Abstract
The Bcl-2 family of proteins regulates mitochondrial outer membrane permeability thereby making life or death decisions for cells. Most of Bcl-2 proteins contain hydrophobic regions that are embedded in intracellular membranes such as mitochondria. These membrane proteins are difficult to express and purify thereby preluding biochemical and biophysical characterizations. Here, we describe a photocrosslinking approach based on in vitro synthesis of Bcl-2 proteins with photo-reactive amino acid analogs incorporated at specific locations. These photo-reactive proteins are reconstituted into liposomal membranes with defined phospholipids or mitochondrial membranes isolated from animals, and their interactions with other Bcl-2 proteins are detected by photocrosslinking.
Keywords: Apoptosis, Mitochondria, Bcl-2, Bax, BH3-only proteins, Photocrosslinking, Membrane protein interaction, Membrane permeabilization
1. Introduction
Mitochondrial outer membrane permeabilization (MOMP) to intermembrane space proteins such as cytochrome c marks the commitment step in a programmed cell death pathway called apoptosis. This step is regulated by the Bcl-2 family of proteins that are dynamic in their expression, localization, and conformation, which dictate their interaction with one another [1–3]. In particular, Bax or Bak containing multiple Bcl-2 homology (BH) regions are monomers in healthy cells located mostly in the cytosol or the mitochondrion, respectively. In apoptotic cells, these proteins form homo-oligomers in the mitochondrial outer membrane (MOM) that induce the formation gigantic pores through which cytochrome c and other mitochondrial proteins are released to the cytosol where they activate proteases and nucleases to dismantle the cell [4–6]. The transition from monomeric (also soluble in the case of Bax) to oligomeric membrane-embedded proteins is activated by Bcl-2 proteins containing only one BH region (BH3; e.g. Bid and Bim) that interact with the multi-BH Bax or Bak in either the cytosol or the mitochondrion [7–12]. The activation is blocked by another group of Bcl-2 proteins (e.g., Bcl-2 and Bcl-XL) that contain multi-BH regions like Bax and Bak, yet, function to inhibit Bax and Bak by sequestering them or their BH3 activators [13–17]. Another group of BH3-only proteins (e.g., Bad and Noxa) bind to Bcl-2, Bcl-XL and other anti-apoptotic family members thereby releasing pro-apoptotic Bax, Bak, and their BH3 activators so they can induce MOMP and cell death [18,19].
In addition to the BH regions, most proteins in the Bcl-2 family have a hydrophobic region at the carboxyl terminus that can insert into membranes [20–24,6,25]. While this sequence is important to the function of respective proteins, it decreases the solubility of recombinant proteins and increases their toxicity to the host cells thereby precluding their expression and purification. In fact, only handful Bcl-2 proteins have been purified as full-length proteins with the native hydrophobic tails. Although biochemical, biophysical and structural characterization of these full-length Bcl-2 proteins has greatly advanced our knowledge about this protein family, investigation of the rest of the family in their native form is warranted not only for a full mechanistic understanding but for a materialization of the promised therapeutic potential [26–30].
Our goal is to produce full-length Bcl-2 proteins that are suitable for investigation of their interactions in a native environment. To achieve this goal, we developed an in vitro system to produce full-length Bcl-2 proteins with a photo-reactive crosslinking probe located at a specific location that are then activated and inserted into membranes, if this is necessary for their interaction and function (see Notes 1–2)[31,32,17]. After the functional complex is formed by the Bcl-2 proteins and their binding partners in dark where the photo-probe remains inert, we activate the probe by light such that it would react with residues in the binding partners and thereby covalently link the two interacting proteins. The resulting photo-adduct is analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by phosphor-imaging as the in vitro synthesized proteins are labeled by [35S]-methionine. The results from these photocrosslinking experiments are used to generate structural models for the Bcl-2 protein complex, which in turn was used to design mutations that would alter the complex and consequently the function of the Bcl-2 proteins. These mutations were tested not only in protein interaction assays but also in function assays to validate the biological relevance of the structural models [31,32,17].
2. Materials
2.1. In Vitro Protein Synthesis Reagents
All solutions are made with nuclease-free ultrapure water (NF H2O) from Milli-Q Direct 8 water purification system (Millipore), and stored at room temperature (~22 °C), unless indicated otherwise.
5x Transcription buffer: 200 mM Tris-HCl (pH 7.9), 30 mM MgCl2, 50 mM NaCl, 10 mM spermidine, 50 mM DTT. Store at −20 °C.
ATP/GTP/CTP/UTP mix: 5 mM each. Store at −20 °C.
RNase inhibitor: 40 U/μl RiboLock (Thermo Scientific). Store at −20 °C.
SP6 RNA polymerase: 200 U/μl (Thermo Fisher). Store at −20 °C.
5x TBE buffer: 450 mM Tris-borate, 10 mM EDTA.
RNA gel-loading buffer: 0.26 g urea, mix with 500 μl of 6x DNA gel-loading buffer (0.25 (w/v) bromophenol blue, 0.25 (w/v) xylene cyanol FF, 30% (v/v) glycerol), 200 μl of 5x TBE buffer, 150 μl NF H2O. Store at 4 °C.
1.6% (w/v) agarose gel: mix 0.4 g agarose with 25 ml of 1x TBE buffer, microwave for 75 s, cool to 37 °C, add 2 μl of 10 mg/ml ethidium bromide and mix, pour and set the gel at room temperature for 20 min.
10x Translation mix: 250 mMHEPES-KOH (pH 7.5), 1.1 M KOAc (pH 7.5), 10 mM Mg(OAc)2, 20 mM glutathione, 0.025% (v/v) Nikkol, 2 mM spermidine, 0.08 mM S-adenosyl-methionine, 10× protease inhibitors (diluted from 200x proteases inhibitors: 10 mg/ml of each Leupeptin, Antipain, Chymostatin and Pepstatin, plus 1.4 mg/ml Aprotinin). Store at −80 °C.
Energy generating system and amino acid mix lacking methionine or lysine or both (EGS–M, –K or –MK): 90 mM HEPES-KOH (pH 7.5), 15 mM ATP, 15 mM GTP, 120 mM phosphocreatine, 0.96 mg/ml creatine phosphokinase, 0.375 mM of each of the 20 amino acids except of methionine and/or lysine to allow incorporation of [35S]Met and/or photo-reactive [14C]Lys. Store at −80 °C.
Wheat Germ Extract (WG): prepare as described [33] using wheat germ from Shawnee Milling Co. in Oklahoma. It contains the translation machinery capable to synthesize protein from messenger RNA (mRNA). Store at −80 °C.
Radioactive [35S]methionine (Perkin Elmer). Concentration varies between batches but typically ~ 40 mCi/ml. Store at −80 °C.
Photo-reactive probe 5-azido-2-nitrobenzoyl (ANB), 4,4-azipentanoyl (AP) or benzophenone (BP)-labeled lysyl-tRNA: yeast εANB-[14C]Lys-tRNALys, εAP-[14C]Lys-tRNALys, or εBP-[14C]Lys-tRNALys, and the control acetylated lysyl-tRNA: yeast εAc-[14C]Lys-tRNALys (tRNA Probes), prepared as described [34,35]. Concentration varies between batches but typically ~15 pmole/μl. Keep in dark. Store at −80 °C.
2.2. Protein Activation, Interaction and Fractionation Reagents
Bax BH3 peptide: contain Bax residues 53-86, prepare as described [36], 10 mM in DMSO, then dilute to 370 μM with NF H2O. Store at −80 °C.
Recombinant His6-tagged Bax protein: prepare as described [37], 9.3 μM in 10 mM HEPES-KOH (pH 7.5), 100 mM NaCl, 20% glycerol. Store at −80 °C.
Liposome: prepare as described [37], contains the following phospholipids (Avanti Polar Lipids) in the mole% typical to the mitochondrial outer membrane [7], 46% phosphatidylcholine, 28% phosphatidylethanolamine, 9% phosphatidylinositol, 9% phosphatidylserine, 7% cardiolipin. Store at 4 °C under argon in sealed microfuge tube.
Buffer A: 25 mM HEPES-KOH (pH 7.5), 500 mM KOAc (pH 7.5), 5 mM Mg(OAc)2. Store at 4 °C.
Sucrose solutions: 2.2, 0.8 or 0.25 M sucrose in buffer A. Store at 4 °C.
2.3. Photocrosslinking and Photo-adduct Enrichment Reagents
DTT: 1 M. Store at −20 °C.
Triton X-100: 10% (v/v)
Imidazole: 50 mM. Store at 4 °C.
Ni2+-chelating agarose: 50% (v/v). Store at 4 °C
Phosphate buffered saline (PBS): 10 mM Na2HPO4, 1.8 mM KH2PO4, 138 mM NaCl, 2.7 mM KCl. Store at 4 °C.
2.4. SDS Polyacrylamide Gel Electrophoresis (SDS-PAGE) Reagents
SDS-PAGE gel-loading buffer: 0.2 M Tris, 13 mM EDTA (pH 8), 26% (v/v) glycerol, 6% (w/v) SDS, 10% (v/v) 2-mercaptonethanol, 0.05% (w/v) bromophenol blue.
SDS-PAGE gel: Resolving gel contains 15% (w/v) acrylamide, 0.4% (w/v) N,N’-methylene-bis-acrylamide, 0.4 M Tris/HCl (pH 8.8), 0.1 (w/v) SDS, 0.02% (v/v) TEMED, 0.06% (w/v) ammonium persulfate. Stacking gel contains 4% (w/v) acrylamide, 0.1% (w/v) N,N’-methylene-bis-acrylamide, 0.06 M Tris/HCl (pH 6.8), 0.1 (w/v) SDS, 0.36 M sucrose, 0 05% (v/v) TEMED, 0.05% (w/v) ammonium persulfate. Gel size = length 13 cm × width 17 cm × thickness 0.7 mm. Well size = 17 × 5 × 0.7 mm (hold up to 40 μl sample). Store at 4 °C.
SDS-PAGE buffer: 50 mM Tris, 400 mM glycine, 0.125% (w/v) SDS.
De-stain solution: 35% (v/v) methanol, 10% (v/v) HO Ac.
Glycerol: 5% (v/v).
2.5. Equipment and Facility
Eppendrof centrifuge with rotor and adapter for 1.5 and 0.5 ml microfuge tubes.
Beckman Optima Max Ultracentrifuge with TLA100.2 rotor.
Dark room with dim red light.
Light source for photocrosslinking: Oriel 500-watt mercury arc lamp assembly.
Vertical gel electrophoresis apparatus: GibcoBRL, model V16.
Power supply: Bio-Rad, model PowerPac 3000.
Gel dryer: Bio-Rad, model 583.
Vacuum pump: Precision Scientific, model DD90.
Phosphor-imager: Fujifilm FLA-9000 multipurpose image scanner.
Phosphor-imaging plate, Fujifilm BAS storage phosphor screen, type MS, 20 × 25 cm.
UV-transilluminator: Spectroline, model TE-312S
Vacuum concentrator: Thermo Scientific, model SpeedVAC
3. Methods
3.1. In Vitro Protein Synthesis
In vitro transcription: Messenger RNAs (mRNAs) encoding proteins of the Bcl-2 family are synthesized in vitro from the corresponding DNA template using SP6 RNA polymerase as described [38] with the following modifications. The cDNA of a Bcl-2 family gene is inserted into a plasmid after SP6 promoter and a 5’-untranslated region plus start site optimized for producing mRNAs with high translation efficiency (e.g., pSPUTK from Stratagene) [39]. The plasmid amplified in E. coli DH5α cells and purified using QIAprep Spin Miniprep kit (Qiagen) is linearized by a restriction enzyme that cleaves the DNA after the stop codon of the Bcl-2 gene producing 5’-overhang or blunt end. The linear DNA is purified by phenol/chloroform extraction and ethanol/NaOAc precipitation, and suspended in nuclease-free ultrapure water (NF H2O). A typical in vitro transcription reaction is shown in Table 1, in which SP6 RNA polymerase synthesizes a Bax mRNA from the linear DNA template. The Bax mRNA is precipitated by ethanol/NaOAc, washed by 70% ethanol, dried in SpeedVAC vacuum concentrator, suspended in 10 mM Tris-HCl (pH 7.6), 1 mM EDTA, 0.3 U/μl RNase inhibitor, frozen in liquid nitrogen, and stored at −80 °C.
In vitro translation: Proteins of the Bcl-2 family, each with a photo-reactive Lys residue incorporated into a specific site for crosslinking and radio-active [35S]Met residues for detection of the protein and photo-adduct, are produced from the corresponding mRNA using a wheat germ extract-based in vitro translation system. A typical in vitro translation producing an εANB-[14C]Lys and [35S]Met-labeled Bax protein is given in Table 2 and explained in Notes 3–22.
Table 1.
In vitro transcription mix for one sample of 100 μl with the following reagents added to a microfuge tube at room temperature in the following order.
| Stock solution concentration | Volume added (μl) | Final concentration | |
|---|---|---|---|
| NF H20 | 28 | ||
| 5x Transcription buffer | 5x | 20 | 1x |
| ATP/GTP/CTP/UTP mix | 5 mM each | 25 | 1.25 mM each |
| BSA | 1 μg/μl | 5 | 0.05 μg/μl |
| RNase inhibitor | 40 U/μl | 1.25 | 0.5 U/μl |
| Mix gently. | |||
| Linearized Bax DNA | 0.25 μg/μl | 20 | 0.05 μg/μl |
| SP6 RNA polymerase | 200 U/μl | 0.75 | 1.5 U/μl |
| Total volume = 100 μl | |||
Mix gently. Incubate at 37 °C for 2 h. Remove 1 μl from the reaction, mix with 4 μl of NF H2O and 5 μl of 2x RNA gel-loading buffer, heat at 65 °C for 2 min. Load to agarose gel, run with 1x TAE buffer at 90 V for 1 h. Visualize RNA band on UV-transilluminator.
Table 2.
In vitro translation master mix (M) for 20 samples of 25 μl with the following reagents added to a microfuge tube on ice in the following order.
| Stock solution concentration | Volume added (μl) | Final concentration | |
|---|---|---|---|
| NF H20 | 307.5 | ||
| 10x translation mix | 10x | 50 | 1x |
| EGS-M or -K or -MK | 40 | 2 μl/25 μl | |
| RNase inhibitor | 40 U/μl | 2.5 | 5 U/25 μl |
| Wheat germ extract | 40 | 2 μl/25 μl | |
| Mix gently. Pre-incubate at 26°C for 5 min to complete translation of residual endogenous untranslated mRNA before addition of mRNA of interest, [35S]Met and aa-tRNA. | |||
| [35S]Met | 40 μCi/μl | 20 | 1.6 μCi/μl |
| Transfer the sample to dark room with red light to avoid activation of the photo-reactive ANB probe. All of the following procedures prior to photocrosslinking (3.3) are performed in dark or under red light. | |||
| εANB-[14C]Lys-tRNALys | 15 pmole/μl | 20 | 15 pmol/25 μl |
| Total volume = 20 sample × 24 μl = 480 μl (see Note 5) | |||
Mix gently. Add 24.0 μl of master mix to each of the 18 microfuge tubes. Then add 1 μl of each mRNA sample to its designated tube and mix gently. Incubate at 26 °C for 15-60 min depending on the mRNA length. Stop translation by adding 0.2 μg/μl of cycloheximide and incubate at 26 °C for 5 min.
3.2. Protein Activation, Interaction and Fractionation
The in vitro synthesized photo-reactive Bax protein (here 11 μl of εANB-[14C]Lys and [35S]Met-labeled Bax R37K single-Lys mutant) is mixed with a BH3 activator (here 40 μM of Bax BH3 peptide), membranes (here liposomes containing mitochondrial characteristic phospholipids, total at 4.5 mM), and a binding partner (here 1.5 μM of His6-tagged Bax protein). The resulting mix in a total volume of 22 μl is incubated at 37 °C for 90 min.
The liposome-bound Bax proteins are separated from the soluble and aggregated proteins by a sucrose gradient centrifugation. (A) Mix the 22-μl sample with 94 μl of buffer A and 184 ul of 2.2 M sucrose solution gently, (B) transfer the mix to a Beckman centrifuge tube (thick wall, polypropylene, 1.0 ml, 11 × 34 mm), (C) overlay the mix with 500 μl of 0.8 M and then 200 μl of 0.25 M sucrose solutions carefully without disturbing the solution below, (D) centrifuge the sucrose gradient in Beckman Optima Max Ultracentrifuge with TLA100.2 rotor at 100,000 rpm, 4 °C for 3 h, (E) collect a 250-μl fraction from the top containing the liposome-bound Bax proteins.
3.3. Photocrosslinking and Photo-adduct Enrichment
The membrane-bound Bax proteins (here the εANB-[l4C]Lys and [35S]Met-labeled Bax and the His6-Bax), and the control samples lacking the ANB label or the His6-Bax, are placed in microfuge tubes in an ice-water bath, and exposed to the light from a mercury arc lamp for 5-10 min. Upon illumination with 320-370 nm light, the photo-reactive probe ANB forms a highly reactive nitrene. If εAP or εBP-[14C]Lys is in the Bax protein, it will form a highly reactive carbine or triplet biradical, respectively, upon photolysis. Whatever the highly reactive species is and wherever it is attached (here the nitrene generated from the ANB attached to the ε-amino group of Lys37 in Bax), it will crosslink to whatever molecular species is in close proximity, including those in the Bax binding partner (here the His6-Bax protein). For the “minus light” control sample 10 mM DTT is added to inactivate the photo-reactive probe on ice for 5-10 min. Hereafter, all procedures are conducted under room light.
To enrich the photo-adduct between the [35S]Met-labeled Bax and the His6-Bax proteins, (A) add 1% (v/v) Triton X-100, 5 mM imidazole and 25 μl of 50% (v/v) Ni2+-chelating agarose to each 250-μl photolyzed or control sample, and rotate at 4 °C for overnight, (B) wash the resin three times with 500 μl of buffer A, 1% (v/v) Triton X-100, 5 mM imidazole, and one time with 500 μl of PBS, (C) elute the Ni2+-bound proteins (here the His6-Bax, the [35S]Met-labeled Bax that bind to the His6-Bax, and the photo-adduct that contains the His6-Bax) from the resin by incubating with 35 μl of SDS-PAGE gel-loading buffer at 65 °C for 30 min.
3.4. SDS-PAGE
Load the protein samples to the wells in a SDS-PAGE gel on a vertical gel electrophoresis apparatus. Run the proteins with SDS-PAGE buffer through the stacking gel at 15 mA for ~1 h, then through the resolving gel at 30 mA for ~3 h.
Shake the gel in de-stain solution for 40 min. Wash the gel three times in double distilled H2O. Soak the gel in 5% (v/v) glycerol for 10 min.
Dry the gel in gel drier for 1 h.
3.5. Phosphor-Imaging
Expose dry gel to Fujifilm phosphor-imaging plate for a few hours or overnight to detect the [35S]Met-labeled Bax protein, or for several days to one week to detect the [35S]Met-labeled photo-adduct or the [14C]Lys-labeled Bax protein.
Scan the phosphor-imaging plate using Fujifilm FLA-9000 multipurpose image scanner controlled by Fujifilm ImageReader FLA-9000 program with the following settings: method = IP, laser = LD685 nm, filter = IP.
Analyze the phosphor-image using Fujifilm Multi Gauge program.
4. Notes
Choice of the photo-reactive probe and the aminoacyl-tRNA (aa-tRNA) to which the probe is attached should be made after a careful consideration of the biological system under investigation and the question to be addressed by the crosslinking approach. Three photo-reactive probes, 5-azido-2-nitrobenzoyl (ANB), 4,4-azipentanoyl (AP), and benzophenone (BP), in combination with three aminoacyl-tRNAs, Lys-tRNALys, Lys-tRNAamb (recognizing the amber stop codon), and Cys-tRNACys, are available at tRNA Probes by December 2017. After then, one may obtain these reagents through collaborations with the laboratories that are able to prepare them, or prepare them as described [34,35]. Upon request the corresponding author can provide an unpublished detail protocol for preparation of these aa-tRNAs. After incorporation into a protein during in vitro translation, a probe will photocrosslinking to whatever molecular moieties is in close proximity, because upon illumination with 320-370 nm light, these photo-reactive probes will form highly reactive species, nitrene, carbene, and triplet biradical, respectively. These species are electrophiles that rapidly react with any nearby heteroatoms possessing nonbonding electron pairs (S, O, N, etc.), double bonds (C=C, etc.), or even single bonds (C-H, etc.) [40]. The resulting photo-adducts are analyzed to identify the protein’s binding partners in a particular assembly. The choice between Lys and Cys for attachment of the photo-reactive probe can be made according to the following factors: the number of Lys or Cys residues in the target protein, whether mutations of Lys or Cys to other amino acid residues alter the structure, localization and function of the protein, and whether the Lys or Cys substitution of a residue in or near the binding site in the protein for the partner affects the complex formation. If the target protein contains many Lys or Cys residues or mutation of any of them alter the protein structure, interaction with the partner, or function, one can introduce an amber stop codon (UAG) into the target mRNA that can be recognized by the suppressor tRNA (tRNAamb) acylated by a photo-reactive Lys or Cys to produce a derivative of the target protein with a photo-reactive probe position in or near the binding site for the partner.
The function of the photo-reactive protein derivative must be determined to ensure the photo-adduct formed later detect a biologically relevant assembly with the binding partner. The first question to be answered by the functional assay is whether the Lys-null or Cys-null mutant and the single-Lys or single-Cys mutant function like the wild type protein. The next question is if the single photo-reactive probe-labeled protein derivative remains active. For Bcl-2 proteins, the first question is relatively easy because a variety of functional assays are available. For example, one can express a Bax mutant in bax-/- bak-/- cells and perform apoptosis assays to determine if the mutant functions like the wild type protein [17]. One also can synthesize the mutant protein using the same in vitro system that is used to produce the photo-reactive derivative, and perform in vitro MOMP assay with the mitochondria isolated from bak-/- mouse liver that lack both Bak and Bax proteins by monitoring cytochrome c release in the presence of BH3 activators such as Bid or Bim [6]. The second question is difficult because the photo-reactive probe is only incorporated into a fraction (typically 25%) of proteins produced in the in vitro system because of the competition from the endogenous Lys-tRNAs or Cys-tRNAs for the ribosomes translating the mRNA with the Lys or Cys codons. Unless one can separate the photo-reactive protein from the native one, the activity detected in an assay such as the in vitro MOMP assay will be the sum from both proteins. This would not be the case if one incorporate the photo-reactive probe via the aminoacyl-supressor tRNA that recognize the amber stop codon because all of the full-length proteins produced will contain the probe-labeled amino acid residue. Unless the truncated protein resulted from termination at the amber codon is also functional, the readout from an activity assay with the total translation products will be for the probe-labeled protein.
Handling and use of mRNA and aa-tRNA: mRNAs and aa-tRNAs are very labile, both chemically and enzymatically. Ribonucleases are secreted by humans, and are transferred in the finger oil to any surface we touch. It is therefore essential to avoid any contact between an mRNA or tRNA solution and any surface that may have been touched by someone or any solution that has a nuclease contamination. In our experience, when transcriptions or translations do not work, it is invariably because of a nuclease contamination that degrades the mRNA and tRNA. Thus, stringent nuclease-free solutions and techniques are absolutely the most critical requirement for achieving successful transcription, translation and probe incorporation. In addition to being extremely sensitive to ribonucleases, aa-tRNAs are susceptible to chemical hydrolysis. An amino acid is covalently attached to the tRNA by an ester bond to form an aminoacyl-tRNA. The aminoacyl ester bond is hydrolyzed in aqueous solutions to release the amino acid from the tRNA, thereby irreversibly degrading aa-tRNAs that are chemically modified on their amino acid side chain because the tRNA synthetase in the in vitro translation mix will not recognize a modified amino acid. The rate of hydrolysis (deacylation) is increased when the sample temperature, pH, or diol concentration (e.g., glycerol or sucrose; see [41]) is increased. To minimize deacylation, stock aa-tRNA solutions are buffered at pH 5.0 and stored in 1 mM KOAc (pH 5.0), 2 mM Mg(OAc)2. Any aa-tRNA solution must be quick-frozen in liquid nitrogen as soon as possible, thawed as few times as possible, and kept on ice during and after thawing. It is best to minimize the warming of an aa-tRNA solution, even during thawing. We typically place a frozen aa-tRNA solution in an ice bucket to thaw, and if necessary, warm the microfuge tube by rolling it between thumb and finger until the solution thaws. We add aa-tRNAs last or next to last to a translation incubation to minimize their exposure to possible nuclease contaminants and the elevated temperatures of the translation. This protocol design is especially important for photocrosslinking experiments, since one wants to make the translations as complete as possible before having to turn out the light and add the photo-reactive aa-tRNAs to samples under red light conditions. Tubes containing photo-reactive aa-tRNA solutions are wrapped in aluminum foil to minimize premature photolysis. These tubes should only be opened in a dark room (turning off typical laboratory room lights is not sufficient) and under stringent red-light conditions (e.g., no cell phone and computer screen lights).
Micropipets with plastic tips do not deliver volumes accurately to 0.1 μl. Hence, one should avoid creating protocols that call for adding volumes less than 1 μl if possible. For that reason, we prepare a “Master Mix” (M) that provides the components common to each incubation in a set of n samples, where n is large enough to require component additions larger than 1 μl. This maximizes our chances of examining samples that have uniform concentrations of various materials.
If our experiment contains 18 separate 25-μl incubations, then M should be prepared for 20 incubations because the calculated M for 18 cannot be distributed into 18 separate tubes without loss.
Each component that is to be added to every sample in the assay should be included in M, so that all tubes in the assay receive the same amount of the component. In the hypothetical experiment shown in Table 2, different samples are to receive different mRNAs (e.g., Bax mRNAs each with a lysine codon at different positions). Thus, the total M volume must be adjusted for the volume of components that will be added separately to each individual sample tube. For example, in the above example, each sample is to receive 1 μl of an mRNA prep, so the volume of the assay provided by M will be 24 μl. Control samples not receiving mRNA should receive 1 μl of the buffer in which the mRNA is suspended. If samples were also to receive 1 μl of different aa-tRNA solutions, then one has to reduce the total M volume to (20 × 23 μl) to take account of the aa-tRNA that is added to tubes separately. Control samples not receiving aa-tRNA would instead receive 1 μl of the buffer in which the aa-tRNA is stored. Other components would be handled similarly if they were not added to M.
Each experiment should have both a positive and negative control so one can properly interpret the results. For example, the experiment shown in Figure 1C contains 4 samples: (i) “the sample” in lane 2 contains the product translated from Bax R37K mRNA in the presence of [35S]Met and εANB-[14C]Lys-tRNALys; (ii) “the minus light control” in lane 3 contains the same product as (i) but does not expose to light until after the photo-reactive probe is inactivated by DTT; (iii) “the minus photo-reactive probe control” in lane 1 contains the product translated from the same mRNA in the presence of [35S]Met, and instead of εANB-[14C]Lys-tRNALys, unlabeled lysine is added as part of the EGS-M solution; (iv) “the minus His6-Bax control” in lane 4 contains the same product as (i) but unlike the samples (i-iii), it does not incubate with the His6-Bax protein prior to photolysis.
Since strong reducing agents will chemically inactivate photo-reactive probes, but ribosomes must be in a reducing atmosphere to function, we use 2 mM glutathione, a weaker reducing agent than DTT in the translation for photocrosslinking.
The total amount of translation obtained and its accuracy is critically dependent on the final Mg2+ concentration, [Mg2+], in the incubation, as is the efficiency of suppression at amber codons by the suppressor tRNA. The optimum [Mg2+] can only be determined by experiment, and one has to routinely optimize the translation with respect to [Mg2+] (as well as other components; see below) for each different WG preparation and each different batch of mRNA. During the past few years, various constructs, WG preps, and amber suppression requirements have dictated final [Mg2+] concentrations ranging from about 1.0 to 3.5 mM Mg(OAc)2.
All components that contribute Mg2+ to the incubation must be accounted for in calculating the [Mg2+] in a sample. Wheat germ extract (WG) contains: 40 mM HEPES (pH 7.5), 100 mM KOAc (pH 7.5), 5 mM Mg(OAc)2, 4 mM glutathione. aa-tRNA stock solutions contain: 1 mM KOAc (pH 5.0), 2 mM Mg(OAc)2.
-
One should use moles to determine the proper amount of stock solution of Mg(OAc)2 to add to M to compensate for the Mg2+ added into the incubation with various materials. For example, if the optimization assays that one did indicated that the WG and mRNA one is using give maximal translation at a final [Mg2+] of 3.0 mM when each 25-μl assay contains 2 μl of WG, 1 μl of mRNA, and 1 μl of aa-tRNA, then the [Mg2+] in the 10x translation mix stock solution that needs to be added to M is determined as follows:
Moles = MV = (molarity in moles/liter)(volume in liters)
Total moles needed in M for 20 incubations of 25 μl = MMVM = (3.0 mM)(20 × 25 μl) = 1500 mM•μl
MMVM = moles added with WG + moles added with aa-tRNA + moles from 10× stock
= MWGVWG + Maa-tRNA Vaa-tRNA + MstockVstock
= (5 mM)(20 × 2 μl) + (2 mM)(20 × 1 μl) + (Mstock mM)(20 × 2.5 μl)
1500 mM•μl = 200 mM•μl + 40 mM•μl + (Mstock mM)(50 μl)
Mstock = 25.2 mM
The volume of KOAc stock solution that must be added to M is determined in the same way as in Note 11, based on the results of assays designed to determine the optimal [K+] required for translation by this WG and mRNA. During the past few years, the optimal KOAc concentrations for various mRNAs and WG preps have ranged from 90–140 mM.
The buffer concentration needs to be high enough to maintain the pH during the course of a translation and experiment. Hence, an excess of buffer is usually best unless the ionic strength gets too high. We typically use 25 mM HEPES-KOH (pH 7.5).
The amount of [35S]Met added depends upon the purpose of the experiment. If the goal is solely to assess translation yields or optimizations, then a final [35S]Met concentration of 0.1-0.5 μCi/μl is sufficient to get results in a few hours or overnight (e.g. Figure 1B, lanes 10-12). For photocrosslinking experiments, the [35S]Met concentration typically increases to 1.0-2.0 μCi/μl to get results in a few days or one week.
The version of EGS to be used in M is dependent on whether [35S]Met or εANB-[14C]Lys-tRNALys will be added to the sample. If only [35S]Met will be added, use EGS-M. If only εANB-[14C]Lys-tRNALys will be added, use EGS-K. If both will be added, use EGS-MK.
The optimal amount of a tRNA to use in a translation has to be determined experimentally, and the amount varies with different tRNAs. Most 25-μl translations for photocrosslinking receive about 15 pmoles of εANB-[14C]Lys-tRNALys. As shown in Figure 1B, the incorporation of this and other photo-reactive [14C]Lys or acetylated-[14C]Lys into wild type Bax that has nine Lys residues or a Bax mutant that has only one Lys residue can be detected on SDS-PAGE gel by phosphor-imaging of the 14C radiation. Based on the intensity of protein bands, the extent of photo-reactive Lys incorporation is in the following order: the acetylated εAc-[14C]Lys ≈ εANB-[14C]Lys > εAP-[14C]Lys ≈ εBP-[14C]Lys.
The translation efficiency of different mRNAs varies substantially, and is dependent upon their sequence, length, and tendency to form secondary structure. Sometimes, but not always, pre-incubating mRNAs at 37-67°C improves translation efficiency, and accuracy (i.e., producing the full-length polypeptide instead of a truncated one begun from an internal Met or ended by a secondary structure that blocks translation before the stop codon). The translation efficiency also varies from one mRNA prep to the next due to the amount of mRNA transcribed and/or to the residual salt in the sample, so it is best to optimize translation for each mRNA prep. Our 25-μl samples typically receive 1-2 μl of mRNA.
The short 5-min pre-incubation of the WG prior to the addition of the mRNA, [35S]Met or εANB-[14C]Lys-tRNA, if any, is designed to complete translation of any residual mRNA fragments in the WG before the addition of radioactive amino acids that could be incorporated into pre-existing nascent chains instead of the protein coded by the added mRNA.
The optimal time of translation for a given mRNA should be determined experimentally. In many cases, synthesis of short mRNAs reaches a maximum in 15-20 min, and additional incubation time is unnecessary and probably deleterious. In other cases, hairpins in the mRNA and/or low mRNA concentrations may slow down ribosome elongation and/or initiation sufficiently to require a longer translation time.
The order in which components are added to M is important. NF H2O should be added first so that the mixing of concentrated salts does not cause precipitation. Chemicals are normally added to M before biochemical components to ensure that the enzymes, proteins, etc., are added to a solution that has a pH, ionic strength, and metal ion concentrations that are not far from the final desired values (this minimizes protein denaturation and precipitation).
Solutions containing proteins should not be vortexed at all, much less vigorously, because the fluid shear stress and the increased exposure to surface tension due to the increase in solution surface area denatures proteins and hence will reduce the activity of translation. Instead, gently rotate tubes to mix or mix by sucking the solution slowly into and out of a large micropipet tip.
Researchers have different styles. We found it best to do all the thinking ahead of time, so that when we began the experiment, we could concentrate solely on what our hands were doing. While writing the protocol the night before doing the experiment, we could think through the experiment in our head and determine whether we could do what we had planned. This also gave us time to contemplate whether we had all of the proper controls and to double-check all of our calculations without being in a hurry. Such non-rushed thinking is particularly important when in vitro assays are more complicated.
Figure 1.
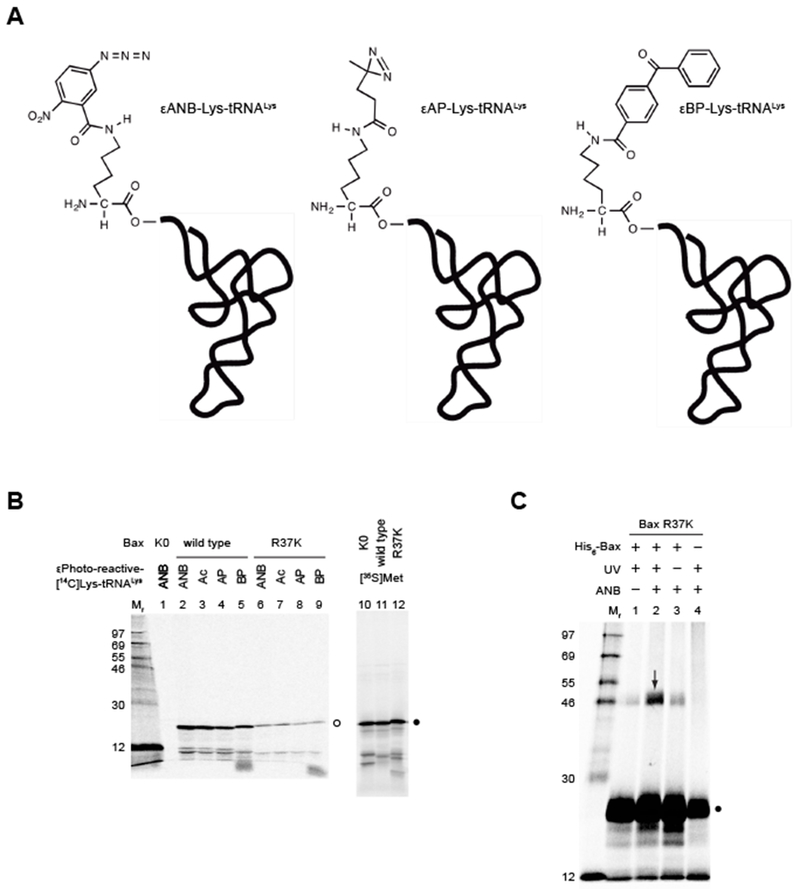
Synthesis and crosslinking of photo-reactive Bax protein. (A) Structures of photo-reactive εANB-[14C]Lys-tRNALys, εAP-[14C]Lys-tRNALys and εBP-[14C]Lys-tRNALys. (B) Synthesis of photo-reactive Bax protein. The wild type and Lys-null (K0) or single-Lys (R37K) mutant proteins are synthesized using wheat germ extract-based in vitro translation system in the presence of photo-reactive probe ANB, AP or BP-labeled [14C]Lys-tRNALys, or the acetylated (Ac) [14C]Lys-tRNALys, or [35S]Met. The resulting proteins are precipitated in trichloroacetic acid, washed by acetone-HCl, dried in vacuum, solubilized in SDS-PAGE gel-loading buffer, and analyzed by SDS-PAGE. The radioactive isotope, either the 14C in the photo-reactive or acetylated Lys or the35S in Met, labeled Bax proteins are detected in the gel by phosphor-imaging. The open circle indicates the 14C-labeled protein bands, among which the wild type Bax in lanes 2-5 contains nine Lys residues thereby displaying higher intensity than the single-Lys Bax mutant in lanes 6-9. As expected, the Lys-null mutant is not labeled by the 14C, and hence, invisible in lane 1. In contrast, the corresponding 35S-labeled protein bands in lanes 10-12 indicated by the close circle display similar intensity because they contain the same number of Met residues. Standard proteins are in lane Mr with their relative molecular mass (Mr) indicated. (C) Crosslinking of photo-reactive Bax protein to His6-tagged Bax protein. The in vitro synthesized Bax R37K protein with a single photo-reactive ANB probe attached to Lys37 and [35S]Met residues is mixed with the purified recombinant His6-Bax protein, activated by Bax BH3 peptide, and targeted to liposomal membranes containing the MOM-characteristic phospholipids. The membrane-bound proteins are fractionated and exposed to ultraviolet (UV) light that induces photocrosslinking. The photo-adduct between [35S]Met-Bax and His6-Bax is enriched on Ni2+-chelating resin, eluted into SDS-PAGE gel-loading buffer, analyzed by SDS-PAGE, and visualized by phosphor-imaging. The arrow in lane 2 indicates the photo-adduct, which is not or less detected in the control reaction lacking either ANB probe (lane 1) or UV irradiation (lane 3) or His6-Bax protein (lane 4). The close circle indicates the monomeric [35S]Met-Bax proteins. Standard proteins are in lane Mr.
Acknowledgements
This work was supported by the United States National Institutes of Health grants (R01GM062964, and P20GM103640), Oklahoma Center for the Advancement of Science and Technology grant (HR16-026), and Presbyterian Health Foundation grant (GRF00000125) to J.L.
5. References
- 1.Leber B, Lin J, Andrews DW (2007) Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 12 (5):897–911. doi: 10.1007/s10495-007-0746-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leber B, Lin J, Andrews DW (2010) Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene 29 (38):5221–5230. doi: 10.1038/onc.2010.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi X, Kale J, Leber B, Andrews DW (2014) Regulating cell death at, on, and in membranes. Biochim Biophys Acta 1843 (9):2100–2113. doi: 10.1016/j.bbamcr.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 4.Czabotar PE, Lessene G, Strasser A, Adams JM (2014) Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15 (1):49–63. doi: 10.1038/nrm3722 [DOI] [PubMed] [Google Scholar]
- 5.Cosentino K, Garcia-Saez AJ (2017) Bax and Bak Pores: Are We Closing the Circle? Trends Cell Biol 27 (4):266–275. doi: 10.1016/j.tcb.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Subramaniam S, Kale J, Liao C, Huang B, Brahmbhatt H, Condon SG, Lapolla SM, Hays FA, Ding J, He F, Zhang XC, Li J, Senes A, Andrews DW, Lin J (2016) BH3-in-groove dimerization initiates and helix 9 dimerization expands Bax pore assembly in membranes. EMBO J 35 (2):208–236. doi: 10.15252/embj.201591552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD (2002) Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111 (3):331–342. doi:S009286740201036X [pii] [DOI] [PubMed] [Google Scholar]
- 8.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW (2008) Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135 (6):1074–1084. doi:S0092–8674(08)01439–6 [pii] 10.1016/j.cell.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 9.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD (2010) BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell 40 (3):481–492. doi:S1097-2765(10)00795-1 [pii] 10.1016/j.molcel.2010.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH (2010) BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science 330 (6009):1390–1393. doi: 10.1126/science.1190217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, Huang DC, Kluck RM, Adams JM, Colman PM (2013) Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152 (3):519–531. doi: 10.1016/j.cell.2012.12.031 [DOI] [PubMed] [Google Scholar]
- 12.Moldoveanu T, Grace CR, Llambi F, Nourse A, Fitzgerald P, Gehring K, Kriwacki RW, Green DR (2013) BID-induced structural changes in BAK promote apoptosis. Nat Struct Mol Biol 20 (5):589–597. doi: 10.1038/nsmb.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW (1997) Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275 (5302): 983–986 [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW (2003) The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 19 (3):341–352. doi:S1074761303002346 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Billen LP, Kokoski CL, Lovell JF, Leber B, Andrews DW (2008) Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol 6 (6):e147. doi:07-PLBI-RA-3722 [pii] 10.1371/journal.pbio.0060147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aranovich A, Liu Q, Collins T, Geng F, Dixit S, Leber B, Andrews DW (2012) Differences in the mechanisms of proapoptotic BH3 proteins binding to Bcl-XL and Bcl-2 quantified in live MCF-7 cells. Mol Cell 45 (6):754–763. doi: 10.1016/j.molcel.2012.01.030 [DOI] [PubMed] [Google Scholar]
- 17.Ding J, Mooers BH, Zhang Z, Kale J, Falcone D, McNichol J, Huang B, Zhang XC, Xing C, Andrews DW, Lin J (2014) After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization. J Biol Chem 289 (17):11873–11896. doi: 10.1074/jbc.M114.552562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH (2006) Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 8 (12):1348–1358. doi:ncb1499 [pii] 10.1038/ncb1499 [DOI] [PubMed] [Google Scholar]
- 19.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315 (5813):856–859. doi: 10.1126/science.1133289 [DOI] [PubMed] [Google Scholar]
- 20.Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G (1999) Solution structure of Bid, an intracellular amplifier of apoptotic signaling. Cell 96:615–624 [DOI] [PubMed] [Google Scholar]
- 21.Suzuki M, Youle RJ, Tjandra N (2000) Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103 (4):645–654 [DOI] [PubMed] [Google Scholar]
- 22.Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW (2004) During apoptosis Bcl-2 changes membrane topology at both the endoplasmic reticulum and mitochondria. Mol Cell 14 (4):523–529. doi:S1097276504002631 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, Andrews DW (2005) Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J 24 (12):2096–2103. doi:7600675 [pii] 10.1038/sj.emboj.7600675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westphal D, Dewson G, Menard M, Frederick P, Iyer S, Bartolo R, Gibson L, Czabotar PE, Smith BJ, Adams JM, Kluck RM (2014) Apoptotic pore formation is associated with in-plane insertion of Bak or Bax central helices into the mitochondrial outer membrane. Proc Natl Acad Sci U S A 111 (39):E4076–4085. doi: 10.1073/pnas.1415142111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao C, Zhang Z, Kale J, Andrews DW, Lin J, Li J (2016) Conformational Heterogeneity of Bax Helix 9 Dimer for Apoptotic Pore Formation. Scientific reports 6:29502. doi: 10.1038/srep29502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten mJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435 (7042):677–681. doi:nature03579 [pii] 10.1038/nature03579 [DOI] [PubMed] [Google Scholar]
- 27.Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES, Walensky LD (2012) Direct and selective small-molecule activation of proapoptotic BAX. Nat Chem Biol 8 (7):639–645. doi: 10.1038/nchembio.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cory S, Roberts AW, Colman PM, Adams JM (2016) Targeting BCL-2-like Proteins to Kill Cancer Cells. Trends Cancer 2 (8):443–460. doi: 10.1016/j.trecan.2016.07.001 [DOI] [PubMed] [Google Scholar]
- 29.Delbridge AR, Grabow S, Strasser A, Vaux DL (2016) Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer 16 (2):99–109. doi: 10.1038/nrc.2015.17 [DOI] [PubMed] [Google Scholar]
- 30.Niu X, Brahmbhatt H, Mergenthaler P, Zhang Z, Sang J, Daude M, Ehlert FGR, Diederich WE, Wong E, Zhu W, Pogmore J, Nandy JP, Satyanarayana M, Jimmidi RK, Arya P, Leber B, Lin J, Culmsee C, Yi J, Andrews DW (2017) A Small-Molecule Inhibitor of Bax and Bak Oligomerization Prevents Genotoxic Cell Death and Promotes Neuroprotection. Cell Chem Biol 24 (4):493–506 e495. doi: 10.1016/j.chembiol.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Zhu W, Lapolla SM, Miao Y, Shao Y, Falcone M, Boreham D, McFarlane N, Ding J, Johnson AE, Zhang XC, Andrews DW, Lin J (2010) Bax forms an oligomer via separate, yet interdependent, surfaces. J Biol Chem 285 (23):17614–17627. doi: 10.1074/jbc.M110.113456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding J, Zhang Z, Roberts GJ, Falcone M, Miao Y, Shao Y, Zhang XC, Andrews DW, Lin J (2010) Bcl-2 and Bax interact via the BH1–3 groove-BH3 motif interface and a novel interface involving the BH4 motif. J Biol Chem 285 (37):28749–28763. doi: 10.1074/jbc.M110.148361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erickson AH, Blobel G (1983) Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol 96:38–50 [DOI] [PubMed] [Google Scholar]
- 34.Johnson AE, Woodward WR, Herbert E, Menninger JR (1976) Nepsilon-acetyllysine transfer ribonucleic acid: a biologically active analogue of aminoacyl transfer ribonucleic acids. Biochemistry 15 (3):569–575 [DOI] [PubMed] [Google Scholar]
- 35.Krieg UC, Walter P, Johnson AE (1986) Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci U S A 83 (22):8604–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan C, Dlugosz PJ, Peng J, Zhang Z, Lapolla SM, Plafker SM, Andrews DW, Lin J (2006) Auto-activation of the apoptosis protein Bax increases mitochondrial membrane permeability and is inhibited by Bcl-2. J Biol Chem 281 (21):14764–14775. doi: 10.1074/jbc.M602374200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kale J, Chi X, Leber B, Andrews D (2014) Examining the molecular mechanism of bcl-2 family proteins at membranes by fluorescence spectroscopy. Methods Enzymol 544:1–23. doi: 10.1016/B978-0-12-417158-9.00001-7 [DOI] [PubMed] [Google Scholar]
- 38.Gurevich VV, Pokrovskaya ID, Obukhova TA, Zozulya SA (1991) Preparative in vitro mRNA synthesis using SP6 and T7 RNA polymerases. Anal Biochem 195 (2):207–213 [DOI] [PubMed] [Google Scholar]
- 39.Falcone D, Andrews DW (1991) Both the 5’ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol Cell Biol 11 (5):2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunner J (1993) New photolabeling and crosslinking methods. Annu Rev Biochem 62:483–514. doi: 10.1146/annurev.bi.62.070193.002411 [DOI] [PubMed] [Google Scholar]
- 41.Johnson AE, Adkins HJ (1984) Glycerol, sucrose, and other diol-containing reagents are not inert components in in vitro incubations containing aminoacyl-tRNA. Anal Biochem 137 (2):351–359 [DOI] [PubMed] [Google Scholar]


