Summary
Ca2+ is a ubiquitous, dynamic and pluripotent second messenger with highly context-dependent roles in complex cellular processes such as differentiation, proliferation, and cell death [1]. These Ca2+ signals are generated by Ca2+-permeable channels located on the plasma membrane (PM) and endoplasmic reticulum (ER) and shaped by PM- and ER-localized pumps and transporters. Differences in the expression of these Ca2+ homeostasis proteins contribute to cell and context-dependent differences in the spatiotemporal organization of Ca2+ signals and, ultimately, cell fate. This review focuses on the Early Growth Response (EGR) family of zinc finger transcription factors and their role in the transcriptional regulation of Stromal Interaction Molecule (STIM1), a critical regulator of Ca2+ entry in both excitable and non-excitable cells.
Graphical Abstract
Section 1: Introduction
Store-operated Ca2+ entry (SOCE) is a ubiquitous Ca2+ entry pathway, with important roles in both excitable and non-excitable cells. The principal means of SOCE to the cytosol is via Ca2+-Release Activated Ca2+ (CRAC) channels formed by Orai proteins and activated by Stromal Interaction Molecule (STIM). STIM is a single-pass membrane protein located primarily in the ER membrane that senses ER Ca2+ content via its EF hand domains. Following a drop in ER Ca2+, STIM translocates within the ER membrane toward the PM where it associates directly with Orai1 (reviewed in [2-4]). Although STIM1 and Orai1 are the primary mediators of SOCE, each has isoforms with high sequence homology and similar overall structural organization. Hence, similar to STIM1, STIM2 has an ER Ca2+ sensor and a capacity to activate CRAC channels, although it is prone to activation with minimal ER Ca2+ depletion [5-8] and activates CRAC with less efficiency [7, 9]. Further, both STIM proteins can activate Orai2 and Orai3 [10-12], which share biophysical characteristics with Orai1, but poorly described physiological roles. The regulation of STIM1 and Orai1 expression contributes directly to Ca2+ signaling in non-excitable cell types. Thus, transcriptional control of Ca2+ homeostasis proteins is critical to understanding how Ca2+ signals contribute to development and disease.
Section 2: Zinc finger transcription factors EGR and WT1
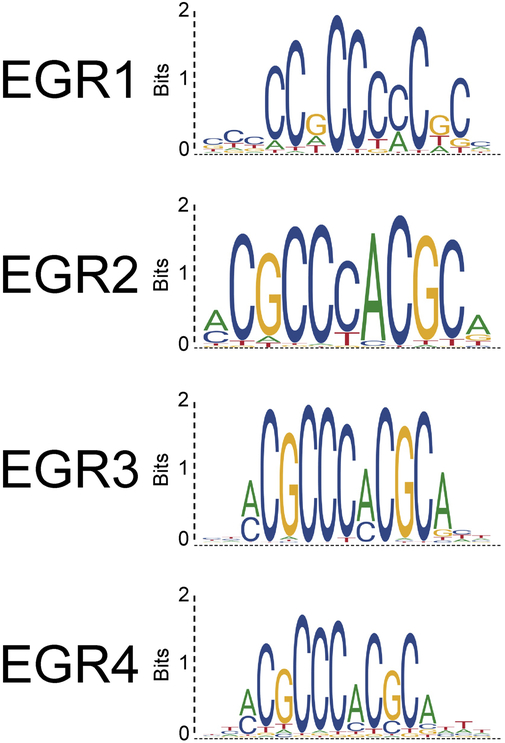
Early Growth Response (EGR) proteins are a family of transcription factors that bind to the GC-rich regions of the promoters of hundreds of genes in multiple cell types, including leukocytes from both the lymphoid and myeloid lineages, fibroblasts, neurons, and endothelial cells. The EGR genes are located on separate chromosomes (5q31 for EGR1, 10q21 for EGR2, 8p21 for EGR3 and 2p13 for EGR4), but are dynamically upregulated together in response to a variety of cell stimuli, including growth and pro-differentiation factors [13]. The four EGR proteins have highly conserved zinc finger domains, and generally share the recognition sequence GCG (G/T)GG GCG [14-16], although some differences in binding affinities for specific nucleotides at each position exist (Fig 1). Further, isoform-specific flanking regions and cell type-specific expression support distinct functions for each protein, which can both positively and negatively regulate target gene expression [17]. For example, EGR1 and EGR4 exhibit the capacity to drive STIM1 expression while EGR2 and EGR3 do not [18]. This is similar to the link between EGR1 and EGR4 for control of Luteinizing Hormone expression [19], indicative of greater targeting similarity between these two family members.
Figure 1: Homology of EGR consensus sequences.
Binding profile of EGR family members based on reported experimental data generated by chromatin immunoprecipitation with next generation DNA sequencing. Source: JASPAR database.
Although Wilms Tumor Suppressor 1 (WT1) exhibits substantial overlap in its consensus targets with EGR1, it is not considered a member of the EGR family. Unlike EGRs which are immediate early genes, WT1 expression is primarily developmentally regulated [20, 21]. Over 20 isoforms of WT1 exist due to multiple transcriptional start sites, alternative splicing, and RNA editing. The most striking difference between these isoforms results from an alternative splice site in exon 9 that leads to the insertion of a lysine-threonine-serine (KTS) sequence between the 3rd and 4th zinc fingers. This has ramifications for their DNA binding affinities. Whereas KTS- splice variants readily bind DNA and act as transcription factors, KTS+ WT1 isoforms have a low affinity for DNA and regulate RNA processing [22, 23]. The extent to which the RNA splicing and gene transcription functions of WT1 overlap is not well established, although we have observed opposing regulation of STIM1 expression between the KTS+ and KTS- forms of WT1 (data not shown).
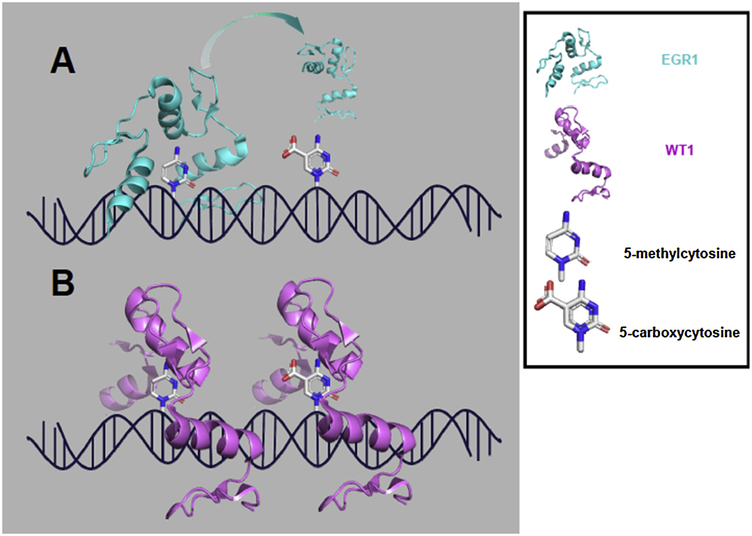
While WT1 and EGR1 share many transcriptional targets, WT1 frequently binds to and represses gene targets that EGR1 upregulates [24]. The frequently opposing effects of EGR1 and WT1 on their shared targets reflects important differences in how the proteins identify targets and recruit transcriptional machinery. They also exhibit differences in their sensitivity to epigenetic modifications. Briefly, within the mammalian genome, 70 to 80% of cytosines found within CpG sites (stands for 5’-C-phosphate-G-3’) (CpG) sites are methylated. Given the EGR1 consensus sequence, methylation is common and has been correlated with reduced EGR1 binding and downregulation of EGR1 target genes in rodent models of cardiomyopathy and bladder cancer [25, 26]. However, X-ray crystallography and affinity studies showed that the EGR1 and WT1 zinc finger domains were able to accommodate fully methylated DNA, and their binding kinetics were insensitive to methylation [27, 28]. Rather, EGR1 and WT1 differ in their ability to bind oxidized forms of cytosine, generally considered intermediate states in epigenetic reprogramming. Hence, a glutamate residue within EGR1 precludes binding to 5-carboxylcytosine, whereas the homologous residue in WT1 is a glutamine which favors binding to carboxylated cytosine (Fig. 2) [28]. This preferential binding to 5-carboxylcytosine is potentially informative for understanding how EGR1/WT1 targets could diverge in neoplasias, as elevated 5-carboxylcytosine has been observed in breast cancer and gliomas and may serve as a distinct epigenetic mark in its own right [29, 30]. The effect of methylation on other EGR isoforms will be equally important to understand how EGR transcriptional activity interacts with epigenetic marks.
Figure 2: EGR1/WT1 binding to methylation marks.
(A) EGR1 can accommodate and bind DNA with 5-methylcytosine but not 5-carboxylcytosine (PDB: 4R2A); (B) WT1 can accommodate both stages of cytosine oxidation (PDB: 4R2E).
Although EGR1 and WT1 are largely insensitive to methylation, their binding may be subject to regulation by, or competition with, other zinc finger transcription factors. These include Sp1, which is a ubiquitously expressed zinc finger DNA-binding transcription factor whose recognition sequence overlaps with EGR1 [31, 32]. The sequence of Sp1/EGR1 binding sites contributes to the binding affinity of the transcription factors, and therefore the extent to which a downstream gene may be regulated by EGR1 and/or Sp1 [33]. Conversely, EGR proteins, with the exception of EGR4, share a repressor binding domain, and are inhibited by NGFI-A binding (NAB) proteins NAB1 and NAB2 [34]. NAB2, which binds to EGR1 and regulates EGR1 target gene expression, is itself regulated by EGR1, forming a negative feedback loop by which EGR1 activity is controlled [35-38]. Whereas NAB1 inhibits EGR1, the role of NAB2 is more nuanced. In the context of T cell activation, NAB2 is upregulated downstream of costimulation and acts as a coactivator of EGR1-mediated IL-2 expression [39]. A better understanding of how these transcription factors simultaneously target genes with overlapping binding sites is an intriguing and important question for future studies [33, 40-42].
EGR1 and WT1 are implicated across cell type and developmental stage. This reflects the several layers of regulation required for the function in the isoforms and splice variants that are expressed in a cell type-specific manner, as well as in differences in DNA binding, and interactions with epigenetic marks, and the expression levels of repressor proteins and other transcription factors.
Section 3: Regulation of EGR expression
EGRs mediate many of the expression changes during diverse types of cell stimulation, including cytokines, mitogens, or oxidative stress [43-45]. EGR1 activity is largely effected by inducing EGR1 upregulation and by post-translational modifications [17, 46-48]. The human EGR1 promoter contains 5 serum response elements (SREs) and a cAMP response element (CRE) [49, 50]. As further reviewed by Veyrac et al., a number of transcription factors can be recruited to the EGR1 promoter in different cell types [51]. EGR1 is able to regulate its own transcriptional activity in a negative feedback loop, both at the expression level by binding to its promoter, and by inducing the expression of its repressor NAB2 [17]. Elk-1, which is activated by phosphorylation downstream of ERK, JNK, and p38 MAPK signaling pathways, associates with serum factors at SREs [52, 53]. EGR1 is also regulated by PI3K/Akt, which phosphorylates and disrupts the DNA-binding ability of FoxO1, a negative regulator of EGR1 expression [54].
In pre-synaptic neurons, EGR1 expression is repressed by transcriptional repressor CtBP1 until neuronal activity relieves repression by CtBP1 [55]. In cells exposed to UVB-induced genotoxic stress, NF-κB binds to the EGR1 promoter to initiate an apoptosis response [56]. Additionally, during T cell activation, EGR1-induced upregulation of STIM1 promotes increases in cytosolic Ca2+ content [18] which also may contribute to EGR1 upregulation (reviewed in [57]). This is supported by the observation that chelation of cytoplasmic Ca2+ by BAPTA-AM downregulates EGR1 expression, and that it may be required for recruiting transcriptional machinery to SREs and CRE within the EGR1 promoter [58-60]. Additional support for the role of Ca2+ signals in EGR1 upregulation came from investigations in vascular smooth muscle revealing mediatory roles of CaMKII and CREB in Angiotensin II-mediated EGR1 expression [61].
EGR1 may be modified post-translationally by phosphorylation, glycosylation and sumoylation with distinct functional outcomes. Low levels of EGR1 phosphorylation in quiescent cells have been shown to contribute to decreased EGR1 stability [17]. In contrast, phosphorylation by casein kinase II negatively modulated EGR1’s DNA binding activity [62]. This likely reflects multiple phosphorylation sites on EGR1, each with a distinct impact on EGR1 function. Phosphorylation of EGR1 by Akt has been revealed as a requirement for sumoylation, a critical precursor to EGR1 ubiquitination and degradation [63]. O-glycosylation of the EGR1 transactivation domain has also been shown [64], although the functional implications were not determined. Future investigations dissecting the coordination of these disparate post-translational mechanisms are needed to understand how the stability and targeting of EGR1 may be regulated through crosstalk mechanisms with other signaling pathways.
EGR4 is also upregulated in response to trophic factors in an ERK1/2-dependent manner that has been characterized in the context of neuronal development and spermatogenesis [65-67]. While EGR4 appears to compensate for EGR1 in some cases, SRF binding to the EGR4 promoter was shown to occur in neuronal cells [68] supported by indirect evidence that serum may regulate the EGR4 promoter generated in Jurkat T cells [69]. Interestingly, the EGR4 promoter is regulated by both EGR1 and 4, with EGR4 opposing its own transcription and EGR1 modestly enhancing EGR4 expression [70]. Hence, despite the many similarities between EGR consensus sequences, they are not interchangeable, and their expression can lead to mutually opposing outcomes.
Section 4: WT1 and EGR1/4 regulate STIM1 expression and function
Our investigations over the last 10 years have revealed modulation of Ca2+ homeostasis due to EGR1-, EGR4- and WT1-mediated control of STIM1 expression. Our first observation ultimately leading us in this direction was that WT1-expressing Wilms Tumor cells exhibit decreased STIM1 expression and SOCE [71]. Using a combination of knockdown and overexpression strategies in HEK293 and G401 cells we subsequently established that EGR1 positively regulates STIM1 expression and WT1 suppresses it [71]. Recognizing that EGRs are primarily immediate early genes, subsequent work focused on dynamic control of STIM1 expression using T cells as a model system (Figure 4) [18, 72]. In Jurkat T cells, we observed that EGR1 and STIM1 are both upregulated within 2 hours of TCR engagement by either the lectin PHA [72] or anti-CD3/CD28 antibodies [18]. However, EGR1 knockdown failed to eliminate STIM1 upregulation, leading to the discovery that EGR1 and EGR4 cooperatively regulate STIM1 expression in T cells [18]. Hence, loss of STIM1 expression was only observed when both EGR1 and EGR4 were knocked down. Further, while chromatin immunoprecipitation (ChIP) analysis showed that TCR engagement leads to EGR1 binding to DNA, EGR4 knockdown decreased EGR1 binding to the STIM1 promoter [18]. These observations reveal a cooperative model by which EGR1 and EGR4 control STIM1 expression during T cell activation.
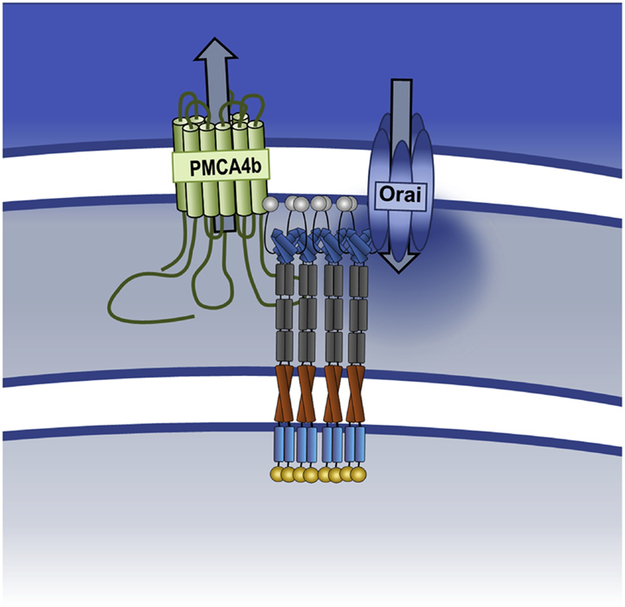
Figure 4: STIM1-mediated control of Ca2+ entry and clearance.
ER Ca2+ depletion causes STIM1 to oligomerize and extend towards the PM to activate Ca2+ entry via Orai channels. STIM1 also interacts with PMCA through its serine/threonine-rich domain causing decreased function.
Because WT1-mediated STIM1 downregulation led to SOCE suppression, our preliminary hypothesis was that EGR-mediated STIM1 upregulation would lead to increased SOCE. However, comparisons of SOCE magnitude in resting and activated T cells revealed only marginal differences in SOCE [18, 72]. In retrospect, this is perhaps not surprising since, although required for SOCE, STIM1 upregulation generally does not increase SOCE unless accompanied by coincident changes in Orai1 expression [10, 73-75]. We also observed PMCA4 upregulation upon T cell activation and significant inhibition of Ca2+ clearance [72]; we have since attributed PMCA4 upregulation to EGR1/4-dependent transcription based upon the identification of two EGR binding sites by ChIP within the PMCA4 promoter [18]. EGR1/4-mediated upregulation of both STIM1 and PMCA4) [18] is accompanied by the translocation of both proteins towards the immunological synapse (IS) where STIM1 and PMCA4 colocalize [72, 76-78] (Fig. 3). Although this only minimally affects the initial rate of Ca2+ clearance, attenuation of the “slow” phase of Ca2+ clearance was observed. Notably, this could be mimicked by STIM1 overexpression without T cell activation and blocked by partial inhibition of SOCE through STIM1 knockdown or pharmacological inhibition [72]. Although which PMCA4 domains are required for STIM1 binding were not determined, the proline/serine-rich domain of STIM1 was required for PMCA inhibition and co-immunoprecipitation. Hence, STIM1-mediated control of Ca2+ entry and clearance are independent events that can be effectively separated under different conditions. This inhibition of PMCA4 by STIM1 facilitates increases in intracellular Ca2+ levels sufficient to engage NFAT transcription, a necessary requirement of T cell activation [18, 79]. Thus, STIM1 sustains elevated Ca2+ levels in activated T cells using multiple mechanisms, namely, via Orai1 activation and PMCA4 regulation (Fig. 4).
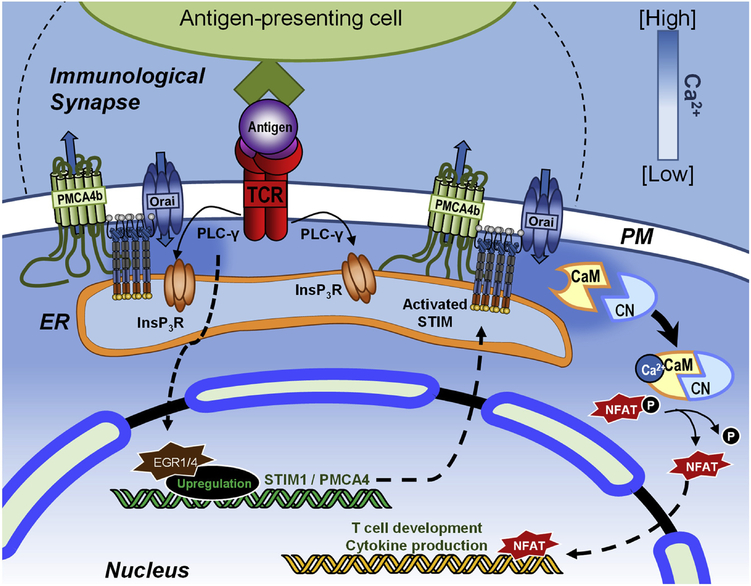
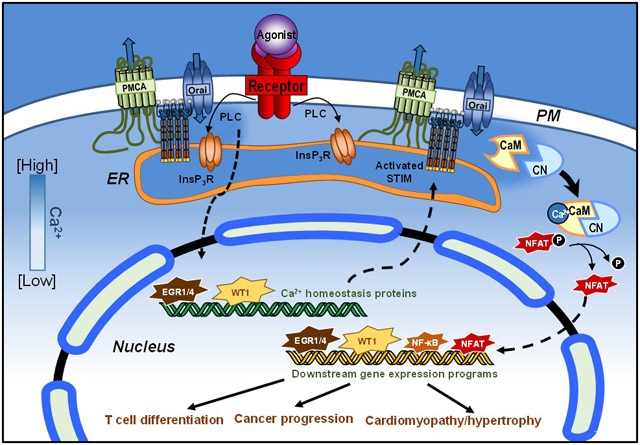
Figure 3: EGR1-mediated regulation of Ca2+ signaling in T cell activation.
The TCR is activated upon engagement with its antigenic peptide presented by an antigen-presenting cell. With TCR engagement, phospholipase Cγ (PLCγ) is activated, leading to the rapid release of ER Ca2+ through the InsP3R and the entry of Ca2+ through Orai1 Over time, the zinc finger transcription factors EGR1 and EGR4 are activated, driving the expression of Ca2+ homeostasis proteins including STIM1 and PMCA4 to facilitate long-term Ca2+ signals. These sustained Ca2+ signals contribute to CaM/CN activation, critical for driving NFAT translocation to the nucleus where it facilitates the expression of T cell differentiation and proliferation gene programs.
EGR1 has previously been shown to be upregulated following strong TCR activation, leading to disruption of development from the αβ to the γδ lineage [80]. Further, Lohoff et al. reported that EGR1 is preferentially expressed in Th2 cells and upregulates IL-4 expression downstream of NFAT activation [44]. The extent to which EGR1/4 regulate STIM1 expression to shape T cell activation and differentiation is under active investigation by our group.
Another component of the intracellular signaling pathway under EGR transcriptional control is the sarco/endoplasmic reticulum ATPase 2 isoform (SERCA2). SERCA family proteins are ATP-driven pumps that drive Ca2+ from the cytosol to the ER. SERCA pumps concentrate at ER-PM junctions upon store-depletion to facilitate store-refilling upon engagement of Orai channels by STIM proteins. Of the three SERCA family members, SERCA1 is expressed exclusively in fast-twitch muscle while SERCA2 and 3 are ubiquitously expressed [81]. The relationship between EGR1 and SERCA2 is still somewhat controversial. A link between EGR1 and SERCA2 was first demonstrated in the context of doxorubicin-induced cardiomyopathy, whereby EGR1 was upregulated downstream of the p44/42 MAPK pathway and a concurrent decrease in SERCA2 expression was observed [82]. While this was initially attributed to direct EGR1 transcriptional regulation, another group found no evidence of functional EGR1 binding sites in the SERCA2 promotor [83]. More recently a study in mitogen-activated protein kinase (MAPK)-activated protein kinase 2/3 knock-out mice found a decrease in EGR1 expression driven by MK2, correlated with an increase in SERCA2a in cardiomyocytes [84]. The group found that SERCA2 expression is promoted by Sp1 binding to its promoter, with EGR1 occluding these sites to act as a negative regulator, under the control of MK2. Therefore, EGR1 may negatively regulate SERCA2 expression through an indirect mechanism. Conversely, NFAT positively regulates SERCA2 expression through a similar indirect mechanism [85, 86]. How this may affect T cell activation is not well established, although increased ER Ca2+ uptake via SERCA2 would support the increased Ca2+ cycling necessary for the Ca2+ oscillations required for T cell activation [87].
EGR1 has also been shown to downregulate sodium-calcium exchanger (NCX), a plasma membrane protein that uses the electrochemical gradient of sodium to extrude one Ca2+ for every three sodium ions imported. ChIP analysis determined that EGR1 could directly bind the promoter of NCX1 and EGR1 over-expression in cardiomyocytes lead to decreased NCX1 expression [88]. EGR1 has been shown to be upregulated during cardiac hypertrophy [99-102] and the corresponding loss of NCX- and SERCA2-mediated Ca2+ clearance in cardiomyocytes would lead to increased cytosolic Ca2+ levels and enhanced NFAT activity, thereby contributing to the myocardial thickening that defines this disease.
Section 5: Cooperativity of transcription factors in T cell activation
EGR family members frequently interact with other transcription factors, leading to fundamental differences in the genes that are regulated. For example, the C-terminal region of zinc finger III in EGR4 directly interacts with the NF-κB family member p65, leading to robust activation of the IL-2 promoter [89]. There is also growing evidence of a Ca2+-dependent feedback loop between NF-κB and STIM1/Orai1 expression in which 1) SOCE drives IκB degradation [90], 2) Ca2+ regulates NF-κB nuclear localization via p65 S536 phosphorylation [90] and 3) NF-κB is recruited to binding sites in the promoters of STIM1 and Orai1 [91-93]. Since both EGR and NF-κB both regulate STIM1 expression and yet are themselves regulated by Ca2+, their co-activation would be expected to be relatively common. If so, one might expect EGR-NF-κB complexes to be similarly common, fundamentally altering gene expression patterns in cell types of interest.
Whereas EGR1 and EGR4 contribute primarily to early steps in T cell activation, EGR2 and EGR3 serve as negative regulators of effector differentiation [39]. Hence, Egr2−/−Egr3−/− lymphocytes exhibit impaired AP-1 activation during antigen-induced activation, resulting in lethal autoimmune syndrome [94]. Further, EGR2/3 inhibit T-bet-mediated IFN-γ production, facilitating clonal expansion and suppressing effector differentiation [95-97]. Consistent with this immunosuppressive role, EGR2 drives TGF-β expression during induction of LAG3+ regulatory T cells (Tregs); in the absence of EGR2, loss of LAG3+ Tregs leads to loss of protection in a mouse model of systemic lupus erythematosus (SLE) [98]. Human LAG3+ Tregs also suppress antibody production and SLE is correlated with both decreased EGR2 expression and decreased LAG3+ Treg numbers [99, 100]. While the molecular mechanisms controlling EGR2/3 expression are not fully elucidated, NF-κB binding to its recognition sequence in the proximal promoter of EGR2 has been observed [89, 101]. Further, Ca2+ dependent expression of EGR2 and EGR3 has been observed [39]. Hence, despite their fundamentally different roles, the EGR family members share some of the mechanisms by which they are regulated. It seems likely that signaling by different EGRs is primarily due to temporal control of EGR isoforms, with EGR1 and EGR4 being expressed during early events followed by negative regulation by EGR2 and EGR3 at later time points.
Section 6: Dysregulation of SOCE in cancer
As mitogen-activated transcription factors, EGRs have a multifaceted role in tumorigenesis and progression, including as regulators of SOCE components. Our group has shown that melanoma cells expressing known oncogene Wnt5a exhibit decreased SOCE and increased invasive ability compared to non-Wnt5a-expressing cells [102]. Further, SOCE promotes apoptotic cell death in prostate cancer [103, 104], providing a potential explanation for why loss of SOCE might be favorable for tumor progression. However, SOCE has been shown to have oncogenic properties as well, enhancing tumor angiogenesis through the production of vascular endothelial growth factor (VEGF) in cervical cancer [105], and cyclooxygenase-2 (COX-2) [106] and nuclear factor of activated T cells (NFAT) [107] in a variety of pathophysiological responses. Furthermore, upregulation of STIM1 and Orai1 has been reported in patients exhibiting a variety of cancers including lung, liver, and colorectal cancers among others [103]. Here, we will address potential some of these seeming contradictions to better define the pleiotropic roles of Ca2+ signals in cancer initiation and progression.
Interestingly, EGR/WT1 transcription factors also play dual roles in cancer progression. EGR1 upregulation is downstream of some established oncogenic pathways such as TGF-β, NF-κB, and MAPKs, however its contributions to cancer remain poorly defined [108, 109]. Although WT1 was originally described as a pro-apoptotic tumor suppressor, WT1 contributes to cancer progression in some contexts [110-114]. We previously showed that WT1 interferes with Ca2+ homeostasis by blocking expression of STIM1 in Wilms tumor cells [71]. Interestingly, aberrant expression of the STIM1-independent Ca2+ channel (CaV2.3) is associated with Wilms tumor relapse through MAPK-related pathways [115]. Further, decreased EGR levels have been correlated with limited response to chemotherapy in patients with Wilms tumor [116]. Therefore, in Wilms tumor, it appears both WT1 expression and loss of Ca2+ signals are associated with tumor suppression, whereas both EGR1 and increased Ca2+ signals are associated with tumor resilience.
EGR/WT1 transcription factors are implicated in other cancers in both tissue-specific and tissue-agnostic roles. In cervical cancer, EGR1 has been shown to promote tumor progression through downregulating telomerase [117]. In hepatocellular carcinoma (HCC), EGR1 downregulation reduces pro-apoptotic and growth inhibiting pathways [108, 118]. However, EGR1 also contribute to chemotherapy resistance by regulating microtubule function and autophagy [119-121]. Interestingly, although it frequently acts in opposition to EGR1, WT1 expression in HCC cell lines was also associated with chemotherapy resistance [122]. In several cancers, including ovarian and colon cancer, EGR1 has been shown to play a role in EMT and metastasis [123, 124]. WT1 plays an intriguing role in renal cell carcinoma (RCC). In a study on RCC, WT1 induced a hybridized state of EMT and MET (EMHT) in which the mesenchymal marker, Snail, was simultaneously expressed with epithelial marker E-cadherin [125]. This exciting finding suggests that EMT and MET are not mutually exclusive, and that WT1 could be an important modulator of the equilibrium between the two states. Interestingly Orai1 expression has been shown to be associated with favorable prognosis in clear cell RCC where genetic and epigenetic alterations in EGR1 expression were associated with survival [126, 127]. However, the extent to which EGR1/WT1-mediated regulation of Ca2+ signaling components contributes to EMT/MET transitions remains unknown.
During both development and tumorigenesis, the expression of EGR/WT1 transcription factors, and the control of Ca2+ signaling machinery are critical. ER- breast cancer exhibits deletion of EGR1 particularly in higher-grade carcinomas [128]. EGR1 deletions also occur in gastric carcinoma and are associated with conferring invasive properties, since they are found primarily in distant metastases [129]. Since EGR deletion attenuates STIM1 expression and interferes with Ca2+ signals [113], these data may be consistent with a link between SOCE suppression and metastatic properties. In prostate cancer, this may be the case since Orai1 and androgen receptor regulate each other’s expression in a negative feedback loop and STIM1 and Orai1 expression were inversely correlated with Gleason score [130]. On the other hand, STIM1 and Orai1 were linked to focal adhesion formation in breast cancer where SOCE suppression decreased focal adhesions leading to increased metastasis in vivo [131]. For the purposes of illustration, the context-specific roles of EGR1, WT1 and SOCE discussed in this section are shown in figure 5. Further investigations delineating the features of these distinct diseases that contribute to differential roles of these factors are needed to better understand the relationship between Ca2+ signals and cell growth, migration, invasion and survival.
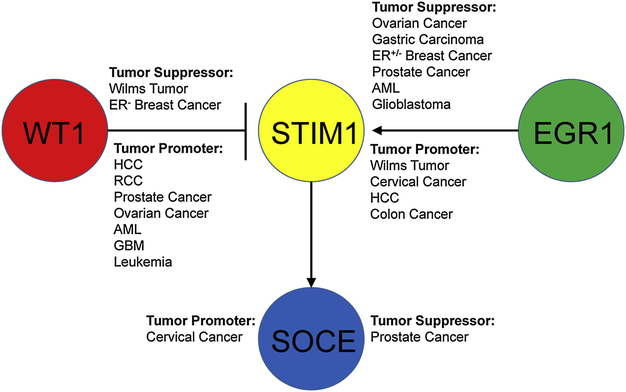
Figure 5: Dual and opposing roles of WT1 and EGR1 in cancer progression.
WT1 and EGR1 play opposing roles in regulating STIM1 during cancer progression. WT1 acts in a tumor suppressing nature in Wilms tumor and ER+ breast cancer, but in a tumor-promoting in ER- breast cancer, and other cancers. EGR1 acts as a tumor promoting nature in both ER+/− breast cancer and other types of cancer.
Section 7: WT1 and EGR1 in cardiovascular and pulmonary pathophysiology
An important regulator of vascular disease is the inflammatory immune response induced upon vascular injury. EGR1 is strongly up-regulated in macrophages and other immune cells during wound healing [132] and vascular injury [133], the latter of which is strongly associated with cardiovascular disease [134]. Outlined below are some of the strides made in recent years in understanding the role of EGR1 in vascular disease.
EGR1 is strongly associated with promoting atherosclerosis progression [135]. In smooth muscle cells from both LDL-receptor deficient [136] and apolipoprotein E (ApoE)-null mice [137]. EGR1 is upregulated in atherosclerotic lesions. Further, statins inhibit EGR1 expression in atherosclerotic lesions [138], further supporting the existence of a link between EGR1 expression and atherosclerosis. EGR1 expression in vascular smooth muscle cells (VSMCs) is primarily driven by NADPH-dependent heme oxidation, amongst other things [139, 140]. These observations support the notion EGR1 target genes are involved in cellular responses to oxidative conditions. This is pertinent because atherosclerotic plaques frequently produce hypoxic conditions, exacerbating damage and inflammation in the vasculature [141]. Following hypoxic conditions, multiple tissue types upregulate EGR1, including neural tissue, cells in peri-infarct regions, and renal cells [135, 142-145]. There is evidence to suggest that this is mediated at least in part through regulation of TGFβ signaling by EGR1, in the context of pulmonary fibrosis, and through cyclin D and EGF signaling in pulmonary adventitial fibroblasts during vascular remodeling [146-148]. These studies establish the beginning of an understanding into how EGR1 regulates vascular remodeling following hypoxic injury.
Similar to what we have described in neoplastic disease, EGR1 is upregulated under ischemic conditions, such as during lung or myocardial reperfusion injury, and has been implicated in numerous aspects of these conditions. In some contexts, EGR1 could be seen as beneficial following some types of cardiovascular injury. For example, EGR1-mediated promotion of angiogenesis improves cardiac remodeling in a rat model of heart failure concurrent with dietary obesity [149]. However, despite its role in vascular remodeling, EGR1 is involved in many pro-inflammatory processes and can drive disease progression. In rodent models of cardiac reperfusion injury following myocardial infarct, treatments that reduce infarct size act by suppressing EGR1 expression [150, 151]. Thus, the ability to repress EGR1 inhibits multiple means of damage from cardiac infarct, including subsequent inflammation, apoptosis, and autophagy. Furthermore, in a mouse model of lung transplant, EGR1 deletion improved graft function by attenuating neutrophil recruitment [152]. Of note, downregulating EGR1 in cardiomyocytes also appears to be beneficial. Rat cardiomyocytes treated in vivo and in vitro with Ca2+ channel blockers had significantly reduced damage. Interestingly, WT1 is also expressed in response to hypoxia and ischemia, however this aspect of vascular injury is only beginning to be elucidated in vivo [153, 154]. This suggests WT1 and EGR1 work in separate pathways to induce hypoxia and ischemia; future studies focused on delineating their roles are needed to better understand how their actions are coordinated, particularly considering the mutually exclusive nature of EGR/WT1-mediated gene transcription.
Section 8: EGR1 and WT1 in cardiomyopathy
In cardiomyocytes, Ca2+ is utilized in both excitation-contraction coupling and non-excitable signaling. However, it is aberrant non-excitable Ca2+ signaling that has been identified in the etiology of cardiomyopathies in response to pathological stimuli, such as aberrant adrenergic signaling or volume overload [155-157].
As discussed in section 4, SERCA2 expression is negatively regulated by EGR1. Reduced replenishment of the sarcoplasmic reticulum (SR) Ca2+ store by SERCA had long been recognized as contributing to abnormal Ca2+ cycling in cardiomyocytes [158-160]. The predominant splice variant in cardiomyocytes, SERCA2a, is downregulated during heart failure, and loss of SERCA2a activity in animal models leads to heart failure [161, 162]. Because of this, the regulation of SERCA2a activity has been developed as a target in the treatment of heart failure. Recent clinical trials, notably the Ca2+ Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) phase I and II trials, demonstrated mixed success with a high dose of a SERCA2a-expressing adeno-associated viral vector. While the smaller scale CUPID phase I trial saw fewer cardiovascular events, the CUPID phase II trial did not see significant differences in time-to-recurrent heart failure-related hospitalization, or time to terminal event [163]. Future approaches targeting SERCA2a activity have also looked at regulating SERCA2a inhibitor phospholamban, or increasing SERCA2a stability through increasing sumoylation [164, 165].
In recent years, our understanding of EGR1 as a prominent marker of cardiac hypertrophy has expanded considerably [166]. In addition to regulating SERCA2a expression, EGR1 upregulation induces transcription of the miR-99 family and downregulates Akt signaling in an in vivo model of pathologic hypertrophy [167]. The consequences of both EGR1 upregulation and Akt downregulation are consistent with the reemergence of fetal heart gene expression programs, and the loss of normal adult heart signaling during pathologic cardiac hypertrophy [168]. Further in a mouse model of heart failure, NAB1, which negatively regulates EGR1 function, was upregulated, facilitating compensatory protection against cardiac hypertrophy [169]. Intriguingly, WT1, which has principally been described in the context of cardiac development, is also upregulated following myocardial infarction [129, 154]. Cardiac-resident colony-forming cells thought to aid in tissue repair following infarction originate from WT1-positive cells [170, 171]. Thus, the role of WT1 in differentiation appears to persist to a limited degree in the adult heart.
Cumulatively, these studies establish EGR1 as a driver of apoptosis and an inflammatory response after injury to endothelial cells, or during the chronic development of a cardiomyopathy. As a corollary, the repression of EGR1 signaling, by NAB1, WT1, or others, have shown protective effects, and are appealing targets to prevent damage and perhaps encourage healthy remodeling and restore cardiac function.
Conclusions
In non-excitable cells, the generation of Ca2+ signals begins with the production of InsP3 and depletion of the ER store and is maintained by mechanisms controlling the movements of Ca2+ across the plasma membrane. The ability of Ca2+ signals to modulate the expression of transcription factors such as EGRs, NFAT and NF-κB is now well understood. However, it is increasingly clear that the expression of these Ca2+ homeostasis proteins themselves is regulated in response to receptor activation by a host of transcription factors including EGRs, NFAT and NF-κB. How these differences shape long-term Ca2+ responses over time are less clear. While complex, future investigations addressing these questions may lead to new insights into fundamental mechanisms driving T cell activation, cancer progression and cardiovascular disease.
Highlights.
EGR family members and WT1 translocate to shared DNA consensus sites, where differences in binding to epigenetic marks and binding partners regulate transcriptional activity.
During T cell activation, gene expression changes are enabled by EGR1/4- and WT1-mediated regulation of the expression and function of proteins involved in Ca2+ homeostasis such as STIM1, STIM2, PMCA4 and SERCA.
The roles of EGR1/4 and WT1 in inflammation contributes to cardiovascular pathologies
EGR1/4-mediated control of Ca2+ homeostasis can contribute to tumor formation and progression through crosstalk with multiple pathways.
References
- [1].Berridge MJ, Lipp P, Bootman MD, The versatility and universality of calcium signalling, Nat Rev Mol Cell Biol, 1 (2000) 11–21. [DOI] [PubMed] [Google Scholar]
- [2].Hogan PG, The STIM1-ORAI1 microdomain, Cell Calcium, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Soboloff J, Rothberg BS, Madesh M, Gill DL, STIM proteins: dynamic calcium signal transducers, Nat Rev Mol Cell Biol, 13 (2012) 549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Prakriya M, Lewis RS, Store-Operated Calcium Channels, Physiol Rev, 95 (2015) 1383–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brandman O, Liou J, Park WS, Meyer T, STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels, Cell, 131 (2007) 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stathopulos PB, Zheng L, Ikura M, Stromal Interaction Molecule (STIM) 1 and STIM2 Calcium Sensing Regions Exhibit Distinct Unfolding and Oligomerization Kinetics, J Biol Chem, 284 (2009) 728–732. [DOI] [PubMed] [Google Scholar]
- [7].Zhou Y, Mancarella S, Wang Y, Yue C, Ritchie M, Gill DL, Soboloff J, The short N-terminal domains of STIM1 and STIM2 control the activation kinetics of Orai1 channels, J Biol Chem, 284 (2009) 19164–19168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M, Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry, Cell, 135 (2008) 110–122. [DOI] [PubMed] [Google Scholar]
- [9].Wang X, Wang Y, Zhou Y, Hendron E, Mancarella S, Andrake MD, Rothberg BS, Soboloff J, Gill DL, Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site, Nature communications, 5 (2014) 3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mercer JC, Dehaven WI, Smyth JT, Wedel B, Boyles RR, Bird GS, Putney JW Jr., Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1, J Biol Chem, 281 (2006) 24979–24990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].DeHaven WI, Smyth JT, Boyles RR, Putney JW Jr., Calcium inhibition and calcium potentiation of Orai1, Orai2, and Orai3 calcium release-activated calcium channels, J Biol Chem, 282 (2007) 17548–17556. [DOI] [PubMed] [Google Scholar]
- [12].Lis A, Peinelt C, Beck A, Parvez S, Monteilh-Zoller M, Fleig A, Penner R, CRACM1, CRACM2, and CRACM3 are store-operated Ca2+ channels with distinct functional properties, Curr Biol, 17 (2007) 794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Skerka C, Decker EL, Zipfel PF, Coordinate expression and distinct DNA-binding characteristics of the four EGR-zinc finger proteins in Jurkat T lymphocytes, Immunobiology, 198 (1997) 179–191. [DOI] [PubMed] [Google Scholar]
- [14].Christy B, Nathans D, DNA binding site of the growth factor-inducible protein Zif268, Proc Natl Acad Sci U S A, 86 (1989) 8737–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nardelli J, Gibson TJ, Vesque C, Charnay P, Base sequence discrimination by zinc-finger DNA-binding domains, Nature, 349 (1991) 175–178. [DOI] [PubMed] [Google Scholar]
- [16].Patwardhan S, Gashler A, Siegel MG, Chang LC, Joseph LJ, Shows TB, Le Beau MM, Sukhatme VP, EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors, Oncogene, 6 (1991) 917–928. [PubMed] [Google Scholar]
- [17].Cao X, Mahendran R, Guy GR, Tan YH, Detection and characterization of cellular EGR-1 binding to its recognition site, J Biol Chem, 268 (1993) 16949–16957. [PubMed] [Google Scholar]
- [18].Samakai E, Hooper R, Martin KA, Shmurak M, Zhang Y, Kappes DJ, Tempera I, Soboloff J, Novel STIM1-dependent control of Ca2+ clearance regulates NFAT activity during T-cell activation, FASEB J, 30 (2016) 3878–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tourtellotte WG, Nagarajan R, Bartke A, Milbrandt J, Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis, Mol Cell Biol, 20 (2000) 5261–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bandiera R, Sacco S, Vidal VP, Chaboissier MC, Schedl A, Steroidogenic organ development and homeostasis: A WT1-centric view, Molecular and cellular endocrinology, 408 (2015) 145–155. [DOI] [PubMed] [Google Scholar]
- [21].Wilm B, Munoz-Chapuli R, The Role of WT1 in Embryonic Development and Normal Organ Homeostasis, Methods Mol Biol, 1467 (2016) 23–39. [DOI] [PubMed] [Google Scholar]
- [22].Montano G, Ullmark T, Jernmark-Nilsson H, Sodaro G, Drott K, Costanzo P, Vidovic K, Gullberg U, The hematopoietic tumor suppressor interferon regulatory factor 8 (IRF8) is upregulated by the antimetabolite cytarabine in leukemic cells involving the zinc finger protein ZNF224, acting as a cofactor of the Wilms' tumor gene 1 (WT1) protein, Leuk Res, 40 (2016) 60–67. [DOI] [PubMed] [Google Scholar]
- [23].Schnerwitzki D, Perner B, Hoppe B, Pietsch S, Mehringer R, Hanel F, Englert C, Alternative splicing of Wilms tumor suppressor 1 (Wt1) exon 4 results in protein isoforms with different functions, Developmental biology, 393 (2014) 24–32. [DOI] [PubMed] [Google Scholar]
- [24].Madden SL, Cook DM, Morris JF, Gashler A, Sukhatme VP, Rauscher FJ 3rd, Transcriptional repression mediated by the WT1 Wilms tumor gene product, Science, 253 (1991) 1550–1553. [DOI] [PubMed] [Google Scholar]
- [25].Ogishima T, Shiina H, Breault JE, Terashima M, Honda S, Enokida H, Urakami S, Tokizane T, Kawakami T, Ribeiro-Filho LA, Fujime M, Kane CJ, Carroll PR, Igawa M, Dahiya R, Promoter CpG hypomethylation and transcription factor EGR1 hyperactivate heparanase expression in bladder cancer, Oncogene, 24 (2005) 6765–6772. [DOI] [PubMed] [Google Scholar]
- [26].Chen M, Xiong F, Zhang L, Promoter methylation of Egr-1 site contributes to fetal hypoxia-mediated PKCepsilon gene repression in the developing heart, American journal of physiology, 304 (2013) R683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zandarashvili L, White MA, Esadze A, Iwahara J, Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1, FEBS Lett, 589 (2015) 1748–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X, Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence, Genes & development, 28 (2014) 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Neri F, Incarnato D, Krepelova A, Rapelli S, Anselmi F, Parlato C, Medana C, Dal Bello F, Oliviero S, Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics, Cell reports, (2015). [DOI] [PubMed] [Google Scholar]
- [30].Eleftheriou M, Pascual AJ, Wheldon LM, Perry C, Abakir A, Arora A, Johnson AD, Auer DT, Ellis IO, Madhusudan S, Ruzov A, 5-Carboxylcytosine levels are elevated in human breast cancers and gliomas, Clinical epigenetics, 7 (2015) 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Philipsen S, Suske G, A tale of three fingers: the family of mammalian Sp/XKLF transcription factors, Nucleic Acids Res, 27 (1999) 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].O'Connor L, Gilmour J, Bonifer C, The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease, The Yale journal of biology and medicine, 89 (2016) 513–525. [PMC free article] [PubMed] [Google Scholar]
- [33].Alfonso-Jaume MA, Mahimkar R, Lovett DH, Co-operative interactions between NFAT (nuclear factor of activated T cells) c1 and the zinc finger transcription factors Sp1/Sp3 and Egr-1 regulate MT1-MMP (membrane type 1 matrix metalloproteinase) transcription by glomerular mesangial cells, Biochem J, 380 (2004) 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J, NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex, J Biol Chem, 281 (2006) 15129–15137. [DOI] [PubMed] [Google Scholar]
- [35].Buitrago M, Lorenz K, Maass AH, Oberdorf-Maass S, Keller U, Schmitteckert EM, Ivashchenko Y, Lohse MJ, Engelhardt S, The transcriptional repressor Nab1 is a specific regulator of pathological cardiac hypertrophy, Nat Med, 11 (2005) 837–844. [DOI] [PubMed] [Google Scholar]
- [36].Ehrengruber MU, Muhlebach SG, Sohrman S, Leutenegger CM, Lester HA, Davidson N, Modulation of early growth response (EGR) transcription factor-dependent gene expression by using recombinant adenovirus, Gene, 258 (2000) 63–69. [DOI] [PubMed] [Google Scholar]
- [37].Kumbrink J, Gerlinger M, Johnson JP, Egr-1 induces the expression of its corepressor nab2 by activation of the nab2 promoter thereby establishing a negative feedback loop, J Biol Chem, 280 (2005) 42785–42793. [DOI] [PubMed] [Google Scholar]
- [38].Thiel G, Muller I, Rossler OG, Expression, signaling and function of Egr transcription factors in pancreatic beta-cells and insulin-responsive tissues, Molecular and cellular endocrinology, 388 (2014) 10–19. [DOI] [PubMed] [Google Scholar]
- [39].Collins S, Lutz MA, Zarek PE, Anders RA, Kersh GJ, Powell JD, Opposing regulation of T cell function by Egr-1/NAB2 and Egr-2/Egr-3, European journal of immunology, 38 (2008) 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu M, Wang X, Peng Y, Shen S, Li G, Egr-1 regulates the transcription of NGX6 gene through a Sp1/Egr-1 overlapping site in the promoter, BMC molecular biology, 15 (2014) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Snyder R, Thekkumkara T, Interplay between EGR1 and SP1 is critical for 13-cis retinoic acid-mediated transcriptional repression of angiotensin type 1A receptor, Journal of molecular endocrinology, 50 (2013) 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cui MZ, Penn MS, Chisolm GM, Native and oxidized low density lipoprotein induction of tissue factor gene expression in smooth muscle cells is mediated by both Egr-1 and Sp1, J Biol Chem, 274 (1999) 32795–32802. [DOI] [PubMed] [Google Scholar]
- [43].McMahon SB, Monroe JG, The role of early growth response gene 1 (egr-1) in regulation of the immune response, Journal of leukocyte biology, 60 (1996) 159–166. [DOI] [PubMed] [Google Scholar]
- [44].Lohoff M, Giaisi M, Kohler R, Casper B, Krammer PH, Li-Weber M, Early growth response protein-1 (Egr-1) is preferentially expressed in T helper type 2 (Th2) cells and is involved in acute transcription of the Th2 cytokine interleukin-4, J Biol Chem, 285 (2010) 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pagel JI, Deindl E, Disease progression mediated by egr-1 associated signaling in response to oxidative stress, International journal of molecular sciences, 13 (2012) 13104–13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yu J, Zhang SS, Saito K, Williams S, Arimura Y, Ma Y, Ke Y, Baron V, Mercola D, Feng GS, Adamson E, Mustelin T, PTEN regulation by Akt-EGR1-ARF-PTEN axis, Embo J, 28 (2009) 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bae MH, Jeong CH, Kim SH, Bae MK, Jeong JW, Ahn MY, Bae SK, Kim ND, Kim CW, Kim KR, Kim KW, Regulation of Egr-1 by association with the proteasome component C8, Biochim Biophys Acta, 1592 (2002) 163–167. [DOI] [PubMed] [Google Scholar]
- [48].Huang RP, Fan Y, deBelle I, Ni Z, Matheny W, Adamson ED, Egr-1 inhibits apoptosis during the UV response: correlation of cell survival with Egr-1 phosphorylation, Cell death and differentiation, 5 (1998) 96–106. [DOI] [PubMed] [Google Scholar]
- [49].Silverman ES, Collins T, Pathways of Egr-1-mediated gene transcription in vascular biology, The American journal of pathology, 154 (1999) 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lee HJ, Mignacca RC, Sakamoto KM, Transcriptional activation of egr-1 by granulocyte-macrophage colony-stimulating factor but not interleukin 3 requires phosphorylation of cAMP response element-binding protein (CREB) on serine 133, J Biol Chem, 270 (1995) 15979–15983. [DOI] [PubMed] [Google Scholar]
- [51].Veyrac A, Besnard A, Caboche J, Davis S, Laroche S, The transcription factor Zif268/Egr1, brain plasticity, and memory, Progress in molecular biology and translational science, 122 (2014) 89–129. [DOI] [PubMed] [Google Scholar]
- [52].Cavigelli M, Dolfi F, Claret FX, Karin M, Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation, EMBO J, 14 (1995) 5957–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gregg J, Fraizer G, Transcriptional Regulation of EGR1 by EGF and the ERK Signaling Pathway in Prostate Cancer Cells, Genes Cancer, 2 (2011) 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cabodi S, Morello V, Masi A, Cicchi R, Broggio C, Distefano P, Brunelli E, Silengo L, Pavone F, Arcangeli A, Turco E, Tarone G, Moro L, Defilippi P, Convergence of integrins and EGF receptor signaling via PI3K/Akt/FoxO pathway in early gene Egr-1 expression, Journal of cellular physiology, 218 (2009) 294–303. [DOI] [PubMed] [Google Scholar]
- [55].Ivanova D, Dirks A, Montenegro-Venegas C, Schone C, Altrock WD, Marini C, Frischknecht R, Schanze D, Zenker M, Gundelfinger ED, Fejtova A, Synaptic activity controls localization and function of CtBP1 via binding to Bassoon and Piccolo, EMbO J, 34 (2015) 1056–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Thyss R, Virolle V, Imbert V, Peyron JF, Aberdam D, Virolle T, NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death, Embo J, 24 (2005) 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE, CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity, The Journal of experimental medicine, 188 (1998) 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mayer SI, Willars GB, Nishida E, Thiel G, Elk-1, CREB, and MKP-1 regulate Egr-1 expression in gonadotropin-releasing hormone stimulated gonadotrophs, Journal of cellular biochemistry, 105 (2008) 1267–1278. [DOI] [PubMed] [Google Scholar]
- [59].Rossler OG, Thiel G, Thrombin induces Egr-1 expression in fibroblasts involving elevation of the intracellular Ca2+ concentration, phosphorylation of ERK and activation of ternary complex factor, BMC molecular biology, 10 (2009) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sugimoto T, Stewart S, Guan KL, The calcium/calmodulin-dependent protein phosphatase calcineurin is the major Elk-1 phosphatase, J Biol Chem, 272 (1997) 29415–29418. [DOI] [PubMed] [Google Scholar]
- [61].Simo-Cheyou ER, Tan JJ, Grygorczyk R, Srivastava AK, STIM-1 and ORAI-1 channel mediate angiotensin-II-induced expression of Egr-1 in vascular smooth muscle cells, Journal of cellular physiology, 232 (2017) 3496–3509. [DOI] [PubMed] [Google Scholar]
- [62].Jain N, Mahendran R, Philp R, Guy GR, Tan YH, Cao X, Casein kinase II associates with Egr-1 and acts as a negative modulator of its DNA binding and transcription activities in NIH 3T3 cells, J Biol Chem, 271 (1996) 13530–13536. [DOI] [PubMed] [Google Scholar]
- [63].Manente AG, Pinton G, Tavian D, Lopez-Rodas G, Brunelli E, Moro L, Coordinated sumoylation and ubiquitination modulate EGF induced EGR1 expression and stability, PloS one, 6 (2011) e25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Rexach JE, Clark PM, Hsieh-Wilson LC, Chemical approaches to understanding O-GlcNAc glycosylation in the brain, Nat Chem Biol, 4 (2008) 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang HY, Hsieh PF, Huang DF, Chin PS, Chou CH, Tung CC, Chen SY, Lee LJ, Gau SS, Huang HS, RBFOX3/NeuN is Required for Hippocampal Circuit Balance and Function, Scientific reports, 5 (2015) 17383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ludwig A, Uvarov P, Pellegrino C, Thomas-Crusells J, Schuchmann S, Saarma M, Airaksinen MS, Rivera C, Neurturin evokes MAPK-dependent upregulation of Egr4 and KCC2 in developing neurons, Neural plasticity, 2011 (2011) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hogarth CA, Mitchell D, Small C, Griswold M, EGR4 displays both a cell- and intracellular-specific localization pattern in the developing murine testis, Dev Dyn, 239 (2010) 3106–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cooper SJ, Trinklein ND, Nguyen L, Myers RM, Serum response factor binding sites differ in three human cell types, Genome research, 17 (2007) 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Holst C, Skerka C, Lichter P, Bialonski A, Zipfel PF, Genomic organization, chromosomal localization and promoter function of the human zinc-finger gene pAT133, Human molecular genetics, 2 (1993) 367–372. [DOI] [PubMed] [Google Scholar]
- [70].Zipfel PF, Decker EL, Holst C, Skerka C, The human zinc finger protein EGR-4 acts as autoregulatory transcriptional repressor, Biochim Biophys Acta, 1354 (1997) 134–144. [DOI] [PubMed] [Google Scholar]
- [71].Ritchie MF, Yue C, Zhou Y, Houghton PJ, Soboloff J, Wilms tumor suppressor 1 (WT1) and early growth response 1 (EGR1) are regulators of STIM1 expression, J Biol Chem, 285 (2010) 10591–10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ritchie MF, Samakai E, Soboloff J, STIM1 is required for attenuation of PMCA-mediated Ca2+ clearance during T-cell activation, EMBO J, 31 (2012) 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Peinelt C, Vig M, Koomoa DL, Beck A, Nadler MJ, Koblan-Huberson M, Lis A, Fleig A, Penner R, Kinet JP, Amplification of CRAC current by STIM1 and CRACM1 (Orai1), Nat Cell Biol, 8 (2006) 771–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Soboloff J, Spassova MA, Tang XD, Hewavitharana T, Xu W, Gill DL, Orai1 and STIM reconstitute store-operated calcium channel function, J Biol Chem, 281 (2006) 20661–20665. [DOI] [PubMed] [Google Scholar]
- [75].Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD, Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity, Proc Natl Acad Sci U S A, 103 (2006) 9357–9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Barr VA, Bernot KM, Srikanth S, Gwack Y, Balagopalan L, Regan CK, Helman DJ, Sommers CL, Oh-Hora M, Rao A, Samelson LE, Dynamic movement of the calcium sensor STIM1 and the calcium channel Orai1 in activated T-cells: puncta and distal caps, Molecular biology of the cell, 19 (2008) 2802–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lioudyno MI, Kozak JA, Penna A, Safrina O, Zhang SL, Sen D, Roos J, Stauderman KA, Cahalan MD, Orai1 and STIM1 move to the immunological synapse and are up-regulated during T cell activation, Proc Natl Acad Sci U S A, 105 (2008) 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Quintana A, Pasche M, Junker C, Al-Ansary D, Rieger H, Kummerow C, Nunez L, Villalobos C, Meraner P, Becherer U, Rettig J, Niemeyer BA, Hoth M, Calcium microdomains at the immunological synapse: how ORAI channels, mitochondria and calcium pumps generate local calcium signals for efficient T-cell activation, EMBO J, 30 (2011) 3895–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hogan PG, Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion, Cell Calcium, 63 (2017) 66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kreslavsky T, Gleimer M, Garbe AI, von Boehmer H, alphabeta versus gammadelta fate choice: counting the T-cell lineages at the branch point, Immunological reviews, 238 (2010) 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Periasamy M, Kalyanasundaram A, SERCA pump isoforms: their role in calcium transport and disease, Muscle & nerve, 35 (2007) 430–442. [DOI] [PubMed] [Google Scholar]
- [82].Arai M, Yoguchi A, Takizawa T, Yokoyama T, Kanda T, Kurabayashi M, Nagai R, Mechanism of doxorubicin-induced inhibition of sarcoplasmic reticulum Ca(2+)-ATPase gene transcription, Circ Res, 86 (2000) 8–14. [DOI] [PubMed] [Google Scholar]
- [83].Brady M, Koban MU, Dellow KA, Yacoub M, Boheler KR, Fuller SJ, Sp1 and Sp3 transcription factors are required for trans-activation of the human SERCA2 promoter in cardiomyocytes, Cardiovascular research, 60 (2003) 347–354. [DOI] [PubMed] [Google Scholar]
- [84].Scharf M, Neef S, Freund R, Geers-Knorr C, Franz-Wachtel M, Brandis A, Krone D, Schneider H, Groos S, Menon MB, Chang KC, Kraft T, Meissner JD, Boheler KR, Maier LS, Gaestel M, Scheibe RJ, Mitogen-activated protein kinase-activated protein kinases 2 and 3 regulate SERCA2a expression and fiber type composition to modulate skeletal muscle and cardiomyocyte function, Mol Cell Biol, 33 (2013) 2586–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Anwar A, Taimor G, Korkususz H, Schreckenberg R, Berndt T, Abdallah Y, Piper HM, Schluter KD, PKC-independent signal transduction pathways increase SERCA2 expression in adult rat cardiomyocytes, Journal of molecular and cellular cardiology, 39 (2005) 911–919. [DOI] [PubMed] [Google Scholar]
- [86].Prasad AM, Inesi G, Regulation and rate limiting mechanisms of Ca2+ ATPase (SERCA2) expression in cardiac myocytes, Molecular and cellular biochemistry, 361 (2012) 85–96. [DOI] [PubMed] [Google Scholar]
- [87].Lewis RS, Calcium oscillations in T-cells: mechanisms and consequences for gene expression, Biochem Soc Trans, 31 (2003) 925–929. [DOI] [PubMed] [Google Scholar]
- [88].Wang C, Dostanic S, Servant N, Chalifour LE, Egr-1 negatively regulates expression of the sodium-calcium exchanger-1 in cardiomyocytes in vitro and in vivo, Cardiovascular research, 65 (2005) 187–194. [DOI] [PubMed] [Google Scholar]
- [89].Wieland GD, Nehmann N, Muller D, Eibel H, Siebenlist U, Suhnel J, Zipfel PF, Skerka C, Early growth response proteins EGR-4 and EGR-3 interact with immune inflammatory mediators NF-kappaB p50 and p65, J Cell Sci, 118 (2005) 3203–3212. [DOI] [PubMed] [Google Scholar]
- [90].Liu X, Berry CT, Ruthel G, Madara JJ, MacGillivray K, Gray CM, Madge LA, McCorkell KA, Beiting DP, Hershberg U, May MJ, Freedman BD, T Cell Receptor-induced Nuclear Factor kappaB (NF-kappaB) Signaling and Transcriptional Activation Are Regulated by STIM1- and Orai1-mediated Calcium Entry, J Biol Chem, 291 (2016) 8440–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].DebRoy A, Vogel SM, Soni D, Sundivakkam PC, Malik AB, Tiruppathi C, Cooperative signaling via transcription factors NF-kappaB and AP1/c-Fos mediates endothelial cell STIM1 expression and hyperpermeability in response to endotoxin, J Biol Chem, 289 (2014) 24188–24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Eylenstein A, Schmidt S, Gu S, Yang W, Schmid E, Schmidt EM, Alesutan I, Szteyn K, Regel I, Shumilina E, Lang F, Transcription Factor NF-kappaB Regulates Expression of Poreforming Ca2+ Channel Unit, Orai1, and Its Activator, STIM1, to Control Ca2+ Entry and Affect Cellular Functions, J Biol Chem, 287 (2012) 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Perego M, Maurer M, Wang JX, Shaffer S, Muller AC, Parapatics K, Li L, Hristova D, Shin S, Keeney F, Liu S, Xu X, Raj A, Jensen JK, Bennett KL, Wagner SN, Somasundaram R, Herlyn M, A slow-cycling subpopulation of melanoma cells with highly invasive properties, Oncogene, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, Symonds AL, Wang P, The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells, Immunity, 37 (2012) 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Singh R, Miao T, Symonds ALJ, Omodho B, Li S, Wang P, Egr2 and 3 Inhibit T-bet-Mediated IFN-gamma Production in T Cells, J Immunol, 198 (2017) 4394–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Miao T, Symonds ALJ, Singh R, Symonds JD, Ogbe A, Omodho B, Zhu B, Li S, Wang P, Egr2 and 3 control adaptive immune responses by temporally uncoupling expansion from T cell differentiation, The Journal of experimental medicine, 214 (2017) 1787–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Omodho B, Miao T, Symonds ALJ, Singh R, Li S, Wang P, Transcription factors early growth response gene (Egr) 2 and 3 control inflammatory responses of tolerant T cells, Immunity, inflammation and disease, 6 (2018) 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Morita K, Okamura T, Inoue M, Komai T, Teruya S, Iwasaki Y, Sumitomo S, Shoda H, Yamamoto K, Fujio K, Egr2 and Egr3 in regulatory T cells cooperatively control systemic autoimmunity through Ltbp3-mediated TGF-beta3 production, Proc Natl Acad Sci U S A, 113 (2016) E8131–E8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Okamura T, Sumitomo S, Morita K, Iwasaki Y, Inoue M, Nakachi S, Komai T, Shoda H, Miyazaki J, Fujio K, Yamamoto K, TGF-beta3-expressing CD4+CD25(−)LAG3+ regulatory T cells control humoral immune responses, Nature communications, 6 (2015) 6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Myouzen K, Kochi Y, Shimane K, Fujio K, Okamura T, Okada Y, Suzuki A, Atsumi T, Ito S, Takada K, Mimori A, Ikegawa S, Yamada R, Nakamura Y, Yamamoto K, Regulatory polymorphisms in EGR2 are associated with susceptibility to systemic lupus erythematosus, Human molecular genetics, 19 (2010) 2313–2320. [DOI] [PubMed] [Google Scholar]
- [101].Nafez S, Oikawa K, Odero GL, Sproule M, Ge N, Schapansky J, Abrenica B, Hatherell A, Cadonic C, Zhang S, Song X, Kauppinen T, Glazner GW, Grilli M, Czubryt MP, Eisenstat DD, Albensi BC, Early growth response 2 (Egr-2) expression is triggered by NF-kappaB activation, Mol Cell Neurosci, 64 (2015) 95–103. [DOI] [PubMed] [Google Scholar]
- [102].Hooper R, Zhang X, Webster M, Go C, Kedra J, Marchbank K, Gill DL, Weeraratna AT, Trebak M, Soboloff J, Novel Protein Kinase C-Mediated Control of Orai1 Function in Invasive Melanoma, Mol Cell Biol, 35 (2015) 2790–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Xie J, Pan H, Yao J, Zhou Y, Han W, SOCE and cancer: Recent progress and new perspectives, International journal of cancer, 138 (2016) 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Flourakis M, Lehen'kyi V, Beck B, Raphael M, Vandenberghe M, Abeele FV, Roudbaraki M, Lepage G, Mauroy B, Romanin C, Shuba Y, Skryma R, Prevarskaya N, Orai1 contributes to the establishment of an apoptosis-resistant phenotype in prostate cancer cells, Cell Death Dis, 1 (2010) e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen YF, Chiu WT, Chen YT, Lin PY, Huang HJ, Chou CY, Chang HC, Tang MJ, Shen MR, Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis, Proc Natl Acad Sci U S A, 108 (2011) 15225–15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Hernandez GL, Volpert OV, Iniguez MA, Lorenzo E, Martinez-Martinez S, Grau R, Fresno M, Redondo JM, Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: roles of the nuclear factor of activated T cells and cyclooxygenase 2, The Journal of experimental medicine, 193 (2001) 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Armesilla AL, Lorenzo E, Gomez del Arco P, Martinez-Martinez S, Alfranca A, Redondo JM, Vascular endothelial growth factor activates nuclear factor of activated T cells in human endothelial cells: a role for tissue factor gene expression, Mol Cell Biol, 19 (1999) 2032–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Magee N, Zhang Y, Role of early growth response 1 in liver metabolism and liver cancer, Hepatoma research, 3 (2017) 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chapman NR, Perkins ND, Inhibition of the RelA(p65) NF-kappaB subunit by Egr-1, J Biol Chem, 275 (2000) 4719–4725. [DOI] [PubMed] [Google Scholar]
- [110].Rivera MN, Haber DA, Wilms' tumour: connecting tumorigenesis and organ development in the kidney, Nature reviews, 5 (2005) 699–712. [DOI] [PubMed] [Google Scholar]
- [111].Yang L, Han Y, Suarez Saiz F, Minden MD, A tumor suppressor and oncogene: the WT1 story, Leukemia, 21 (2007) 868–876. [DOI] [PubMed] [Google Scholar]
- [112].Takahashi M, Yang XJ, Lavery TT, Furge KA, Williams BO, Tretiakova M, Montag A, Vogelzang NJ, Re GG, Garvin AJ, Soderhall S, Kagawa S, Hazel-Martin D, Nordenskjold A, Teh BT, Gene expression profiling of favorable histology Wilms tumors and its correlation with clinical features, Cancer Res, 62 (2002) 6598–6605. [PubMed] [Google Scholar]
- [113].Ritchie MF, Zhou Y, Soboloff J, WT1/EGR1-mediated control of STIM1 expression and function in cancer cells, Frontiers in bioscience, 16 (2011) 2402–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Menke AL, Shvarts A, Riteco N, van Ham RC, van der Eb AJ, Jochemsen AG, Wilms' tumor 1-KTS isoforms induce p53-independent apoptosis that can be partially rescued by expression of the epidermal growth factor receptor or the insulin receptor, Cancer Res, 57 (1997) 1353–1363. [PubMed] [Google Scholar]
- [115].Natrajan R, Little SE, Reis-Filho JS, Hing L, Messahel B, Grundy PE, Dome JS, Schneider T, Vujanic GM, Pritchard-Jones K, Jones C, Amplification and overexpression of CACNA1E correlates with relapse in favorable histology Wilms' tumors, Clin Cancer Res, 12 (2006) 7284–7293. [DOI] [PubMed] [Google Scholar]
- [116].Wittmann S, Wunder C, Zirn B, Furtwangler R, Wegert J, Graf N, Gessler M, New prognostic markers revealed by evaluation of genes correlated with clinical parameters in Wilms tumors, Genes, chromosomes & cancer, 47 (2008) 386–395. [DOI] [PubMed] [Google Scholar]
- [117].Akutagawa O, Nishi H, Kyo S, Terauchi F, Yamazawa K, Higuma C, Inoue M, Isaka K, Early growth response-1 mediates downregulation of telomerase in cervical cancer, Cancer science, 99 (2008) 1401–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Rui L, Energy metabolism in the liver, Comprehensive Physiology, 4 (2014) 177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Peng WX, Pan FY, Liu XJ, Ning S, Xu N, Meng FL, Wang YQ, Li CJ, Hypoxia stabilizes microtubule networks and decreases tumor cell chemosensitivity to anticancer drugs through Egr-1, Anatomical record, 293 (2010) 414–420. [DOI] [PubMed] [Google Scholar]
- [120].Peng WX, Wan YY, Gong AH, Ge L, Jin J, Xu M, Wu CY, Egr-1 regulates irradiation-induced autophagy through Atg4B to promote radioresistance in hepatocellular carcinoma cells, Oncogenesis, 6 (2017) e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Peng WX, Xiong EM, Ge L, Wan YY, Zhang CL, Du FY, Xu M, Bhat RA, Jin J, Gong AH, Egr-1 promotes hypoxia-induced autophagy to enhance chemo-resistance of hepatocellular carcinoma cells, Exp Cell Res, 340 (2016) 62–70. [DOI] [PubMed] [Google Scholar]
- [122].Perugorria MJ, Castillo J, Latasa MU, Goni S, Segura V, Sangro B, Prieto J, Avila MA, Berasain C, Wilms' tumor 1 gene expression in hepatocellular carcinoma promotes cell dedifferentiation and resistance to chemotherapy, Cancer Res, 69 (2009) 1358–1367. [DOI] [PubMed] [Google Scholar]
- [123].Ahmad SS, Glatzle J, Bajaeifer K, Buhler S, Lehmann T, Konigsrainer I, Vollmer JP, Sipos A, Ahmad SS, Northoff H, Konigsrainer A, Zieker D, Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer, International journal of oncology, 43 (2013) 586–590. [DOI] [PubMed] [Google Scholar]
- [124].Kim J, Kang HS, Lee YJ, Lee HJ, Yun J, Shin JH, Lee CW, Kwon BM, Hong SH, EGR1-dependent PTEN upregulation by 2-benzoyloxycinnamaldehyde attenuates cell invasion and EMT in colon cancer, Cancer letters, 349 (2014) 35–44. [DOI] [PubMed] [Google Scholar]
- [125].Sampson VB, David JM, Puig I, Patil PU, de Herreros AG, Thomas GV, Rajasekaran AK, Wilms' tumor protein induces an epithelial-mesenchymal hybrid differentiation state in clear cell renal cell carcinoma, PloS one, 9 (2014) e102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Hansson ML, Behmer S, Ceder R, Mohammadi S, Preta G, Grafstrom RC, Fadeel B, Wallberg AE, MAML1 acts cooperatively with EGR1 to activate EGR1-regulated promoters: implications for nephrogenesis and the development of renal cancer, PloS one, 7 (2012) e46001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lkhagvadorj S, Kim JH, Oh SS, Lee MR, Jung JH, Chung HC, Cha SK, Eom M, Orai1 Expression Is Closely Related with Favorable Prognostic Factors in Clear Cell Renal Cell Carcinoma, Journal of Korean medical science, 31 (2016) 879–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ronski K, Sanders M, Burleson JA, Moyo V, Benn P, Fang M, Early growth response gene 1 (EGR1) is deleted in estrogen receptor-negative human breast carcinoma, Cancer, 104 (2005) 925–930. [DOI] [PubMed] [Google Scholar]
- [129].Kobayashi D, Yamada M, Kamagata C, Kaneko R, Tsuji N, Nakamura M, Yagihashi A, Watanabe N, Overexpression of early growth response-1 as a metastasis-regulatory factor in gastric cancer, Anticancer research, 22 (2002) 3963–3970. [PubMed] [Google Scholar]
- [130].Xu Y, Zhang S, Niu H, Ye Y, Hu F, Chen S, Li X, Luo X, Jiang S, Liu Y, Chen Y, Li J, Xiang R, Li N, STIM1 accelerates cell senescence in a remodeled microenvironment but enhances the epithelial-to-mesenchymal transition in prostate cancer, Scientific reports, 5 (2015) 11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yang S, Zhang JJ, Huang XY, Orai1 and STIM1 are critical for breast tumor cell migration and metastasis, Cancer cell, 15 (2009) 124–134. [DOI] [PubMed] [Google Scholar]
- [132].Rios-Doria J, Kuefer R, Ethier SP, Day ML, Cleavage of beta-catenin by calpain in prostate and mammary tumor cells, Cancer Res, 64 (2004) 7237–7240. [DOI] [PubMed] [Google Scholar]
- [133].Brett A, Pandey S, Fraizer G, The Wilms' tumor gene (WT1) regulates E-cadherin expression and migration of prostate cancer cells, Molecular cancer, 12 (2013) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Fraizer GC, Eisermann K, Pandey S, Brett-Morris A, Bazarov A, Nock S, Ghimirey N, Kuerbitz SJ, Functional Role of WT1 in Prostate Cancer, in: van den den Heuvel-Eibrink MM (Ed.) Wilms Tumor, Brisbane (AU), 2016. [PubMed] [Google Scholar]
- [135].Santoiemma PP, Powell DJ Jr., Tumor infiltrating lymphocytes in ovarian cancer, Cancer biology & therapy, 16 (2015) 807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Miller-Hodges E, Hohenstein P, WT1 in disease: shifting the epithelial-mesenchymal balance, The Journal of pathology, 226 (2012) 229–240. [DOI] [PubMed] [Google Scholar]
- [137].Andersson C, Oji Y, Ohlson N, Wang S, Li X, Ottander U, Lundin E, Sugiyama H, Li A, Prognostic significance of specific anti-WT1 IgG antibody level in plasma in patients with ovarian carcinoma, Cancer medicine, 3 (2014) 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Davis FM, Azimi I, Faville RA, Peters AA, Jalink K, Putney JW Jr., Goodhill GJ, Thompson DW, Roberts-Thomson SJ, Monteith GR, Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent, Oncogene, 33 (2014) 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Rossi G, Minervini MM, Carella AM, Melillo L, Cascavilla N, Wilms' Tumor Gene (WT1) Expression and Minimal Residual Disease in Acute Myeloid Leukemia, in: van den Heuvel-Eibrink MM (Ed.) Wilms Tumor, Brisbane (AU), 2016. [PubMed] [Google Scholar]
- [140].Watts JM, Dumitriu B, Hilden P, Kishtagari A, Rapaport F, Chen C, Ahn J, Devlin SM, Stein EM, Rampal R, Levine RL, Young N, Tallman MS, Telomere length and associations with somatic mutations and clinical outcomes in acute myeloid leukemia, Leuk Res, 49 (2016) 62–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Marsch E, Sluimer JC, Daemen MJ, Hypoxia in atherosclerosis and inflammation, Curr Opin Lipidol, 24 (2013) 393–400. [DOI] [PubMed] [Google Scholar]
- [142].Gubits RM, Burke RE, Casey-McIntosh G, Bandele A, Munell F, Immediate early gene induction after neonatal hypoxia-ischemia, Brain Res Mol Brain Res, 18 (1993) 228–238. [DOI] [PubMed] [Google Scholar]
- [143].Bonn SE, Sjolander A, Lagerros YT, Wiklund F, Stattin P, Holmberg E, Gronberg H, Balter K, Physical activity and survival among men diagnosed with prostate cancer, Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 24 (2015) 57–64. [DOI] [PubMed] [Google Scholar]
- [144].Cheng JC, Chang HM, Leung PC, Epidermal growth factor induces human oviductal epithelial cell invasion by down-regulating E-cadherin expression, J Clin Endocrinol Metab, 97 (2012) E1380–1389. [DOI] [PubMed] [Google Scholar]
- [145].He J, Yu JJ, Xu Q, Wang L, Zheng JZ, Liu LZ, Jiang BH, Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy, Autophagy, 11 (2015) 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Siegel RL, Miller KD, Jemal A, Cancer Statistics, 2017, CA: a cancer journal for clinicians, 67 (2017) 7–30. [DOI] [PubMed] [Google Scholar]
- [147].Lee CG, Cho SJ, Kang MJ, Chapoval SP, Lee PJ, Noble PW, Yehualaeshet T, Lu B, Flavell RA, Milbrandt J, Homer RJ, Elias JA, Early growth response gene 1-mediated apoptosis is essential for transforming growth factor beta1-induced pulmonary fibrosis, The Journal of experimental medicine, 200 (2004) 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Faouzi M, Hague F, Potier M, Ahidouch A, Sevestre H, Ouadid-Ahidouch H, Down-regulation of Orai3 arrests cell-cycle progression and induces apoptosis in breast cancer cells but not in normal breast epithelial cells, Journal of cellular physiology, 226 (2011) 542–551. [DOI] [PubMed] [Google Scholar]
- [149].Suda M, Shimizu I, Yoshida Y, Hayashi Y, Ikegami R, Katsuumi G, Wakasugi T, Yoshida Y, Okuda S, Soga T, Minamino T, Inhibition of dipeptidyl peptidase-4 ameliorates cardiac ischemia and systolic dysfunction by up-regulating the FGF-2/EGR-1 pathway, PloS one, 12 (2017) e0182422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mudaliar H, Rayner B, Billah M, Kapoor N, Lay W, Dona A, Bhindi R, Remote ischemic preconditioning attenuates EGR-1 expression following myocardial ischemia reperfusion injury through activation of the JAK-STAT pathway, International journal of cardiology, 228 (2017) 729–741. [DOI] [PubMed] [Google Scholar]
- [151].Kim TD, Jung HR, Seo SH, Oh SC, Ban Y, Tan X, Min Kim J, Hyun Lee S, Koh DS, Jung H, Park YJ, Ran Yoon S, Doh J, Ha SJ, Choi I, Greenberg PD, MicroRNA-150 modulates intracellular Ca (2+) levels in naive CD8(+) T cells by targeting TMEM20, Scientific reports, 7 (2017) 2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Kawakami A, Fisher DE, The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology, Laboratory investigation, 97 (2017) 649–656. [DOI] [PubMed] [Google Scholar]
- [153].Stanisz H, Stark A, Kilch T, Schwarz EC, Muller CS, Peinelt C, Hoth M, Niemeyer BA, Vogt T, Bogeski I, ORAI1 Ca(2+) channels control endothelin-1-induced mitogenesis and melanogenesis in primary human melanocytes, The Journal of investigative dermatology, 132 (2012) 1443–1451. [DOI] [PubMed] [Google Scholar]
- [154].Rios-Doria J, Day KC, Kuefer R, Rashid MG, Chinnaiyan AM, Rubin MA, Day ML, The role of calpain in the proteolytic cleavage of E-cadherin in prostate and mammary epithelial cells, J Biol Chem, 278 (2003) 1372–1379. [DOI] [PubMed] [Google Scholar]
- [155].Dustin ML, The cellular context of T cell signaling, Immunity, 30 (2009) 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Crabtree GR, Calcium, calcineurin, and the control of transcription, J Biol Chem, 276 (2001) 2313–2316. [DOI] [PubMed] [Google Scholar]
- [157].Gwathmey JK, Copelas L, MacKinnon R, Schoen FJ, Feldman MD, Grossman W, Morgan JP, Abnormal intracellular calcium handling in myocardium from patients with end-stage heart failure, Circ Res, 61 (1987) 70–76. [DOI] [PubMed] [Google Scholar]
- [158].Davia K, Davies CH, Harding SE, Effects of inhibition of sarcoplasmic reticulum calcium uptake on contraction in myocytes isolated from failing human ventricle, Cardiovascular research, 33 (1997) 88–97. [DOI] [PubMed] [Google Scholar]
- [159].Qi M, Shannon TR, Euler DE, Bers DM, Samarel AM, Downregulation of sarcoplasmic reticulum Ca(2+)-ATPase during progression of left ventricular hypertrophy, Am J Physiol, 272 (1997) H2416–2424. [DOI] [PubMed] [Google Scholar]
- [160].Hasenfuss G, Reinecke H, Studer R, Meyer M, Pieske B, Holtz J, Holubarsch C, Posival H, Just H, Drexler H, Relation between myocardial function and expression of sarcoplasmic reticulum Ca(2+)-ATPase in failing and nonfailing human myocardium, Circ Res, 75 (1994) 434–442. [DOI] [PubMed] [Google Scholar]
- [161].Andersson KB, Birkeland JA, Finsen AV, Louch WE, Sjaastad I, Wang Y, Chen J, Molkentin JD, Chien KR, Sejersted OM, Christensen G, Moderate heart dysfunction in mice with inducible cardiomyocyte-specific excision of the Serca2 gene, Journal of molecular and cellular cardiology, 47 (2009) 180–187. [DOI] [PubMed] [Google Scholar]
- [162].Zarain-Herzberg A, Afzal N, Elimban V, Dhalla NS, Decreased expression of cardiac sarcoplasmic reticulum Ca(2+)-pump ATPase in congestive heart failure due to myocardial infarction, Molecular and cellular biochemistry, 163-164 (1996) 285–290. [DOI] [PubMed] [Google Scholar]
- [163].Hulot JS, Salem JE, Redheuil A, Collet JP, Varnous S, Jourdain P, Logeart D, Gandjbakhch E, Bernard C, Hatem SN, Isnard R, Cluzel P, Le Feuvre C, Leprince P, Hammoudi N, Lemoine FM, Klatzmann D, Vicaut E, Komajda M, Montalescot G, Lompre AM, Hajjar RJ, Investigators A-H, Effect of intracoronary administration of AAV1/SERCA2a on ventricular remodelling in patients with advanced systolic heart failure: results from the AGENT-HF randomized phase 2 trial, European journal of heart failure, 19 (2017) 1534–1541. [DOI] [PubMed] [Google Scholar]
- [164].Fechner H, Suckau L, Kurreck J, Sipo I, Wang X, Pinkert S, Loschen S, Rekittke J, Weger S, Dekkers D, Vetter R, Erdmann VA, Schultheiss HP, Paul M, Lamers J, Poller W, Highly efficient and specific modulation of cardiac calcium homeostasis by adenovector-derived short hairpin RNA targeting phospholamban, Gene Ther, 14 (2007) 211–218. [DOI] [PubMed] [Google Scholar]
- [165].Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, Park WJ, Hajjar RJ, SUMO1-dependent modulation of SERCA2a in heart failure, Nature, 477 (2011) 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Khachigian LM, Early growth response-1 in cardiovascular pathobiology, Circ Res, 98 (2006) 186–191. [DOI] [PubMed] [Google Scholar]
- [167].Ramasamy S, Velmurugan G, Rekha B, Anusha S, Shanmugha Rajan K, Shanmugarajan S, Ramprasath T, Gopal P, Tomar D, Karthik KV, Verma SK, Garikipati VNS, Sudarsan R, Egr-1 mediated cardiac miR-99 family expression diverges physiological hypertrophy from pathological hypertrophy, Exp Cell Res, 365 (2018) 46–56. [DOI] [PubMed] [Google Scholar]
- [168].Liggett SB, Kelly RJ, Parekh RR, Matkovich SJ, Benner BJ, Hahn HS, Syed FM, Galvez AS, Case KL, McGuire N, Odley AM, Sparks L, Kardia SL, Dorn GW 2nd, A functional polymorphism of the Galphaq (GNAQ) gene is associated with accelerated mortality in African-American heart failure, Human molecular genetics, 16 (2007) 2740–2750. [DOI] [PubMed] [Google Scholar]
- [169].Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D, Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells, Cell metabolism, 2 (2005) 21–33. [DOI] [PubMed] [Google Scholar]
- [170].Maus M, Cuk M, Patel B, Lian J, Ouimet M, Kaufmann U, Yang J, Horvath R, Hornig-Do HT, Chrzanowska-Lightowlers ZM, Moore KJ, Cuervo AM, Feske S, Store-Operated Ca(2+) Entry Controls Induction of Lipolysis and the Transcriptional Reprogramming to Lipid Metabolism, Cell metabolism, 25 (2017) 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP, Decoding of cytosolic calcium oscillations in the mitochondria, Cell, 82 (1995) 415–424. [DOI] [PubMed] [Google Scholar]