestered by iron binding proteins such as transferrin, lactoferrin, and hemoglobin [4]. To overcome this iron deficiency, A. fumigatus synthesizes and secretes low-molecular-weight iron chelators, called siderophores, to scavenge iron from the host. Siderophores are synthesized via non-ribosomal peptide synthetases and contain iron-chelating functional groups such as carboxylates, catecholates, and hydroxamates [4, 5].
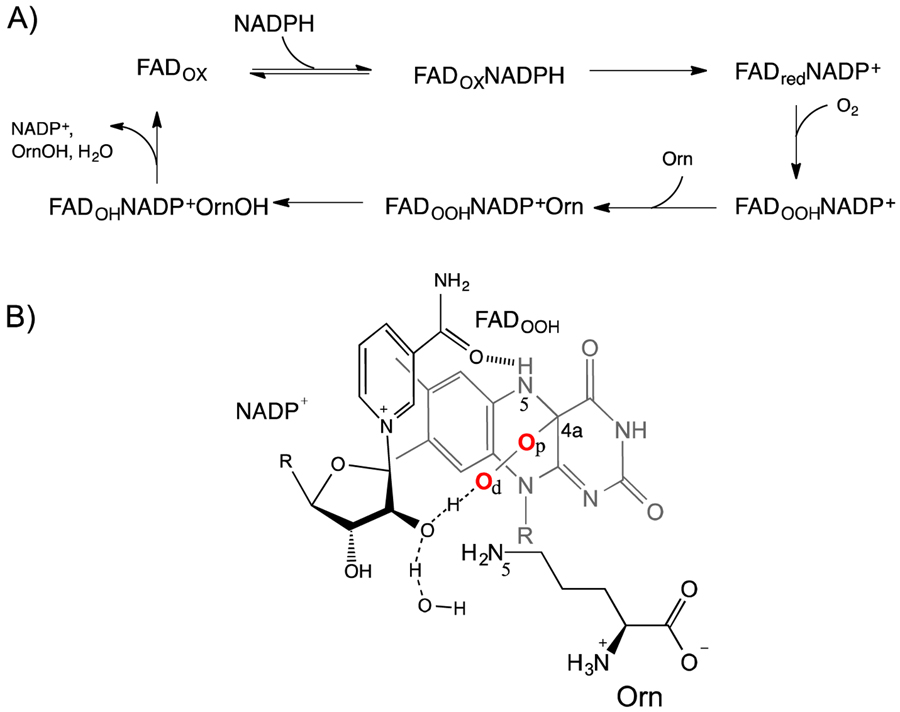
The hydroxamate functional groups of siderophores are commonly derived from the hydroxylation of the terminal amino group of the amino acids L-lysine and L-ornithine (Orn), a reaction catalyzed by flavin-dependent monooxygenases [6] [7]. In A. fumigatus, the N5-ornithine hydroxylase (SidA) has been shown to be essential for pathogenesis [8]. This enzyme is not present in humans, making it an ideal drug target. The catalytic cycle of SidA includes reduction of the flavin adenine nucleotide (FAD) by NADPH, followed by reaction with molecular oxygen to form C4a-hydroperoxyflavin (FADOOH), which is the hydroxylating species. This intermediate is very stable in SidA with a half-life of ~30 min. After Orn binding, the distal oxygen of the FADOOH is transferred to Orn to form HO-N5-Orn (Scheme 1A)[9–11].
Scheme 1.
A) Sequential kinetic mechanism for SidA. The reaction is initiated by binding of NADPH to SidA with the flavin in its oxidized form (FADox). The rate-limiting step in the reaction is the stereospecific transfer of the pro-R-hydride equivalent from NADPH to yield reduced flavin (FADred) and NADP+. The FADred-NADP+ complex reacts with molecular oxygen forming the C4a-hydroperoxyflavin (FADOOH). In this complex, NADP+ is essential for stabilization of the FADOOH. After ornithine (Orn) binding, hydroxylation occurs very quickly. The last step in the reaction is the release of products. B) Predicted structure of the FADOOH /NADP/ornithine complex. Protonation of the distal oxygen (Od) is mediated by proton transfer of the 2’OH of the NADPH ribose. The hydroxylation mechanism involves the transfer of the Od to the N5-Orn atom.
Perhaps the most interesting mechanistic aspect of the SidA reaction is that after the reduction step, NADP+ remains bound throughout the catalytic cycle [10]. This has been shown to be essential for efficient catalysis by stabilizing the FADOOH and thus minimizing the release of reactive oxygen species [12]. It has been proposed that after FAD reduction, the NADP+ swings into a conformation that promotes stabilization of the FADOOH through interaction with the 2’-OH of the nicotinamide ribose and hydrogen bonding with the H-N5-FAD (Scheme 1B) [11, 13]. This crucial role for NADP(H) appears to be a common mechanistic feature among flavin-dependent monooxygenases [11, 14–17].
SidA is specific for Orn and is almost 100% coupled, meaning that for every one molecule of oxygen consumed, a nearly stoichiometric amount of HO-N5-Orn is formed [10, 11, 18]. The strict substrate specificity of SidA differs from related members of Class B monooxygenases, which can hydroxylate a large number of substrates [19, 20]. SidA forms a complex between FADOOH and Orn. In contrast, other related enzymes, such as mammalian flavinmonooxygenases, do not form a complex with their substrates but, instead, react by a second order mechanism [21].
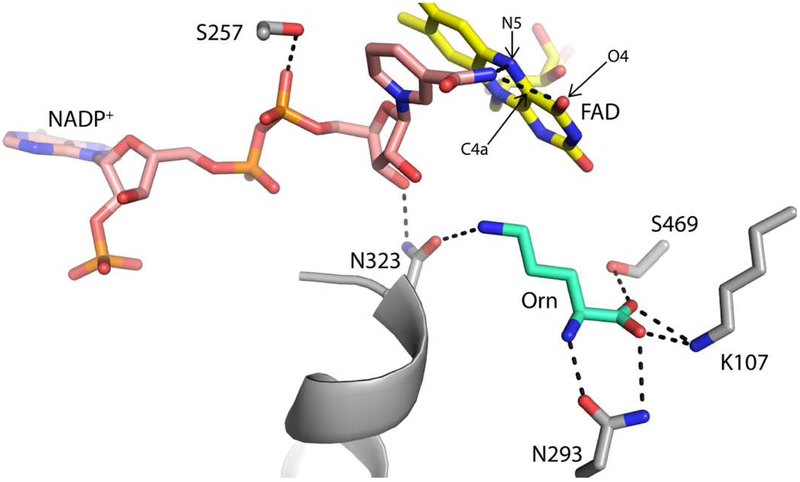
The three-dimensional crystal structure of SidA has been solved with Orn and NADP(H) bound in the active site [11]. In SidA, the binding site of Orn includes ionic interaction with Lys107, and hydrogen bonding interactions with Asn293, Ser469, and Asn323 (Fig. 1). These residues are conserved in related Orn monooxygenases from Pseudomona aeruginosa, PvdA, and from Kutzneria sp. 744, KtzI [22, 23]. These four residues were mutated to Ala in order to determine their effect on Orn binding and role in catalysis.
Fig. 1.
View of the Orn binding site in oxidized SidA (PDB code 4B63) highligting the interactions formed by the four residues mutated in this study (Lys107, Asn293, Asn323, and Ser469).
Materials and Methods
Materials.
Buffers and media were obtained from Fisher Scientific (Pittsburgh, PA). BL21(DE3)-T1R chemically competent cells were obtained from Sigma-Aldrich (St. Louis, MO). NADPH was obtained from EMD4 Biosciences (Billerica, MA). DNA primers were synthesized by Integrated DNA Technologies (Coralville, IA). Plasmid preparation and gel purification kits were obtained from Qiagen (Valencia, CA). Escherichia coli TOP-10 chemically competent cells were obtained from Invitrogen (Carlsbad, CA). Chromatography columns were from GE Healthcare.
Site–directed mutagenesis.
Mutagenesis of residues was performed using the QuikChange (Agilent Technologies) method following manufacturer’s instructions. The wild-type SidA (wtSidA) gene, subcloned into the pET15b plasmid, was used as the template [9]. Primers used in mutagenesis reactions were: forward primer (5’-GCAGATCAGCTTCATCGCGGATCTCG-3’) and reverse primer (5’-CGCGGAGGGTGGCGAGATCCGCGAT-3’) for K107A; forward primer (5’-CTCGCCTTTCGTCGCGGAGATCTTCA-3’) and reverse primer (5’-GGGTTGAAGATCTCCGCGACGAAAG-3’) for N293A; forward primer (5’-GACAAGGCTACCGCGTACTCCGTCGT-3’) and reverse primer (5’-CGGACGACGGAGTACGCGGTAGCCT-3’) for N323A; and forward primer (5’-GACTCCCTGCTGGCCGTCCTCGCCG-3’) and reverse primer (5’-CGGCGAGGACGGCCAGCAGGGAGTC-3’) for S469A. Codons for the mutated residues are underlined. The mutations were confirmed by DNA sequencing at the Virginia Bioinformatics Institute, Core Sequencing Facility.
Protein expression and purification.
All mutant proteins were expressed in E. coli BL21(DE3)-T1R competent cells and purified as previously described [11]. In general, ~25 mg of protein was obtained per liter of media. The purified proteins were stored in 100 mM sodium phosphate and 50 mM NaCl, pH 7.5, at −80 °C at a concentration of ~200 μM (based on flavin content) in 30 μL aliquots. The 6x-His tag was removed from the SidA N323A sample used for crystallization by incubating overnight at 4 °C with thrombin as previously described [11].
Determination of flavin incorporation and enzyme concentration
There were no variations in the flavin spectra for any mutant enzyme (data not shown). Flavin incorporation was determined by measuring protein concentration via the Bradford assay (BioRad) and comparing it to the protein concentration based on the flavin spectra. SidA concentration was determinied based on flavin content using an extinction coeficent of 13.7 mM−1cm−1 at 450 nm [10]. Flavin incorporation was generally close to ~70%. For all of the enzymatic assays presented here the enzyme concentration was determined using the flavin absorbance value at 450 nm.
Steady-state kinetics.
The rate of oxygen consumption was measured using a Hansatech Oxygraph system (Norfolk, England). Reactions consisted of a 1 mL volume of 100 mM sodium phosphate, pH 7.5, at 25 °C. When NADPH was varied (0.01–1.5 mM), Orn was held constant at 15 mM for wtSidA, 200 mM for N293A, 30 mM for N323A, and 500 mM for S469A. When Orn was varied (1– 250 mM), NADPH was kept constant at a concentration of 1 mM for all SidA isoforms. For K107A, activity was tested with 500 mM Orn and 1 mM NADPH. Reactions were initiated by addition of 2 μM enzyme and monitored for 5 min with constant stirring.
Hydroxylated Orn was monitored using a variation of the Csaky iodine oxidation assay [24–26]. The assay solution contained 104 μL of 100 mM sodium phosphate (pH 7.5) with varying concentrations of Orn and NADPH was held constant at 1 mM. Reactions were initiated by addition of 2.0 μM enzyme and incubated for 10 min at 25 °C with constant shaking at 750 rpm.
Pre-steady-state kinetics.
All rapid reaction experiments were carried out at 25 °C using a SX-20 stopped-flow spectrophotometer (Applied Photophysics, Leatherhead, UK) housed inside an anaerobic glove box (Coy, Grass Lake, MI). Oxygen was removed from buffers solutions with five cycles of vacuum and flushing (12 min each) with O2-free argon. The enzyme was made anaerobic with cycles of vacuum (60 s) and flushing with O2 -free argon (5 min) for 1 hr. Solutions containing substrate were made in anaerobic buffer inside the glove box. The stopped-flow was made anaerobic by flushing with 1 mL of anaerobic 100 mM sodium acetate, pH 5.0, containing 100 mM D-glucose and 100 μg/mL glucose oxidase Type-X. The rate constants for flavin reduction were measured in single mixing mode where anaerobic SidA (15 μM after mixing) was mixed with an equal volume of NADPH (15 μM or 100 μM after mixing).
The rate constants for flavin oxidation were measured in double mixing mode. Anaerobic SidA (60 μM before mixing) was first mixed with an equal volume of NADPH (60 μM before mixing). This mixture was allowed to incubate in an aging loop for 60 s until the bound flavin was fully reduced. The reduced SidA-NADP+ complex was then allowed to react with molecular oxygen (300 μM after mixing). Spectra were taken on a logarithmic time scale until the half-reactions were complete.
Synthesis of 4-pro-R-42H-NADPH.
4-pro-R-42H-NADPH was synthesized by the method of Jeong [27] with some minor modifications as described by Dhatwalia et al. [28]. Briefly, 25 mg of NADP+ (5.6 mM final concentration), 480 μL 2-propanol-d8 (1 M final concentration), and 50 units of alcohol dehydrogenase from Thermoanaerobium brockii were added to 6 mL of 25 mM Tris-Cl, pH 9.0. The reaction was allowed to proceed for 30 min at 40 °C until the A260/A340 reached ~2.5. The solution was then filtered with a 30 kDa molecular-weight cutoff filter and rotary evaporated to ~1.5 mL. The sample was then diluted up to 15 mL with 100% ethanol and stored at −20 °C for 30 min. The precipitated sample was then centrifuged for 10 min at 14,100 g at 4 °C. The resulting pellets were resuspended in 25 mM Tris-Cl, pH 9.0, and stored at −20 °C. NADPH was synthesized by the same method as the control to account for any differences with commercial samples.
Analysis of kinetic data.
All data were fitted using KaleidaGraph (Synergy Software, Reading, PA), and the derived kinetic parameters are listed in Tables 1 and 2. Rates of reduction were determined by fitting the decrease in absorbance at 452 nm to a double exponential decay equation (eq. 1).
Table 1.
Steady-state kinetic parameters with Orn as substrate.
| Parameter | Wild-type | N293A | N323A | S469A |
|---|---|---|---|---|
| Oxygen consumption Assay | ||||
| kcat, s−1 | 0.59 ± 0.01 | 0.88 ± 0.08 | 1.06 ± 0.06 | 0.15 ± 0.01 |
| Km(Orn), mM | 0.3 | 18 ± 2 | 15 ± 1 | 21 ± 2 |
| kcat/Km(Orn), M−1s−1 | 540 ± 20 | 50 ± 7 | 71 ± 6 | 7.1 ± 0.8 |
| Km(NADPH), mM | 0.0070 ± 0.0001 | 0.06 ± 0.01 | 0.010 ± 0.003 | 0.025 ± 0.010 |
| kcat/Km(NADPH), M−1s−1 | 84000 ± 2000 | 15000 ± 3000 | 110000 ± 32000 | 6000 ± 2000 |
| Orn hydroxylation assay | ||||
| kcat, s−1 | 0.62 ± 0.02 | 0.53 ± 0.04 | 0.5 ± 0.1 | 0.107 ± 0.009 |
| Km(Orn), mM | 1.0 ± 0.2 | 16 ± 1 | 12 ± 2 | 18± 4 |
| kcat/Km(Orn), M−1s−1 | 600 ± 100 | 33 ± 3 | 42 ± 11 | 6± 1 |
| Coupling, % | 105 ± 4 | 60 ± 7 | 50 ± 10 | 71 ± 8 |
Conditions: 100 mM sodium phosphate, pH 7.5, and 25 °C.
Table 2.
Pre-steady-state kinetic parameters of flavin reduction and oxidation with NADPH.
| Parameter | wtSidA | K107A | N293A | N323A | S469A |
|---|---|---|---|---|---|
| kred1, s−1 | 0.62 ± 0.01 | 0.30 ± 0.01 | 1.15 ± 0.03 | 11.24 ± 0.06 | 0.67 ± 0.03 |
| kred2, s−1 | 0.220 ± 0.005 | 0.079 ± 0.003 | 0.12 ± 0.01 | 0.46 ± 0.01 | 0.057 ± 0.001 |
| kOOH, s−1 | 0.65 ± 0.07 | 1.4 ± 0.1 | 0.49 ± 0.06 | 0.95 ± 0.03 | 0.60 ± 0.07 |
| kox, s−1 | 0.016 ± 0.0002 | 0.0070 ± 0.00002 | 0.012 ± 0.0002 | 0.012 ± 0.0002 | 0.012 ± 0.0004 |
Conditions: 100 mM sodium phosphate, pH 7.5, and 25 °C.
| (1) |
For flavin oxidation studies, the increase in absorbance at 372 nm, which monitors formation of the C4a-hydroperoxyflavin intermediate, was fited to eq. 2, which describes a single exponential rise. The subsequent increase in absorbance at 452 nm, which corresponds to flavin oxidation, was fited to eq. 3, which describes a double exponential rise.
| (2) |
| (3) |
Crystallization of SidA N323A.
SidA N323A crystals were grown under similar conditions as wtSidA [11]. In brief, crystals were obtained using the hanging drop vapor diffusion method aided by microseeding. The reservoir solution contained 0.1 M HEPES, pH 6.6, 1.86 M ammonium sulfate, and 1% (v/v) dioxane. Drops were formed by mixing the reservoir in a 1:1 ratio with 8 mg/mL SidA N323A, which was pre-incubated with 1 mM NADP+ and 100 mM Orn. Crystals were allowed to grow for one month and then flash cooled in liquid nitrogen, with the reservoir solution supplemented with 30% (v/v) glycerol as the cryoprotectant.
Structure determination.
Crystals of the ternary complex of N323A with NADP+ and Orn were analyzed at beamline 24-ID-E of the Advanced Photon Source using an ADSC Quantum 315 detector. The data were processed with XDS [29] and AIMLESS [30] via CCP4i [31]. PHENIX [32] and COOT [33] were used for refinement and model building, respectively. Crystallographic structure refinement was initiated from the coordinates of a wtSidA structure (Protein Data Bank (PDB) entry 4B63). MolProbity was used for structure validation [34]. Data collection and refinement statistics are listed in Table 3.
Table 3.
X-ray Diffraction Data Collection and Refinement of N323A
| Beam line | APS 24-ID-E |
| Space group | I222 |
| Unit cell parameters (Å) | a = 77.1 |
| b = 84.0 | |
| c = 145.1 | |
| Wavelength | 0.979 |
| Resolution (Å) | 72.7–2.10 (2.16–2.10) |
| Observations | 156743 |
| Unique reflections | 27670 |
| Rmerge(I) | 0.131 (0.617) |
| Rmens(I) | 0.144 (0.681) |
| Rpim(I) | 0.058 (0.280) |
| Mean I/σ | 9.3 (2.7) |
| Mean CC1/2 | 0.994 (0.861) |
| Completeness (%) | 99.6 (100.0) |
| Multiplicity | 5.7 (5.6) |
| No. of protein chains | 1 |
| No. of protein residues | 452 |
| No. of protein atoms | 3475 |
| No. of FAD atoms | 53 |
| No. of Orn atoms | 9 |
| No. of NADP+ atoms | 31 |
| No. of water molecules | 180 |
| Rcryst | 0.172 (0.221) |
| Rfreeb | 0.228 (0.282) |
| rmsd bond lengths (Å) | 0.007 |
| rmsd bond angles (°) | 1.04 |
| Ramachandran plotc | |
| Favored (%) | 97.31 |
| Outliers (%) | 0.00 |
| All-atom clashscorec | 3.4 |
| Average B-factor (Å2) | |
| Protein | 28.1 |
| FAD | 26.8 |
| Orn | 25.1 |
| NADP+ | 40.2 |
| Water | 27.8 |
| Coordinate error (Å)d | 0.24 |
| PDB entry | 5CKU |
Values for the outer resolution shell of data are given in parenthesis.
5 % test set.
Generated with MolProbity via PHENIX refine.
Maximum likelihood-based coordinate error estimate reported by PHENIX refine.
Results
Steady-state oxygen consumption.
The four mutant enzymes were studied using an oxygen consumption assay, which measures the steady-state rate of conversion of the reduced enzyme-NADP+ complex (FADredNADP+) to the hydroxylating species, FADOOH. K107A displayed no activity even at high concentrations of substrates (1 mM NADPH and 500 mM Orn). The other three mutant enzymes were active, so steady-state kinetic parameters were determined using NADPH or Orn as the variable substrate. These mutations have a modest effect on kcat (Table 1). The kcat values of N293A and N323A are 1.5- and 1.8-fold higher than wtSidA, respectively. The kcat value for S469A is ~4-fold lower than for wtSidA. In contrast, the mutations have a large effect on the Km value for Orn, which increased ~16-fold, ~14-fold, and ~19-fold for N293A, N323A, and S469A, respectively. This result is consistent with crystal structures showing that these three residues interact with Orn (Fig. 1). The mutations also increase the Km value for NADPH, but the magnitude of the effect is variable. The largest change was observed with N293A, which has a 9-fold greater Km value for NADPH compared to wtSidA. The mutational effect was lower with S469A, which has 3-fold higher Km, whereas the Km value for N323A was unchanged.
Steady-state Orn hydroxylation.
As observed with the oxygen consumption assay, no activity was detected with K107A even at high substrate concentrations of 1 mM NADPH and 500 mM Orn. Kinetic parameters could be obtained for the other three mutant enzymes, and the main effect of these mutations is to increase the Km for Orn. The kcat values for N293A and N323A are essentially unchanged from that of wtSidA, while kcat for S469A was ~6-fold lower (Table 1). The Km values for Orn increased 16-, 12-, and 21- fold for N293A, N323A, and S469A, respectively.
The coupling percentages for the three active mutant enzymes could be calculated as the ratio of the Orn hydroxylation kcat to the oxygen consumption kcat. Results indicate that coupling for all three mutants is lower than coupling for wtSidA (Table 1).
Pre-steady-state kinetics of flavin reduction and oxidation.
The rate constant for flavin reduction (kred) was measured using the stopped-flow spectrophotometer under anaerobic conditions. For wtSidA, 100 μM NADPH is considered a saturating concentration since it has been shown that the KD value is close to1 μM. The rate constants of the mutants with a stoichiometric concentration (15 μM) of NADPH produced similar results as with 100 μM of NADPH, suggesting that the mutations do not have a large effect on the affinity for NADPH. This is consistent with the low, or lack of, effect observed on the Km value for NADPH with these mutants in steady-state assays (Table 1). The time resolved spectral changes during reduction of wtSidA and its mutants occurs in two phases, which we fitted to a double exponential equation (eq 1). The first phase corresponds to flavin reduction, while the second phase corresponds to either product release or a small population of slow reactive enzyme (or a combination of both). Results (Table 2) show that the first phase of reduction for N293A is ~2-fold faster than for wtSidA, while the reduction of K107A is ~2-fold slower. There was no change in the S469A enzyme. N323A, however, displayed an ~18-fold increase in the rate of reduction in the first phase compared to wtSidA.
For the oxidative half-reaction experiments, the reduced form of each enzyme was reacted with 300 μM O2 in the absence of Orn to determine if the rate of formation of the C4a-hydroperoxyflavin (FADOOH) or its stability were affected by any of the mutations. The results in Table 2 show that the rate of formation constant of FADOOH (kOOH) for each mutation is not significantly different from wtSidA, with K107A showing the largest change (only ~2-fold). The rate constant for FADOOH oxidation (kOX) also indicates that the stability of FADOOH is unaffected by each mutation as the kOX values are all similar to wtSidA.
Primary kinetic isotope effect (KIE) with N323A.
In this experiment, the steady state kinetic parameters using the oxygen consumption assay were measured with either 4-pro-R-42H-NADPH or NADPH. The results are expressed as the ratio of kcat or kcat/Km obtained with the proteated substrate to the respective value obtained with the deuterated substrate. These ratios are represented as Dkcat and D(kcat/Km). For wtSidA a Dkcat value of 3.70 ± 0.10 and a D(kcat/Km) value of 4.90 ± 0.50 were determined. These values are consistent with our previous work where it was determined that hydride transfer is partially rate limiting in wtSidA and that the reaction involves the stereospecific transfer of the 4-pro-R-42H-of NADPH [9, 12]. With N323A, a Dkcat value of 2.80 ± 0.10 and a D(kcat/Km) value of 2.70 ± 0.10 were determined.
Three-dimensional structure analysis of N323A.
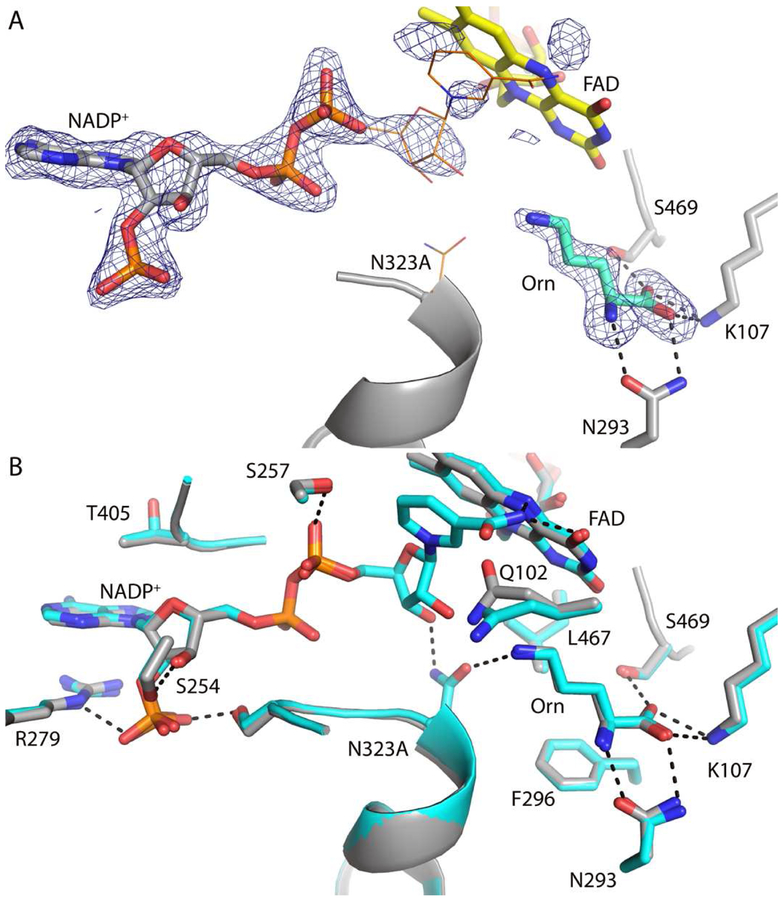
The crystal structure of N323A was solved to provide insight into the 18-fold increased rate of flavin reduction and 15-fold higher Km for Orn observed with this mutant enzyme. The structure of the ternary complex of the oxidized enzyme with NADP+ and Orn was determined at 2.1 Å resolution using the same SidA crystal form that was used for wtSidA structures (Table 3) [11].
The fold of N323A is virtually identical to wtSidA (PDB code 4B63) as evidenced by a root mean square deviation for Cα atoms of only 0.34 Å. Electron density for Orn and the ADP part of NADP+ is strong (Fig. 2A). In contrast, electron density for the nicotinamide riboside (NR) group is very weak, indicating conformational disorder (Fig. 2A). As a result, only the ADP group has been included in the deposited structure, although the NR from wtSidA is included in Fig. 2A to guide the eye. The ligand conformations and interactions with the enzyme in N323A are virtually identical to those of wtSidA, except for a slight deflection of Gln102 in N323A toward the space occupied by the NR in wtSidA (Fig. 2B).
Fig. 2.
Active site of N323A. (A) Electron density for NADP+ and Orn bound to N323A. The mesh represents a simulated annealing Fo-Fc map contoured at 2.5σ. For reference, Asn323 and the NR of NADP+ from wtSidA (PDB entry 4B63) are shown in thin orange bonds. (B) Superposition of N323A (gray) and wtSidA (cyan) (PDB entry 4B63).
Discussion
Flavin-dependent monooxygenases make up a large family of enzymes divided into 8 different classes (A-H) [35]. Members of this family of enzymes are involved in hydroxylation, epoxidation, halogenation, and Baeyer-Villiger type ring expansions [36]. Among the many hydroxylated compounds produced by flavin-monooxygenases are the hydroxamate-containing siderophores (HCS) [10, 26, 35–37]. HCS have been shown to be essential for virulence in Mycobacterium tuberculosis and A. fumigatus [8, 38–40]. The biosynthesis of HCS in A. fumigatus (i.e., triacetylfusarine and hydroxyferricrocin) is initiated by the hydroxylation of ornithine by SidA, a member of the flavin-dependent Class B monooxygenases [6, 41]. The binding site of Orn in SidA is conserved among the other Orn monooxygenases, PvdA and KtzI [22, 37]. The binding site provides these enzymes with specificity for Orn. Although, lysine has been shown to bind to SidA in the same position as Orn, the hydroxylation of this longer amino acid is very inefficient, as it protrudes too far into the active site, destabilizing the FADOOH and leading to uncoupling of the reaction (i.e., formation of hydrogen peroxide instead of hydroxylated product) [11, 42]. Binding of norvaline, which is shorter than Orn, does not lead to hydroxylation or uncoupling since it does not reach the FADOOH [13]. Thus, in addition to providing residues to recognize the main chain of amino acids, the binding site of SidA also evolved a precise specificity for the particular aliphatic chain length of Orn, which allows the enzyme to orient the N5-Orn atom for hydroxylation by the distal oxygen of the FADOOH (Scheme 1B and Fig. 1). This aspect of substrate specificity appears to be enforced by Asn323, which hydrogen bonds to the terminal amino of Orn, and by Gln102, Phe296, and Leu467, which pack against the aliphatic chain of Orn (Fig. 2B).
The roles of four of the Orn-binding residues of SidA were probed using site directed mutagenesis (Fig. 1). The steady-state kinetic data for the four mutant enzymes indicates that they are all required for efficient hydroxylation of Orn. Mutation of Lys107, which forms ionic interactions with the substrate carboxylate, abrogates activity, suggesting essentiality of these interactions for catalysis. The other three mutations affect mainly Orn binding as evidenced by greater than 10-fold increases in Km (Table 1). In addition, S469A exhibits a ~ 6-fold decrease in the kcat value (Table 1). We note that Ser469 is adjacent to the pyrimidine ring of the flavin, so it is possible that this mutation perturbs the flavin conformation. Also, Ser469 is next to Lys107, a residue that is essential for catalysis. It thus appears that this region of the active site is particularly sensitive to perturbations.
To test if the selected Orn binding residues play additional roles in SidA activity, the rate constants for flavin reduction and oxidation were determined in a stopped-flow spectrophotometer. Neither the rate of formation of the FADOOH (kOOH) nor its decay to the oxidized FAD (kOX) were significantly different from the values obtained for wtSidA (Table 2). This is consistent with our previous structural, biochemical, and computational results that suggest that formation and stabilization of the FADOOH is independent of the binding of Orn [13, 42, 43].
wtSidA is highly coupled as it produces one HO-Orn for every molecule of oxygen that is activated to form the FADOOH. All of the mutations show a decrease in coupling. Since the mutations do not destabilize the FADOOH intermediate, we propose that the decrease in coupling may reflect suboptimal binding of Orn to the SidA−FADOOH−NADP+ complex, causing elimination of hydrogen peroxide rather than hydroxylation. Curiously, the pose of Orn in our structure of N323A is virtually identical to that of wtSidA (Fig. 2B), despite the fact that this mutant enzyme displays a greater than 10-fold elevated Km for Orn. This result perhaps suggests that crystal structures of SidA−FAD−NADP+−Orn, in which the flavin is unmodified, do not correctly depict all the details of the Orn pose in the genuine ternary Michaelis complex (SidA− FADOOH –NADP+ −Orn). We note that our attempts to capture the FADOOH in crystallo have so far been unsuccessful [11].
Reaction of NADPH with FADOX with all the mutants tested showed a double exponential decay at 445 nm, similar to what has been observed for wtSidA [9, 43]. Here, the fast phase represents flavin reduction and the slow phase represents either NADP+ release or reaction of a population of enzyme with lower activity. No major change in NADPH binding was observed as the same kred1 values were obtained at 15 μM and 150 μM NADPH (data not shown), indicating that the KD value is in the low micromolar value. This is also consistent with the steady-state results that show no major effects on the Km values for NADPH (Table 1). For N293A, S469A, and K107A the kred1 value is not significantly different from the wtSidA value (Table 3). This was expected as these residues are located away from the NADP(H) binding region (Fig. 1). In contrast, Asn323 is unique in that it interacts with both substrates. Asn323 is within hydrogen bonding distance of the 3’-OH of the nicotinamide ribose of NADP(H); thus, a decrease in NADPH binding affinity or kred1 was expected (Fig. 1). Instead, an 18-fold increase of kred1 was observed. In addition, the primary KIE value for N323A was significantly lower than for wtSidA (2.8 vs 3.7 for Dkcat), consistent with a faster and less rate limiting hydride transfer step in this mutant enzyme.
It has been proposed for wtSidA and other Class B flavin monoxygenases that domain rotation occurs to allow NADP(H) to “slide” into the conformation required for FADOOH formation after hydride transfer [12, 16, 44, 45]. It is possible that Asn323 is involved in guiding the NR group during the reduction before it acquires the position required for FADOOH stabilization (Fig. 1). We have previously shown that mutation of Ser257, which interacts with the pyrophosphate group of NADPH (Fig. 1), also leads to an increase in the rate constant of flavin reduction. Molecular dynamics simulation showed that mutating Ser257 leads to increased motion of the nicotinamide ring permitting sampling of shorter distances between the NADPH- C4 and the N5-FAD [12]. We propose that in the case of N323A, deletion of the hydrogen bond with the ribose 3’-OH similarly leads to higher degrees of freedom of the nicotinamide ring during the sliding process, which increases the probability of hydride transfer. This possibility is supported by the structure of N323A, which suggests higher flexibility for the nicotinamide ring (Fig. 2A).
The SidA flavin monooxygenase catalyzes the hydroxylation of ornithine.
We studied four residues in the ornithine site using mutagenesis and kinetics.
Lys107, which binds the carboxylate of ornithine, is essential for activity.
The three other residues function mainly in stabilizing ornithine.
Asn323 facilitates ornithine binding at the expense of hindering flavin reduction.
Acknowledgements
This work was supported in part by a grant from the National Science Foundation MCB-1021384. We thank Dr. Jonathan Schuermann of the Northeastern Collaborative Access Team for help with X-ray diffraction data collection and processing. Part of this work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357.
ABBREVIATIONS
- SidA
ornithine N5-monooxygenase siderophore A
- wtSidA
wild-type SidA
- Orn
ornithine
- MbtG
lysine N6-monoxygenase from Mycobacterium tuberculosis
- PDB
Protein Data Bank
- NR
nicotinamide riboside
- HCS
hydroxamate-containing siderophores
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Knutsen AP, Bush RK, Demain JG, Denning DW, Dixit A, Fairs A, Greenberger PA, Kariuki B, Kita H, Kurup VP, Moss RB, Niven RM, Pashley CH, Slavin RG, Vijay HM, Wardlaw AJ, J. Allergy. Clin. Immunol 129 (2012) 280–291. [DOI] [PubMed] [Google Scholar]
- [2].Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC, Sci. Transl. Med 4 (2012) 165rv113. [DOI] [PubMed] [Google Scholar]
- [3].Brown GD, Denning DW, Levitz SM, Science (New York, N Y) 336 (2012) 647. [DOI] [PubMed] [Google Scholar]
- [4].Wandersman C, Delepelaire P, Annu. Rev. Microbiol 58 (2004) 611–647. [DOI] [PubMed] [Google Scholar]
- [5].Haas H, Annu. Rev. Phytopathol 46 (2008) 149–87.. [DOI] [PubMed] [Google Scholar]
- [6].van Berkel WJ, Kamerbeek NM, Fraaije MW, J. Biotechnol 124 (2006) 670–689. [DOI] [PubMed] [Google Scholar]
- [7].Dimkpa C, Svatos A, Merten D, Buchel G, Kothe E, Can. J. Microbiol 54 (2008) 163–172. [DOI] [PubMed] [Google Scholar]
- [8].Hissen AH, Wan AN, Warwas ML, Pinto LJ, Moore MM, Infect. Immun 73 (2005) 5493–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Romero E, Fedkenheuer M, Chocklett SW, Qi J, Oppenheimer M, Sobrado P, Biochim. Biophys. Acta 1824 (2012) 850–857. [DOI] [PubMed] [Google Scholar]
- [10].Chocklett SW, Sobrado P, Biochemistry 49 (2010) 6777–6783. [DOI] [PubMed] [Google Scholar]
- [11].Franceschini S, Fedkenheuer M, Vogelaar NJ, Robinson HH, Sobrado P, Mattevi A, Biochemistry 51 (2012) 7043–7045. [DOI] [PubMed] [Google Scholar]
- [12].Shirey C, Badieyan S, Sobrado P, J. Biol. Chem 288 (2013) 32440–32448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Robinson R, Badieyan S, Sobrado P, Biochemistry 52 (2013) 9089–9091. [DOI] [PubMed] [Google Scholar]
- [14].Orru R, Dudek HM, Martinoli C, Torres Pazmino DE, Royant A, Weik M, Fraaije MW, Mattevi A, J. Biol. Chem 286 (2011) 29284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Orru R, Pazmino DE, Fraaije MW, Mattevi A, J. Biol. Chem 285 (2010) 35021–35028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Alfieri A, Malito E, Orru R, Fraaije MW, Mattevi A, Proc. Natl. Acad. Sci. USA 105 (2008) 6572–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Malito E, Alfieri A, Fraaije MW, Mattevi A, Proc. Natl. Acad. Sci. USA 101 (2004) 13157–13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Robinson R, Franceschini S, Fedkenheuer M, Rodriguez PJ, Ellerbrock J, Romero E, Echandi MP, Martin Del Campo JS, Sobrado P, Biochim. Biophys. Acta 1844 (2014) 778–784. [DOI] [PubMed] [Google Scholar]
- [19].Ziegler DM, Drug Metab. Rev 34 (2002) 503–511. [DOI] [PubMed] [Google Scholar]
- [20].Sheng D, Ballou DP, Massey V, Biochemistry 40 (2001) 11156–11167. [DOI] [PubMed] [Google Scholar]
- [21].Beaty NB, Ballou DP, J. Biol. Chem 256 (1981) 4619–4625. [PubMed] [Google Scholar]
- [22].Setser JW, Heemstra JR Jr., Walsh CT, Drennan CL, Biochemistry 53 (2014) 6063–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Olucha J, Meneely KM, Chilton AS, Lamb AL, J. Biol. Chem 286 (2011) 31789–31798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Csaky T, Acta Chem. Scand 2 (1948) 450–454. [Google Scholar]
- [25].Robinson RM, Rodriguez PJ, Sobrado P, Arch. Biochem. Biophys 550–551 (2014) 58–66. [DOI] [PubMed] [Google Scholar]
- [26].Robinson R, Sobrado P, Biochemistry 50 (2011) 8489–8496. [DOI] [PubMed] [Google Scholar]
- [27].Jeong SS, Gready JE, Anal. Biochem 221 (1994) 273–277. [DOI] [PubMed] [Google Scholar]
- [28].Dhatwalia R, Singh H, Solano LM, Oppenheimer M, Robinson RM, Ellerbrock JF, Sobrado P, Tanner JJ, J. Am. Chem. Soc 134 (2012) 18132–18138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kabsch W, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Evans PR, Murshudov GN, Acta Cryst D69 (2013) 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Potterton E, Briggs P, Turkenburg M, Dodson E, Acta Cryst D59 (2003) 1131–1137. [DOI] [PubMed] [Google Scholar]
- [32].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Acta Crystallogr. D Biol. Crystallogr 68 (2012) 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Emsley P, Lohkamp B, Scott WG, Cowtan K, Acta Cryst. D Biol. Crystallogr 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, Acta Crystallogr. D Biol. Crystallogr D66 (2010) 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huijbers MM, Montersino S, Westphal AH, Tischler D, van Berkel WJ, Arch. Biochem. Biophys 544 (2014) 2–17. [DOI] [PubMed] [Google Scholar]
- [36].Walsh CT, Wencewicz TA, Nat. Prod. Rep 30 (2013) 175–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Olucha J, Lamb AL, Bioorg. Chem 39 (2011) 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sassetti CM, Boyd DH, Rubin EJ, Mol. Microbiol 48 (2003) 77–84. [DOI] [PubMed] [Google Scholar]
- [39].Sassetti CM, Rubin EJ, P Proc. Natl. Acad. Sci. USA 100 (2003) 12989–12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Eisendle M, Oberegger H, Zadra I, Haas H, Mol. Microbiol 49 (2003) 359–375. [DOI] [PubMed] [Google Scholar]
- [41].Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, Eisendle M, Wallner A, Arst HN Jr., Haynes K, Haas H, PLoS Pathogens 3 (2007) 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Badieyan S, Bach RD, Sobrado P, J. Org. Chem 80 (2015) 2139–2147. [DOI] [PubMed] [Google Scholar]
- [43].Romero E, Robinson R, Sobrado P, J. Vis. Exp (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Binda C, Robinson RM, Martin Del Campo JS, Keul ND, Rodriguez PJ, Robinson HH, Mattevi A, Sobrado P, J. Biol. Chem 290 (2015) 12676–12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mirza IA, Yachnin BJ, Wang S, Grosse S, Bergeron H, Imura A, Iwaki H, Hasegawa Y, Lau PC, Berghuis AM, J. Am. Chem. Soc 131 (2009) 8848–8854. [DOI] [PubMed] [Google Scholar]