Abstract
The crystal structure of the heterotrimeric quinohemoprotein amine dehydrogenase from Paracoccus denitrificans has been determined at 2.05-Å resolution. Within an 82-residue subunit is contained an unusual redox cofactor, cysteine tryptophylquinone (CTQ), consisting of an orthoquinone-modified tryptophan side chain covalently linked to a nearby cysteine side chain. The subunit is surrounded on three sides by a 489-residue, four-domain subunit that includes a diheme cytochrome c. Both subunits sit on the surface of a third subunit, a 337-residue seven-bladed β-propeller that forms part of the enzyme active site. The small catalytic subunit is internally crosslinked by three highly unusual covalent cysteine to aspartic or glutamic acid thioether linkages in addition to the cofactor crossbridge. The catalytic function of the enzyme as well as the biosynthesis of the unusual catalytic subunit is discussed.
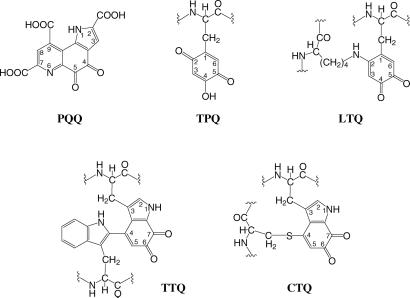
Enzymes that catalyze the oxidation of biological amines use a variety of redox cofactors to temporarily store the reducing equivalents derived from the substrate before passing them on to exogenous one- or two-electron acceptors such as a cytochrome, a cupredoxin, or molecular oxygen. The majority of such enzymes employ a flavin (FMN or FAD) as prosthetic group, but some, such as copper amine oxidase (CAO) and methylamine dehydrogenase (MADH) contain a quinone cofactor (1). In the case of CAO, the cofactor is topaquinone (TPQ) (2) or lysine tyrosylquinone (LTQ) (3), whereas for MADH, the cofactor is tryptophan tryptophylquinone (TTQ) (4) (Fig. 1). All three quinone cofactors are unusual because each of them is derived from one or two amino acid side chains that are encoded by the genome and are chemically modified posttranslationally (5). Another class of quinoenzyme, as represented by methanol dehydrogenase, is involved in the oxidation of alcohols and sugars, and contains pyrroloquinoline quinone (PQQ) as redox cofactor (6). In these enzymes, the PQQ is bound noncovalently and is incorporated from an intracellular or extracellular pool that is maintained by a separate biosynthetic pathway.
Figure 1.
Chemical structures of the quinone cofactors PQQ, TPQ, lysine tyrosylquinone (LTQ), and TTQ. The structure of the newly identified cofactor, cysteine tryptophylquinone (CTQ), is also shown.
The CAO enzymes are found in both eukaryotes and prokaryotes and use molecular oxygen as the exogenous electron acceptor (7). The remaining quinoenzymes, containing TTQ or PQQ, are bacterial in origin and are periplasmic. Neither of them directly reduces molecular oxygen, but rather they feed the substrate-derived electrons one at a time into the terminal oxidase electron transport system of the periplasm, either directly or through the mediation of cupredoxin or cytochrome electron-carrier proteins (1, 8). Some PQQ enzymes are quinohemoproteins, because they contain both PQQ and one or more heme moieties located in the same or different subunits, and undergo intramolecular electron transfer before being reoxidized by an exogenous electron acceptor (9).
The Gram-negative chemoorganotrophic bacterium Paracoccus denitrificans contains two amine dehydrogenases in its periplasm, one being the well-known TTQ-containing MADH and the other a quinohemoprotein amine dehydrogenase (QHNDH) that is preferentially induced by growth on n-butylamine (10). QHNDH was reported to be a heterodimer of 96-kDa molecular mass. The large subunit of 60 kDa appeared to contain one heme c per molecule and to stain positively for quinone-dependent redox cycling. The smaller subunit of 36 kDa showed no redox activity. Partial reduction of the enzyme by dithionite gave a semiquinone radical EPR signal similar to that of TTQ in MADH. Cytochrome c550 has been shown to be the natural electron acceptor for QHNDH (11). A similar QHNDH has been isolated from Pseudomonas putida, but apparently with somewhat different subunit composition and with exogenous electron acceptor specificity for azurin (12).
In this paper we describe the crystal structure and gene sequence of QHNDH from P. denitrificans, revealing that it comprises three nonidentical subunits, with the largest containing two heme c groups, and the smallest containing the unusual quinone cofactor cysteine tryptophylquinone (CTQ). More surprisingly, the small subunit contains three additional chemical crosslinks involving a cysteine side chain bridged by a thioether bond to a carboxylic acid side chain, either aspartate or glutamate.
Materials and Methods
Crystal Structure Analysis.
QHNDH was isolated and purified as described previously (10). Microcrystals were obtained by sitting-drop vapor diffusion at 293 K. The protein (≈15 mg⋅ml−1 in 10 mM Tris⋅HCl buffer, pH 7.5) was mixed with an equal volume of reservoir solution (100 mM sodium citrate buffer, pH 5.6/18% PEG 4,000/21% t-butyl alcohol) and allowed to equilibrate. Large crystals (up to 0.5 × 0.5 × 0.4 mm3) were obtained by macroseeding.
X-ray data were collected at ≈100 K from flash frozen crystals, cryo-protected with mineral oil, on an R-axis IV image plate system mounted on a Rikagu RU-200 rotating anode x-ray generator operating at 50 kV and 100 mA. Data were processed with the HKL package (13). Two forms of native crystals (native I and native II) were identified, each with one molecule in the asymmetric unit, but with slightly different cell dimensions. Crystals soaked with ethylmercury phosphate, platinum ethylenediamine dichloride, and methylmercury chloride produced interpretable diffraction changes. Two Fe positions were located from the anomalous difference Patterson map from a highly redundant native data set (native II). Initial phase information for reflections up to 2.6-Å resolution was obtained by multiple isomorphous replacement (MIRAS) using the anomalous contribution of the native data set (native II) and the isomorphous plus anomalous contributions from the three derivatives. The program SOLVE (14) was used for all of the MIRAS calculation. The initial experimental phases were improved by solvent flattening by using the program DM (15) in the CCP4 program suite (16). Results are summarized in Table 1.
Table 1.
Data collection, phasing, and refinement statistics
| Native I | Native II | EMP | PEDD | MMC | |
|---|---|---|---|---|---|
| Resolution, Å | 30–2.04 | 30–2.6 | 30–3.0 | 30–3.0 | 30–3.0 |
| Unit cell | |||||
| a, Å | 99.49 | 99.49 | 99.11 | 98.80 | 98.56 |
| c, Å | 214.49 | 210.38 | 211.58 | 209.16 | 213.07 |
| No. unique reflections | 58,964 | 31,819 | 19,897 | 19,960 | 18,235 |
| Rsym* | 6.2 | 5.1 | 7.6 | 9.4 | 10.2 |
| (Outer shell) | (21.7) | (9.4) | (11.7) | (12.8) | (22.6) |
| Completeness, % | 86.4 | 96.4 | 91.4 | 92.3 | 84.5 |
| (Outer shell) | (50.6) | (77.0) | (67.1) | (77.8) | (47.1) |
| Redundancy | 8.0 | 12.6 | 6.7 | 5.2 | 4.0 |
| I/σ(I) | 24.9 | 44.3 | 28.8 | 28.5 | 11.3 |
| (Outer shell) | (2.1) | (14.8) | (11.4) | (9.5) | (2.0) |
| No. phasing sites | 2 | 4 | 5 | 2 | |
| Phasing power† | |||||
| Centric | 1.00 | 0.64 | 0.73 | 0.84 | |
| Acentric | 1.40 | 0.90 | 0.54 | 0.22 | |
| Mean figure of merit | 0.52 | ||||
| R/Rfree‡, % | 20.2/25.1 | ||||
| No. protein atoms | 6,960 | ||||
| No. nonprotein atoms | 103 | ||||
| No. waters | 642 | ||||
| Average B factors, Å2 | 37.6 | ||||
| rms deviation | |||||
| Bonds, Å | 0.007 | ||||
| Angles, ° | 1.5 |
EMP, ethylmercury phosphate; PEDD, platinum ethylene diamine dichloride; MMC, methylmercury chloride.
Rsym = ∑|Ii − 〈I〉|/∑ Ii, where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity.
Phasing power = 〈FH〉/〈E〉, where FH and E are the heavy atom amplitudes and error.
R = ∑∥Fo| − |Fc∥/∑|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree is calculated using 10% of reflections omitted from the refinement (40).
Side chains in some parts of the initial map could easily be recognized after initial main-chain tracing, and were matched with the genetically obtained amino acid sequence. Most of the sequence could be modeled, except for two loop regions, from this initial map. The programs O (17) and TURBO-FRODO (18) were used for the model building, MAPMAN (19) for map manipulations, and VOIDOO (20) for cavity volume calculations. The initial model was first refined and improved against the native II data set to 2.6 Å in CNS (21). After locating the molecule in the native I unit cell by molecular replacement using AMORE (22), further refinement in CNS was carried out with this latter native data set (native I) to 2.05 Å. During model building, a total of 642 water molecules, 3 molecules of t-butyl alcohol, and 1 sodium ion were identified. A total of six proline residues are in cis conformation. Analysis of the final model in PROCHECK (23) indicated that 85.6% of the dihedral angles of the Ramachandran plot were in the most favored range, 13.7% in the allowed range and 0.8% in the disallowed range.
Chemical Analysis of the γ Subunit and Quinone-Containing Peptide.
The small γ subunit was separated by SDS/PAGE of the purified enzyme denatured for a long period (>15 min) in 2% SDS and 0.1 mM DTT. A protein band of about 9 kDa, detected by quinone-dependent redox cycling (24) after electroblotting onto a poly(vinylidene difluoride) membrane, was eluted from the finely ground gel pieces in 20 mM ammonium bicarbonate containing 8 M urea, dialyzed thoroughly against distilled water, and analyzed for the content of free SH groups with 0.2 mM 5,5′-dithiobis(2-nitorobenzoic acid) by monitoring the increase in absorbance at 412 nm. For reductive carboxymethylation, the γ subunit polypeptide was first incubated for 4 h with 0.5 mM DTT in 50 mM Tris buffer (pH 8.0) containing 6 M guanidine hydrochloride under N2 gas stream, then reacted for 1 h with a freshly prepared solution of iodoacetic acid (1 mM). Amino acid composition of the peptide was determined with an amino acid analyzer after hydrolysis at 110°C for 24 h in 6 M HCl containing 0.2% phenol. For isolation of the quinone-containing peptide, the enzyme was modified with excess p-nitrophenylhydrazine (NPH) to protect the quinone group, and digested with trypsin for 24 h and then with Pronase for 4 h in 0.15 M NH4HCO3 (pH 8.0) and 2.5 M urea. The peptide containing the NPH-quinone hydrazone was isolated by reverse-phase HPLC by monitoring the 464-nm absorbance derived from the hydrazone. Sequence analysis of the peptide with or without prior reductive carboxymethylation was done with an Applied Biosystems 476A protein sequencer. Electrospray ionization (ESI)-MS/MS was carried out on a hybrid quadrupole orthogonal acceleration tandem mass spectrometer (Q-TOF; Micromass, Manchester, U.K.). MS/MS data were processed by the maximum entropy data enhancement program MAXENT 3 (Micromass). The resultant spectra were interpreted by SEQMS (25), a software aid for de novo sequencing by MS/MS.
Gene Cloning and Sequence Analysis.
Each subunit of QHNDH was separated by SDS/PAGE (14% acrylamide) and eletrotransferred onto a poly(vinylidene difluoride) membrane for N-terminal amino acid sequence analysis, and was also digested directly within polyacrylamide gel pieces, soaked in 20 mM ammonium bicarbonate with lysyl endopeptidase at 37°C for 24 h. Internal peptide fragments liberated from each subunit were isolated by HPLC for sequence analysis. Degenerate oligonucleotide primers (26-mer) were designed from the N-terminal and internal nine-residue sequences of the α subunit and used for amplification of a part of the QHNDH gene by the PCR with the P. denitrificans genome as a template. The amplified fragment (about 1.3 kbp) was then used as a probe for Southern hybridization of SalI- or PstI-digested genomic DNA. Genomic libraries were constructed by inserting SalI or PstI fragments (4–6 kbp), positively hybridized with the probe, into the vector pUC18 (Takara, Kyoto) and transformed into Escherichia coli XL1-Blue cells. Colony hybridization using the same probe led to isolation of clones containing a SalI (about 4 kbp) or PstI (about 6 kbp) fragment from about 104 clones. Restriction analysis indicated that the two fragments overlap each other, covering the entire gene for QHNDH. DNA sequence analyses were done in both directions of the two fragments with an Applied Biosystems Prism 377 DNA sequencer.
Results and Discussion
Molecular Architecture.
X-ray crystallography and DNA sequencing have revealed that QHNDH is an αβγ heterotrimer. Although the small γ subunit was not identified previously (10), its existence was confirmed by protein chemistry in the present studies (data not shown, see Materials and Methods). Presumably, the quinone-containing γ subunit associates very tightly with the α subunit to form a protein complex with an apparent molecular mass of 60 kDa and is resistant to complete denaturation. The crystal structure has also revealed the presence of two heme c groups in the α subunit; the reason for electrochemical detection of only one heme c in the previous studies (10) is unclear.
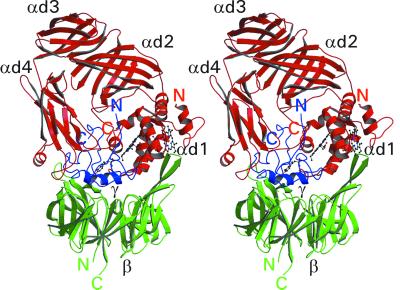
The α subunit consists of 489 amino acids and is folded into four domains (Fig. 2). The first domain (residues 1α–167α) is a diheme cytochrome consisting of two subdomains (αd1a, residues 3α–70α; αd1b, 95α–162α), which are related with each other by approximate twofold symmetry. Surprisingly, whereas subdomain αd1a is a typical c-type cytochrome with a histidine and a methionine iron ligand, both iron ligands of the subdomain αd1b cytochrome are histidine. The two iron atoms are separated by 16.3 Å. The heme rings are within 10 Å of each other, and their planes are tilted by about 45° to each other. The heme of subdomain αd1a is slightly exposed to solvent, where Sγ of Cys-14α forms a thioether linkage to the methylene carbon atom of a heme vinyl group. The other heme group is totally buried and is close to the γ subunit. Domains 2 (residues 168α–272α), 3 (274α–350α), and 4 (354α–480α) of the α subunit are antiparallel β-barrel structures of 8, 7, and 7 β-strands, respectively. In cross section, the α subunit is roughly a horseshoe in shape with the two prongs of the horseshoe projecting down onto the concave β subunit surface and the body of the horseshoe encompassing the γ subunit.
Figure 2.
Stereo ribbon diagram of the full QHNDH heterotrimer. The subunits α, β, and γ and their N and C termini are shown in red, green, and blue, respectively. The four domains of the α subunit (αd1, αd2, αd3, and αd4) and the β and γ subunits are labeled in black. The two heme groups in subunit α and the CTQ cofactor in the γ subunit are also shown. This and subsequent diagrams, except where stated, were prepared by using MOLSCRIPT (41) and rendered by using RASTER3D (42).
The β subunit is a seven-bladed β-propeller containing 337 residues and is the simplest and most regular QHNDH subunit. It provides a platform on which the α and γ subunits rest. The γ subunit covers approximately half of the concave “top” surface of the β subunit, with the quinone cofactor being approximately coincident with the sevenfold pseudosymmetry axis of the β-propeller. In this respect, there is a superficial similarity of the molecular architecture of QHNDH to that of MADH. Both enzymes contain a seven-bladed β-propeller subunit, on which the quinone-containing subunit is positioned. Comparison of the two propeller domains gives an rms deviation of 1.96 Å for 148 equivalent Cα atoms. In contrast, there is absolutely no similarity between the tertiary structures of the quinone-containing subunits of the two enzymes. The γ subunit of QHNDH has a highly crosslinked globular structure with little secondary structure except for a three-turn helix near the middle of the molecule and a two-turn helix close to the C terminus (Fig. 3A). It makes extensive contact with the α and β subunits. Approximately 60% of its 5250 Å2 surface is buried within the protein, 40% through contact with the α subunit and 20% with the β subunit.
Figure 3.
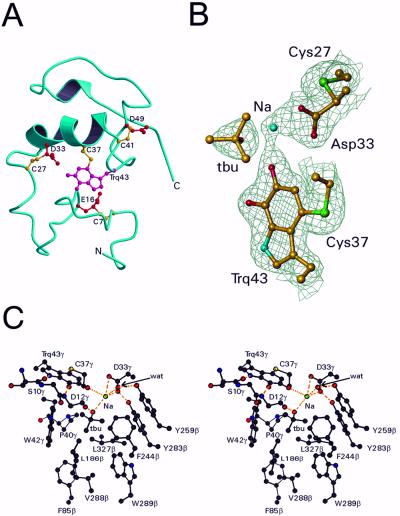
Structure of the γ subunit and active site of QHNDH. (A) The backbone ribbon is blue, the crosslinked side chains of Cys are yellow, and the crosslinked side chains of tryptophylquinone (Trq), Glu, and Asp are red. (B) Electron density around CTQ. Also shown are the putative active site base Asp-33γ, its covalently linked Cys-27γ, the molecule of t-butyl alcohol (tbu) and the sodium ion (Na). Only Cα and side chain atoms are included. The map was computed with coefficients (2Fo − Fc) exp(−iα) where Fc and α, the calculated structure factors and phase angles, were derived from the final refined model. The contours are drawn at 1.2 σ, where σ is the rms value of the electron density. This diagram was prepared by using TURBO-FRODO (18). (C) Stereoview of the active site of QHNDH. See text for details.
The 82-residue γ subunit contains 5 Trp and 4 Cys residues in the sequence deduced from DNA. However, neither free SH groups nor S—S bridges were detected in it by chemical analysis. Surprisingly, all four cysteines are involved in highly unusual covalent crosslinks to other amino acid side chains within the subunit (Fig. 3A). Two are to aspartates, one to glutamate, and one to tryptophan, all in carbon-sulfur thioether linkages with residues that have never been reported before in proteins. The functionally most important crosslink is between Cys-37γ and Trp-43γ, and makes up part of the quinone cofactor, CTQ. The Sγ of Cys-37γ is covalently bound to C-4 of tryptophylquinone (Trq; atom Cɛ2 of tryptophan), analogous to the Trp–Trq crosslink in TTQ (Fig. 1), and an oxygen atom is bound to both C-6 and C-7 (atoms Cη2 and Cζ2) to form an orthoquinone (Fig. 3B). In addition to the covalent link to Cys-37γ, the Trq portion of CTQ forms hydrogen bonds with two main chain atoms, N-1 to Ser-10γ O (2.8 Å) and O-7 to Asp-12γ N (2.6 Å) (Fig. 3C). There is also a close contact between O-6 and Asp-12γ O (2.7 Å). The closest side chain to the Trq group is the carboxylate of Asp-33γ (3.1 Å). This side chain is the only one close enough to the active site to serve a catalytic role in substrate oxidation.
The Sγ of Cys-7γ is covalently bound to the methylene carbon Cγ of Glu-16γ; this crosslink appears to stabilize the intervening polypeptide that sits below and circumscribes Trq-43γ (Fig. 3A). The Sγ of Cys-41γ is covalently bound to Cβ of Asp-49γ; this bond may serve to fix the ends of the polypeptide loop containing Trp-42γ and Trq-43γ. The fourth crosslink, between Sγ of Cys-27γ and the methylene carbon Cβ of Asp-33γ, serves to position Asp-33γ at the active site close to the orthoquinone of CTQ, thereby providing structural stabilization for the only candidate for an active site base in the γ subunit. Interestingly, two of the six cis-prolines in the structure are located in the γ domain, Pro-13γ and Pro-29γ, both located within loops defined by cysteine–carboxylic acid crosslinks. Thus the structure of the γ subunit is unique, containing four internal crosslinks in such a short polypeptide chain, which might otherwise be difficult to fold into a globular structure.
Active Site.
During model building, a large fragment of difference electron density within an internal cavity close to the CTQ cofactor was modeled as t-butyl alcohol, present during crystallization (Fig. 3B). Another strong difference peak, close to the side chain of Asp-33γ, was modeled as a sodium ion on the basis of its interaction with several oxygen atoms. Both t-butyl alcohol and the sodium ion were included in the final refinement and exhibited consistent behavior in the electron density maps. The t-butyl alcohol was modeled so that O-1 donates a hydrogen bond to the carbonyl of Asp-12γ (2.8 Å) and the three methyl groups protrude into the hydrophobic portion of the cavity. The sodium ion is surrounded by the two carboxylate oxygen atoms of Asp-33γ, O-6 of CTQ and O-1 of t-butyl alcohol, at an average distance of 2.37 ± 0.16 Å (Fig. 3C). There is also a water molecule 2.7 Å away from the cation that donates its hydrogen bonds to the hydroxyl of Tyr-259β and the γ-sulfur of Cys-27γ, the latter forming a thioether linkage to the β-carbon of Asp-33γ. The internal cavity has a volume of ≈160 Å3 that is a likely binding site for substrate (Fig. 3C). Except for Trq-43γ and the side chain of Asp-33γ, the cavity is lined by hydrophobic side chains, consistent with the enzyme specificity for aliphatic and aromatic amine substrates. About half of the side chains and protein atoms in contact with the cavity are contained within the γ subunit and the remainder are part of the β subunit.
Chemical Analysis of Quinone-Containing Peptide.
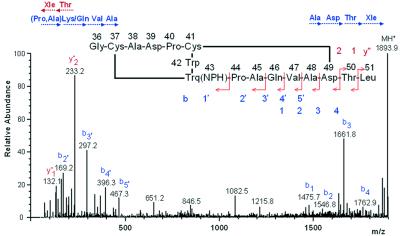
To provide unequivocal evidence for the cofactor and crosslinking structures in the γ subunit, a combination of classical Edman degradation with MS analyses was applied to determine the chemical structure of a peptide containing the quinone group obtained by protease digestion of the NPH-derivatized enzyme. Without the x-ray structure, we had initially encountered difficulties in the interpretation of the results from both analyses. However, with the crystal structure solved at high resolution, the absence of a phenylthiohydantoin derivative at several noncontiguous positions could be explained by the presence of crosslinks within the peptide, covering the region Gly-36γ–Leu-51γ. The ESI-MS/MS spectrum of this peptide (Fig. 4) also clearly shows that the sequence of eight residues from its C terminus can be deduced from the spectrum, but no information could be obtained from the NPH-Trq-43γ upward, because of the crosslink between that residue and Cys-37γ in the peptide. The theoretical mass for the NPH-quinone peptide calculated as a monovalent ion (MH+) is 1897.9 Da, whereas the experimental value was 1893.9 Da, clearly demonstrating the loss of four hydrogen atoms because of the formation of thioether links between Cys-37γ and Trq-43γ and between Cys-41γ and Asp-49γ. Furthermore, the assignment of oxygen atoms for the electron densities protruding from the 6 and 7 positions of the indole ring of Trq-43γ (Fig. 3B) was justified by the mass data.
Figure 4.
ESI-MS/MS spectrum of the NPH-quinone peptide derived from the γ subunit. Fragmentation occurred for the most part along the backbone of a C-terminal portion of the peptide. Amino acids shown in three-letter code denote immonium ions assigned by SEQMS (24). Arrows below the amino acids show the sequences from the N and C termini based on y"m and bl ions, respectively, where m and l denote positions counted from the C and N termini that were produced by cleavage of peptide bonds during MS/MS. The predicted structure of the peptide is shown in the middle with possible cleavage sites.
Catalysis of Substrate Oxidation.
In MADH and CAO, the steady-state reaction is believed to follow a ping-pong mechanism in which the quinone cofactor is reduced to an aminoquinol (the reductive half-reaction) and is then reoxidized by an electron acceptor to regenerate the oxidized quinone form of the cofactor (the oxidative half-reaction) (26, 27). In both enzymes, the reductive half reaction proceeds through formation of two Schiff base intermediates in which the imine nitrogen of the substrate replaces one of the quinone oxygens. Conversion of the substrate Schiff base to the product Schiff base, during which a carbanion intermediate is formed and the cofactor becomes reduced, is catalyzed by proton abstraction by a catalytic base on the enzyme. This base has been identified as an aspartate residue in CAO (28) and is likely to be an aspartate in MADH (29, 30). In QHNDH, the only side chain present in the active site that can act as the catalytic base is Asp-33γ, the others being hydrophobic or aromatic. In MADH, the O-6 of TTQ is almost certainly the site of Schiff base formation with the amine substrate during the reductive half reaction, both because O-7 is hydrogen bonded to a backbone amide group, and therefore sterically inaccessible to substrate (29), and because O-6 is the site of attack by hydrazine inhibitors of the enzyme (31). In QHNDH, O-6 of CTQ is likely to be the site of attack by amine substrates because O-7 is also hydrogen bonded to a backbone amide group and O-6 is closer to the substrate analogue, t-butyl alcohol, in the crystal structure. Therefore, the mechanism of substrate oxidation in QHNDH is likely to be similar to that proposed for MADH (32).
In the oxidative half reaction, two electrons derived from substrate are transferred to the physiological electron acceptor cytochrome c550 (11), probably by means of the two heme c groups bound in domain α1. This electron transfer likely occurs one at a time, forming a semiquinone radical intermediate, as observed by partial reduction of the enzyme by dithionite (10). The most likely site for the interaction of cytochrome c550 with QHNDH is through the solvent-accessible edge of the N-terminal cytochrome heme. Modeling studies have shown that this site would be accessible to cytochrome c550, although no precise binding interactions could be established. The other two redox centers, i.e., the CTQ cofactor in subunit γ and the heme of the second cytochrome, are buried in the protein interior and would be difficult for cytochrome c550 to access.
Gene Structure.
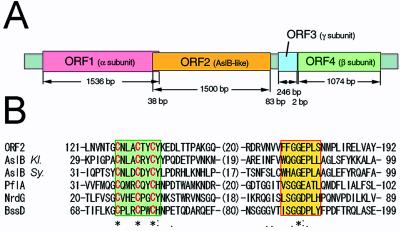
Structural genes coding for the three subunits of QHNDH were cloned in two overlapping restriction fragments from the P. denitrificans genome. There are four ORFs in the region of about 5,000 nt in length (Fig. 5A). The α subunit is encoded in ORF1, consisting of 1,536 nt. Because Edman sequencing of the α subunit yielded a sequence of VTGEEVLQNA, the 23-residue sequence from the putative translation initiator Met (MKPFTRTALVSVSALAFAAPVLA) found in the gene-derived sequence is likely a signal peptide directing translocation of QHNDH into the periplasm (10, 11). The β subunit is encoded in ORF4 of 1,074 nt, in which the 21-residue sequence (MRKSLMLLASAAAMLAGPALA) from the presumed initiator Met similarly appears to function as a signal peptide; Edman sequencing of the β subunit provided a sequence of RDYILAPARPDKLV, adjacent the signal sequence. The ORF for the γ subunit is located at the third small ORF (ORF3, 246 nt), which is separated by 2 nt 5′-upstream from ORF4 for the β subunit, but without a notable signal peptide. A homology search of the protein sequences deposited in the GenBank database failed to detect any protein similar (with >30% identities) to any of the α, β, and γ subunit sequences.
Figure 5.
Gene structure of QHNDH and local homology alignment of ORF2 protein with Radical S-adenosylmethionine (SAM) proteins. (A) Tandem arrangement of four ORFs coding for subunit α, the hypothetical [Fe—S] protein, subunit γ, and subunit β, respectively (from left to right). Nucleotide numbers of each ORF and of intervening noncoding regions are also indicated. (B) Multiple alignment of an N-terminal region of ORF2 protein of P. denitrificans with Radical SAM Proteins registered in GenBank [AslB of Klebsiella (Kl.), gi/114710; AslB of Synechocystis sp. (Sy.), gi/1652765; PflA (pyruvate formate-lyase activating enzyme), gi/6093720; NrdG (anaerobic ribonucleotide reductase activating protein), gi/730195; BssD (benzylsuccinate synthase activating enzyme), gi/3184129]. Aligned residue numbers are shown on both sides of each sequence, and invariant (*), highly (:), and weakly (.) conserved residues are indicated in the bottom line. Cys residues in the [Fe—S]-binding motifs (green box) are shown in red letters, and the putative SAM-binding Gly-rich motif is boxed in yellow.
The second ORF, consisting of 1,500 nt, does not correspond to any subunit of QHNDH. Because an ATG codon, which appears 38 nt 5′-uptream from the termination codon of ORF1, possesses a putative ribosome-binding site (5′-GAGG-3′) at −13 or −10 position, this ATG codon is presumably the translation initiation site for ORF2, overlapping with ORF1 (Fig. 5A). Such an overlap of neighboring ORFs occurs often in operons of prokaryotic genes. This ORF2 potentially encodes a 500-residue protein of about 55 kDa. In a homology search, the deduced amino acid sequence of ORF2 was found to show an overall weak but locally significant similarity with the sequences reported for the aslB gene product from Klebsiella pneumoniae as well as the activating enzymes for pyruvate formate-lyase and anaerobic ribonucleotide reductase. All these proteins are included in a recently described protein superfamily, named “Radical SAM” (S-adenosylmethionine) proteins, catalyzing diverse reactions, such as unusual methylations, isomerization, sulfur insertion, ring formation, anaerobic oxidation, and protein radical formation (33). AslB is required for the oxidation of a specific serine or cysteine residue in sulfatases to posttranslationally generate a formylglycine (2-oxoalanine) cofactor (34), hence designated as an arylsulfatase-activating enzyme. The Cys-rich region predicted to serve as a binding motif for the [Fe—S] cluster and a Gly-rich sequence likely involved in binding of SAM (33) are both conserved in the ORF2 protein (Fig. 5B). This ORF, intervening between the structural genes for α and γ subunits of QHNDH, thus encodes a previously unrecognized member of the Radical SAM proteins superfamily.
Cofactor and Cofactor Subunit Biogenesis.
The γ subunit undergoes three types of posttranslational modification during the maturation of QHNDH. One is the formation of a cysteine-to-tryptophan cross-bridge, the second is the introduction of a pair of oxygen atoms into the tryptophan ring, and the third is the crosslinking of cysteine side chains to the Cβ or Cγ atoms of carboxylic acid side chains. It has been shown that the TPQ cofactor in CAO (35) and the Cys–Tyr crosslinked cofactor in galactose oxidase (36) are both generated by the copper-dependent autocatalytic process of the precursor proteins. On the other hand, the biosynthesis of TTQ in MADH appears to be catalyzed by an enzymatic process, because the absence of one of the genes (mauG) of the 11 genes in the mau operon prevents the maturation of TQQ (37). The biosynthetic process for CTQ in the γ subunit of QHNDH may involve an enzyme-mediated process as well. A likely candidate is the AslB-like [Fe—S] protein encoded in ORF2 of the enzyme genes (Fig. 5), which may oxidize the tryptophan residue (Trp-43γ) to Trq in the γ subunit polypeptide, analogous to the Radical SAM AslB protein oxidizing a Cys or Ser residue to formylglycine in sulfatases (34). Although there is no precedent for a Radical SAM protein to be involved in the biogenesis of protein quinone cofactors, the PqqE protein functioning in PQQ biosynthesis has been recognized as a Radical SAM protein (33). Moreover, the genetic locus of Klebsiella aslB, separated by only 52 nt 5′-upstream from the structural gene for sulfatase (aslA) contained in the same Asl operon, is also very similar to that of ORF2 relative to the γ subunit gene (ORF3), which is separated by 83 nt. It has been speculated that the AslB protein, which lacks a signal peptide, oxidizes the critical serine of the unfolded sulfatase during or shortly after synthesis and that, after cofactor formation the sulfatase polypeptide with a signal peptide, is translocated into the periplasm (34). Interestingly, neither the ORF2 nor the ORF3 gene codes for signal peptides, suggesting that the interaction of the Asl-like protein and the γ subunit occurs within the cytoplasm before the oxidized γ subunit associates with the periplasmic α and β subunits. As for the periplasmic localization, the predicted signal peptides of the α and β subunits (see above) as well as the coding region of the γ subunit do not contain the (S/T)RRXF motif, which is shared by many bacterial proteins containing redox cofactors (including MADH) and recognized by the Tat transport system for their periplasmic translocation (38). Thus it is suggested that QHNDH is transported into the periplasm by a system different from the Tat system.
The cysteine–carboxylic acid as well as the Cys–Trq crosslinking may also require external enzymatic catalysis. Particularly, thioether modifications at methylene carbon atoms of carboxylic acid side chains would require an unusual chemical process to activate the very stable methylene carbon atoms. A possible way is through a radical intermediate. Also in this case, the ORF2 protein may participate, being a member of the Radical SAM superfamily of proteins, many of which can generate a radical species by reductive cleavage of SAM through an unusual [Fe—S] center (39). In this respect, it is noteworthy that carbon–sulfur bond-forming enzymes (biotin and lipoate synthases) and peptidyl glycyl/tyrosyl/thiyl radical-forming enzymes are all Radical SAM proteins that use an intermediate adenosyl radical in their catalytic processes. As to how the correct Cys–Xxx crosslinks are selected, it may be that the ORF2 protein activates the chemical reaction but that the γ subunit provides the crosslinking specificity. For all these reasons, we speculate that the biogenesis of CTQ cofactor and/or the highly unusual crosslinking structure within the γ subunit proceeds through a redox process, involving at least the presumed QHNDH-activating Radical SAM enzyme encoded in the same gene region.
Acknowledgments
We thank Dr. T. Takao for mass spectrometric analysis. This work was supported by U.S. Public Health Service Grant GM31611 and National Science Foundation Grant MCB-0091084 (to F.S.M.), and by Grants-in-Aid for Scientific Research and “Research for the Future” from the Japan Society for the Promotion of Science (to K. Tanizawa).
Abbreviations
- CAO
copper amine oxidase
- CTQ
cysteine tryptophylquinone
- MADH
methanol dehydrogenase
- PQQ
pyrroloquinoline quinone
- QHNDH
quinohemoprotein amine dehydrogenase
- SAM
S-adenosylmethionine
- TPQ
topaquinone
- TTQ
tryptophan tryptophylquinone
- Trq
tryptophylquinone
- ESI
electrospray ionization
- NPH
nitrophenylhydrazine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The gene sequence and the atomic coordinates of QHNDH have been deposited in GenBank (accession no. AB063330) and the Protein Data Bank, www.rcsb.org (PBD ID code 1JJU), respectively.
References
- 1.Anthony C. Biochem J. 1996;320:697–711. doi: 10.1042/bj3200697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janes S M, Mu D, Wemmer D, Smith A J, Kaur S, Maltby D, Burlingame A L, Klinman J P. Science. 1990;248:981–987. doi: 10.1126/science.2111581. [DOI] [PubMed] [Google Scholar]
- 3.Wang S X, Mure M, Medzihradszky K F, Burlingame A L, Brown D E, Dooley D M, Smith A J, Kagan H M, Klinman J P. Science. 1996;273:1078–1084. doi: 10.1126/science.273.5278.1078. [DOI] [PubMed] [Google Scholar]
- 4.McIntire W S, Wemmer D E, Chistoserdov A, Lidstrom M E. Science. 1991;252:817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- 5.Klinman J P, Mu D. Annu Rev Biochem. 1994;63:299–344. doi: 10.1146/annurev.bi.63.070194.001503. [DOI] [PubMed] [Google Scholar]
- 6.Anthony C. In: Principles and Applications of Quinoproteins. Davidson V L, editor. New York: Dekker; 1993. pp. 17–45. [Google Scholar]
- 7.Knowles P F, Dooley D M. In: Metal Ions in Biological Systems. Sigel H, Sigel A, editors. New York: Dekker; 1994. pp. 361–403. [Google Scholar]
- 8.Davidson V L, Jones L H. Anal Chim Acta. 1991;249:235–240. [Google Scholar]
- 9.Matsushita K, Yakushi T, Toyama H, Shinagawa E, Adachi O. J Biol Chem. 1996;271:4850–4857. doi: 10.1074/jbc.271.9.4850. [DOI] [PubMed] [Google Scholar]
- 10.Takagi K, Torimura M, Kawaguchi K, Kano K, Ikeda T. Biochemistry. 1999;38:6935–6942. doi: 10.1021/bi9828268. [DOI] [PubMed] [Google Scholar]
- 11.Takagi K, Yamamoto K, Kano K, Ikeda T. Eur J Biochem. 2001;268:470–476. doi: 10.1046/j.1432-1033.2001.01912.x. [DOI] [PubMed] [Google Scholar]
- 12.Adachi O, Kubota T, Hacisalihoglu A, Toyama H, Shinagawa E, Duine J A, Matsushita K. Biosci Biotechnol Biochem. 1998;62:469–478. doi: 10.1271/bbb.62.469. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowtan K, Main P. Acta Crystallogr D. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 16.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1991. p. 86. [Google Scholar]
- 19.Kleywegt G J, Jones T A. Acta Crystallogr D. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 20.Kleywegt G J, Jones T A. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 23.Morris A L, MacArthur M W, Hutchinson E G, Thornton J M. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 24.Paz M A, Flückiger R, Boak A, Kagan H M, Gallop P M. J Biol Chem. 1991;266:689–692. [PubMed] [Google Scholar]
- 25.Fernandez-de-Cossio J, Gonzalez J, Betancourt L, Besada V, Padron G, Shimonishi Y, Takao T. Rapid Commun Mass Spectrom. 1998;12:1867–1878. doi: 10.1002/(SICI)1097-0231(19981215)12:23<1867::AID-RCM407>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 26.Davidson V L, Brooks H B, Graichen M E, Jones L H, Hyun Y L. Methods Enzymol. 1995;258:176–190. doi: 10.1016/0076-6879(95)58046-8. [DOI] [PubMed] [Google Scholar]
- 27.Mure M, Klinman J P. Methods Enzymol. 1995;258:39–52. doi: 10.1016/0076-6879(95)58036-0. [DOI] [PubMed] [Google Scholar]
- 28.Wilmot C M, Murray J M, Alton G, Parsons M R, Convery M A, Blakeley V, Corner A S, Palcic M M, Knowles P F, McPherson M J, Phillips S E. Biochemistry. 1997;36:1608–1620. doi: 10.1021/bi962205j. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Mathews F S, Davidson V L, Huizinga E G, Vellieux F M, Duine J A, Hol W G. FEBS Lett. 1991;287:163–166. doi: 10.1016/0014-5793(91)80041-z. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Doi M, Durley R C, Chistoserdov A Y, Lidstrom M E, Davidson V L, Mathews F S. J Mol Biol. 1998;276:131–149. doi: 10.1006/jmbi.1997.1511. [DOI] [PubMed] [Google Scholar]
- 31.Huizinga E G, van Zanten B A, Duine J A, Jongejan J A, Huitema F, Wilson K S, Hol W G. Biochemistry. 1992;31:9789–9795. doi: 10.1021/bi00155a036. [DOI] [PubMed] [Google Scholar]
- 32.Davidson V L, editor. Principles and Applications of Quinoproteins. New York: Dekker; 1993. pp. 73–95. [Google Scholar]
- 33.Sofia H J, Chen G, Hetzler B G, Reyes-Spindola J F, Miller N E. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szameit C, Miech C, Balleininger M, Schmidt B, von Figura K, Dierks T. J Biol Chem. 1999;274:15375–15381. doi: 10.1074/jbc.274.22.15375. [DOI] [PubMed] [Google Scholar]
- 35.Matsuzaki R, Fukui T, Sato H, Ozaki Y, Tanizawa K. FEBS Lett. 1994;351:360–364. doi: 10.1016/0014-5793(94)00884-1. [DOI] [PubMed] [Google Scholar]
- 36.Rogers M S, Baron A J, McPherson M J, Knowles P F, Dooley D M. J Am Chem Soc. 2000;122:990–991. [Google Scholar]
- 37.Page D, Ferguson S J. Eur J Biochem. 1993;218:711–717. doi: 10.1111/j.1432-1033.1993.tb18425.x. [DOI] [PubMed] [Google Scholar]
- 38.Berks B C. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 39.Cheek J, Broderick J B. J Biol Inorg Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 40.Kleywegt G J, Brünger A T. Structure (London) 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 41.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 42.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]