Watch a video presentation of this article
Watch the interview with the author
Abbreviations
- apoB
apolipoprotein B
- BASL
British Association for the Study of the Liver
- EU
exposed uninfected
- HCV
hepatitis C virus
- HS‐GAG
heparan sulfate glycosaminoglycan
- LDLR
low‐density lipoprotein receptor
- LVP
lipoviral particle
- OPLS‐DA
orthogonal projections to latent structures discriminant analysis
- SR
spontaneous resolver
- SVR
sustained viral responder
- VLDL
very low density lipoprotein
Hepatitis C virus (HCV) infection is a curable infection in the majority of cases, but an effective vaccine remains elusive. An important explanation for lack of efficacy of vaccine candidates to protect from HCV may relate to the dynamic and complex interactions of HCV with host lipid metabolism. HCV is dependent on interactions with host lipid metabolism for all stages of the viral life cycle (Fig. 1)—from attachment and entry into hepatocytes, to replication and assembly of new viral particles. Production of infectious HCV is dependent on the hepatocytes machinery for export of very low density lipoproteins (VLDLs).1 HCV replication takes place on lipid droplets with colocalization of core protein and nonstructural NS5A to the lipid droplet. Codependency of HCV assembly on VLDL secretion is demonstrated by silencing of VLDL synthesis components apolipoprotein B (apoB), apoE, and microsomal triglyceride transfer protein, all of which inhibit HCV production in Huh 7 cells. Thus, HCV replication and assembly is dependent on host VLDL pathways.1
Figure 1.
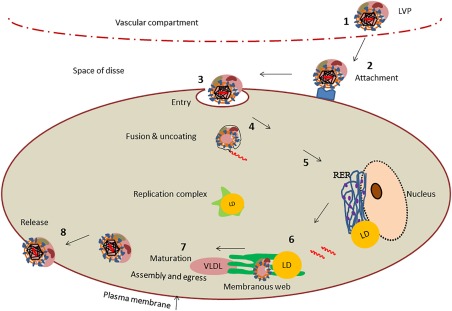
HCV life cycle. (1) HCV circulates in the blood as LVPs. (2) The LVP leaves the circulation through fenestrated endothelia and accesses the hepatocytes via the space of Disse. The LVP interacts with host cell receptor molecules such as heparan sulfate glycosaminoglycan (HS‐GAG), low‐density lipoprotein receptor (LDLR), and others with high affinity for apolipoproteins. (3) The LVP then binds to entry factors enabling clathrin‐mediated endocytosis. (4) Uncoating of viral capsid delivers viral genomic material to cytoplasmic replication site. (5) Successful viral entry through lipoprotein channels leads to translation of polypeptides in endoplasmic reticulum. (6) Replication complex forms +ssRNA strands via −ssRNA intermediates in membranous web, which shows characteristics of lipid rafts. (7) Virions assemble and bud through lipid channels that secrete mature virions via VLDL pathway. (8) Mature virions are released from the cell. LD, lipid droplet; RER, rough endoplasmic reticulum.
Circulating HCV is associated with lipoproteins as complex “lipoviral particles” (LVPs) (Fig. 2).2 LVPs contain both viral RNA and viral proteins, and host apolipoprotein constituents including apoB, apoE, apoA1, and apoC. The structure of LVP has recently been demonstrated by electron microscopy studies.2 Low‐density LVPs have increased infectivity in cell culture, compared with high‐density viral particles,3 and decreased susceptibility to antibody mediated neutralization.4 Concentrations of LVPs in patients with chronic HCV are dynamic and fluctuate with postprandial lipemia.5 LVPs have been correlated with deranged metabolism including insulin resistance, altered fasting triglyceride concentrations, and apoE levels.6 LVPs are detectable in both early acute and chronic HCV infection, with lower concentrations observed in early acute HCV infection that subsequently spontaneously resolved compared with those patients subsequently developing chronic HCV infection.7
Figure 2.
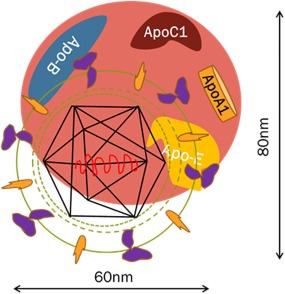
Schematic model of an HCV LVP. HCV circulates in the blood in association with host lipoproteins in a complex called a LVP that contains both viral (envelope, core, RNA) and host lipoprotein constituents including cholesterol, triglycerides, apoB, apoE, and apoC1.2 LVPs are important in HCV attachment and entry, and may mask viral epitopes from antibody‐mediated neutralization.
Growing evidence suggests that the LVP lipid and apolipoprotein components facilitate viral attachment to host cells by binding to cellular lipoprotein receptors. The exchangeable apoE appears to be crucial for infectivity in the attachment step and masking envelope glycoproteins from neutralizing antibodies.8 HCV association with lipoproteins may also be one means by which circulating HCV can avoid immunoglobulin recognition and evade the host's immune surveillance. Given this codependency on host lipid pathways, any mechanism that would disrupt the LVP formation would potentially reduce the viral infectivity and influence outcome after HCV exposure.
Potential resistance to HCV infection has been described in high‐risk injection drug users who do not become HCV‐infected despite a long history of likely exposure. These cases test persistently negative for anti‐HCV antibodies and HCV RNA, and are often referred to as HCV‐exposed uninfected (EU). Many HCV EU cases have demonstrable HCV‐specific T cell responses,9 which serves as further confirmation of HCV exposure. However, these responses are typically weak and transient, and it remains unclear whether they are responsible for any degree of protection from infection or simply an immunological correlate of exposure.
Given the importance of lipid pathways for all stages of the viral life cycle, we have investigated whether variation in the lipidome of apparently HCV‐resistant individuals may be associated with the EU phenotype.
At the British Association for the Study of the Liver (BASL) Annual Meeting in Manchester, we reported on a lipidomics analysis using ultra‐performance liquid chromatography mass spectrometry (UPLC‐MS) comparing lipid profiles in serum from HCV “susceptible” with HCV “resistant” cases.10 Initial data were presented from an analysis of nonfasting serum from HCV‐susceptible cases who were all HCV antibody‐positive. These data were divided into three categories: (1) chronic infection (HCV RNA‐positive), (2) sustained viral responders to previous antiviral treatment (SVRs; HCV RNA‐negative), or (3) spontaneous resolvers (SRs; HCV RNA‐negative). The lipidomes of these groups were compared with HCV‐resistant EU cases.
Principal component analysis and orthogonal projections to latent structures discriminant analysis (OPLS‐DA) were performed on all data and showed that the HCV EU cohort had very distinctly different lipidomic features from all the HCV‐susceptible groups, including chronic infection, SVR, and SR, in both positive and negative ionization modes (Fig. 3).
Figure 3.
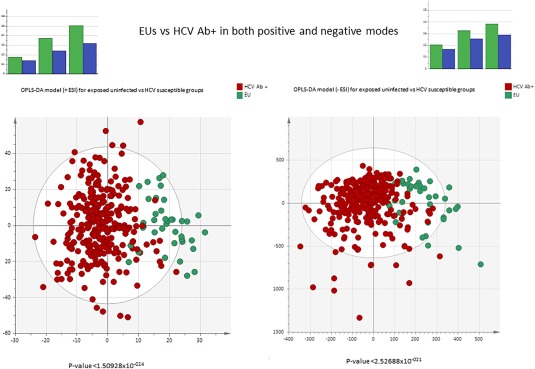
Lipidomics. Multivariate analysis using principal component analysis and OPLS‐DA for HCV‐resistant cases (EU, n = 38) compared with HCV‐susceptible (HCV antibody‐positive, n = 295), including chronic HCV patients genotypes 1 or 3 (HCV RNA‐positive, n = 159), SVRs (n = 100), and SRs (n = 36). Summary of data presented as poster 15 at the BASL Annual Meeting 2016.10
Conclusions
Lipidomics profiling demonstrated that the HCV‐resistant EU phenotype was clearly distinct from HCV‐susceptible individuals, even those who clear viremia, either spontaneously or after antiviral therapy. The HCV–lipid interaction is an essential feature of the virus life cycle with the early formation of LVPs a key step to establish infection in susceptible individuals. The demonstration of clearly distinct lipid profiles in cases apparently resistant to HCV infection provides further support that lipid pathways are crucial for early and persistent HCV infection. Detailed lipid class assignment to identify specific metabolic pathways associated with HCV resistance is ongoing and will require additional validation of identified lipid species. Identification of the precise lipid moieties that distinguish susceptible from resistant phenotypes will help our understanding of this key interaction and may help guide future vaccine design strategies.
†These authors contributed equally to this work.
REFERENCES
- 1. Bassendine MF, Sheridan DA, Felmlee DJ, Bridge SH, Toms GL, Neely RD. HCV and the hepatic lipid pathway as a potential treatment target. J Hepatol 2011;55:1428‐1440. [DOI] [PubMed] [Google Scholar]
- 2. Piver E, Boyer A, Gaillard J, Bull A, Beaumont E, Roingeard P, et al. Ultrastructural organisation of HCV from the bloodstream of infected patients revealed by electron microscopy after specific immunocapture. Gut; 10.1136/gutjnl-2016-311726 [DOI] [PubMed] [Google Scholar]
- 3. Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 2007;9:1089‐1097. [DOI] [PubMed] [Google Scholar]
- 4. Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, et al. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol 2008;82:12020‐12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Felmlee DJ, Sheridan DA, Bridge SH, Nielsen SU, Milne RW, Packard CJ, et al. Intravascular transfer contributes to postprandial increase in numbers of very‐low‐density hepatitis C virus particles. Gastroenterology 2010;139:1774‐1783. [DOI] [PubMed] [Google Scholar]
- 6. Sheridan DA, Bridge SH, Felmlee DJ, Crossey MM, Thomas HC, Taylor‐Robinson SD, et al. Apolipoprotein‐E and hepatitis C lipoviral particles in genotype 1 infection: evidence for an association with interferon sensitivity. J Hepatol 2012;57:32‐8. [DOI] [PubMed] [Google Scholar]
- 7. Sheridan DA, Hajarizadeh B, Fenwick FI, Matthews GV, Applegate T, Douglas M, et al. Maximum levels of hepatitis C virus lipoviral particles are associated with early and persistent infection. Liver Int 2016;36:1774‐1782. [DOI] [PubMed] [Google Scholar]
- 8. Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefevre M, et al. Apolipoprotein E mediates evasion from hepatitis c virus neutralizing antibodies. Gastroenterology 2016;150:206‐217. [DOI] [PubMed] [Google Scholar]
- 9. Thurairajah PH, Hegazy D, Chokshi S, Shaw S, Demaine A, Kaminski ER, et al. Hepatitis C virus (HCV)—specific T cell responses in injection drug users with apparent resistance to HCV infection. J Infect Dis 2008;198:1749‐1755. [DOI] [PubMed] [Google Scholar]
- 10. Shawa IT, Gomez‐Romero M, Pechlivanis A, Felmlee DJ, Crossey M, Holmes E, et al. P15. Serum lipid profiling using ultra‐performance liquid chromatography mass spectrometry (UPLC/MS) discriminates HCV exposed uninfected injection drug users from those susceptible to infection. Poster presented at: British Association for the Study of the Liver (BASL) Annual Meeting; September 7–9, 2016; Manchester, United Kingdom. http://www.baslannualmeeting.org.uk/


