Abstract
Diffuse intrinsic pontine gliomas (DIPGs) are deadly tumors comprising 10%–15% of all childhood CNS cancers. Standard treatment is considered palliative and prognosis is near universal mortality. DIPGs have been classified into genomic subtypes based on histone variants with the lysine to methionine mutation on position 27 of histone tails (K27M). Given the increasing promise of immunotherapy, there have been ongoing efforts to identify tumor-specific antigens to serve as immunologic targets. We evaluated a large cohort of CNS specimens for Wilms’ tumor protein (WT1) expression. These specimens include primary pediatric CNS tumors (n = 38 midline gliomas and n = 3 non-midline gliomas; n = 23 DIPG, n = 10 low-grade gliomas, n = 8 high-grade gliomas), and DIPG primary cells. Here, we report the validation of WT1 as a tumor-associated antigen in DIPGs. We further report that WT1 expression is significantly correlated with specific oncohistone variants, with the highest expression detected in the H3.3K27M subgroup. WT1 expression was absent in all control specimens (n = 21). Western blot assays using DIPG primary cells (n = 6) showed a trend of higher WT1 expression in H3.3K27M cells when compared with H3.1 K27M cells and H3 wildtype cells. Our data are the first indication of the association between WT1 and DIPG, with specific upregulation in those harboring oncohistone H3.3K27M.
Keywords: Brainstem glioma, Childhood midline glioma, Diffuse intrinsic pontine glioma (DIPG), Wilms tumor protein (WT1)
INTRODUCTION
Diffuse intrinsic pontine glioma (DIPG) is a childhood brainstem cancer with a dismal median survival of 9–12 months postdiagnosis (1). Given the location of the tumor and the infiltration of tumor cells with normal tissue, DIPGs are not amenable to meaningful surgical resection. Chemotherapy has thus far shown little evidence of benefit (2, 3), and radiation therapy provides only a temporary clinical stabilization. Clearly, a new treatment approach for children diagnosed with DIPG is necessary as conventional treatments have failed despite 40 years of clinical studies.
Recently, recurrent somatic gain-of-function mutations in the gene encoding the histone H3 protein (substitution of lysine 27 to methionine [p.Lys27Met, K27M]) have been identified (4, 5). This has allowed DIPGs to be classified into 3 main genomic subgroups based on histone variant mutations: H3.1 (HIST1H3B/C, representing 20% of all DIPGs), H3.3 (H3F3A, representing >70% of all DIPGs), and H3 wildtype (H3WT). In addition, we and others have shown that DIPGs exhibit unique protein expression patterns that may provide attractive targets for immunotherapeutic approaches (6–8).
In order to further define molecular pathways associated with H3 subtypes, we performed a literature search and identified Wilms’ tumor protein (WT1) as a protein highly associated with various cancers including adult glioblastomas (9). The WT1 gene encodes the WT1 protein, which is a transcription factor containing 4 zinc-finger DNA binding domains, essential for embryonic development of the spleen, kidneys, gonads, and cardiac vasculature (10). Initial studies described WT1 as a tumor suppressor, as mutations associated with the WT1 gene were identified in a subset of Wilms’ tumors (11). A report by the National Cancer Institute identified WT1 as the protein with the highest potential for cancer immunotherapy (12), and a recent phase I clinical trial showed that WT1 peptide vaccine therapy in adult glioblastomas was safe, which induced cellular and humoral immune response (13).
Here, we assess WT1 upregulation at the mRNA and protein level in DIPGs, focusing on differential expression of WT1 based on H3 oncohistone mutations. Given the emerging role of WT1 in brain cancers, our report provides a comprehensive analysis of WT1 expression, its subcellular localization and dysregulation across DIPGs.
MATERIALS AND METHODS
Biological Specimens
Midline glioma specimens (including tumor and adjacent normal brain tissue) were obtained at autopsy in accordance with Children’s National Health System’s Institutional Review Board approval (IRB #1339). Patient identifiers were removed and specimens were dissected in transverse sections, and alternative sections were frozen or formalin-fixed as described in our previous work (14). Non-midline glioma specimens were obtained at biopsy and/or at autopsy.
Whole-Exome Sequencing and Digital Droplet Polymerase Chain Reaction
Genomic DNA was extracted from postmortem samples. DNA library preparation and sequencing was performed as described in our previous work (15). Digital PCR was performed using the RainDrop Digital PCR system (RainDance Technologies, Inc., Billerica, MA). For 50-μL droplet PCR reactions, 12 μL of pre-amplified DNA in TE buffer, 1× TaqMan Genotyping Master Mix (Life Technologies, Carlsbad, CA), 0.2 μM target probes (H3F3A wildtype-HEX, HIST1H3B wildtype-HEX and mutant-FAM), 0.9 μM of forward and reverse primers and 1× hydrofluorinated droplet stabilizer (RainDance Technologies, Inc.) were added. Tumor tissue DNA and water-only reactions were included as the positive and negative controls, respectively. Emulsions were prepared on the RainDrop Source instrument to produce ∼8 million droplets per 50-μL reaction. Emulsions were then placed on ABI 2720 Thermal Cycler (Applied Biosystems, Waltham, MA) to amplify the targets using initial activation of 95°C for 10 minutes, followed by 45 cycles of 95°C for 30 seconds and 58°C for 2 minutes, and in activation of 98°C for 10 minutes. The reactions were then placed on the RainDrop Sense instrument for signal detection.
Antibodies
Mouse monoclonal anti-WT1 (6F-H2, Abcam, Cambridge, MA), rabbit polyclonal anti-histone H3K27M and rabbit monoclonal anti-tri-methyl-histone H3 (H3K27me3) (C36B11, Cell Signaling Technology, Danvers, MA) were diluted in Bond primary antibody diluent (#AR9352 Leica Biosystems, Buffalo Grove, IL) to concentrations of 1:15, 1:200, and 1:400, respectively. The Bond polymer refined detection kit (Leica Biosystems) was used for secondary detection.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded (FFPE) slides (5 μm). FFPE slides were deparaffinized and antigen retrieval completed by heat-induced epitope retrieval in citrate buffer (pH = 6.0) for WT1 and in tris-EDTA buffer (pH = 8.0) for H3K27M and H3K27me3. Immunostaining was performed by 3, 3′-diaminobenzidine (DAB) detection in the Leica BOND-MAX automated stainer (Leica Biosystems). FFPE slides were probed by hematoxylin and eosin (H&E) and WT1.
WT1 Immunoreactivity Scoring
For IHC scoring, a neuropathologist blinded to diagnosis and histone mutation status reviewed H&E-stained slides of tumor and adjacent normal tissue to assign tumor grades and confirm absence of infiltrating tumor cells in the control tissue. None of the adjacent normal regions showed hypercellularity or tumor infiltration. The WT1 score was assigned to each slide and categorized into the bin values as previously described (16). WT1 expression varied from 0 to 4 with 0 representing expression only at the endothelial cell level; 1 representing <1% of total number of cells expressing WT1; 2 representing 1%–19% of total number of cells expressing WT1; 3 representing 20%–50% of total number of cells expressing WT1; 4 representing >50% of total number of cells expressing WT1.
Western Blot Assays
Frozen tissue samples and cultured cells were lysed in 2% sodium dodecyl sulfate (SDS) containing radioimmunoprecipitation assay (RIPA) buffer (ThermoFisher, Waltham, MA) with a miniature handheld homogenizer (Kimble Chase, Vineland, NJ). Protein concentration was measured with the Pierce bicinochoninic acid assay (BCA) protein assay kit (ThermoFisher). Protein lysates of each patient and cell were loaded into 4%–12% Bis-Tris protein gels (Invitrogen, Waltham, MA) in triplicates and protein electrophoresis performed at 100 V for 90 minutes. K562 cell lysates were used as a positive control in each gel. Protein bands were transferred to nitrocellulose membranes with 250 mA current for 120 minutes. After the transfer, the membrane was blocked with 5% Blotting-grade blocker (Bio-Rad, Hercules, CA) for 1 hour. The membrane was incubated with anti-WT1 primary antibody diluted in 5% blocker (1:200) overnight at 4°C and horseradish peroxidase-conjugated secondary antibody (1:5000) was used to incubate the membrane for 1 hour at room temperature. After washing in PBST, the membrane was incubated for 1 minute with chemiluminescence substrate (PerkinElmer, Waltham, MA) and the bands were detected using ChemiDoc Touch Imaging System (Rio-Rad). Rabbit monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (14C10, Cell Signaling Technology) was used at concentration of 1:1000. The WT1 bands were normalized with GAPDH bands.
Immunofluorescence
Immunofluorescent (IF) staining was performed on FFPE slides. FFPE slides were deparaffinized and antigen retrieval completed by boiling in citrate buffer (pH = 6.0) for 20 minutes. The tissues were then permeabilized in 0.3% Triton X-100 for 15 minutes. Blocking was performed for 1 hour in 5% goat serum. The tissues were incubated with anti-WT1 primary antibody and anti-histone H3 K27M mutant primary antibody overnight at 4°C then incubated with fluorescent tagged secondary antibodies for 1 hour at room temperature. After washing in PBS, the tissues were briefly stained with 4′, 6-diamidino-2-phenylindole (DAPI) and imaged with laser scanning confocal microscope (Zeiss, Oberkochen, MA). DIPG cells and K562 cells were collected (500 000 cells each) and adhered to glass slides by centrifuging the cells in Cytospin funnels at 500 rpm for 5 minutes. To ensure strong adherence, 5 µL of 20% BSA was added to the cells before centrifugation. After the centrifugation, the cells were stained and imaged as described above.
Gene Expression Omnibus
The gene expression profile of pediatric high-grade glioma specimens (n = 139) and pediatric non-CNS tumor specimens (n = 13) from datasets available on the Gene Expression Omnibus (Accession numbers: GSE34824, GSE50161, GSE19578, GSE36245) (4, 17–19) were used to produce an unsupervised hierarchical clustering map using Applied Biosystems Transcriptome Analysis Console (TAC) Software version 4.0.1. Gene expression values of WT1 in pediatric high-grade glioma specimens from datasets available on the Gene Expression Omnibus (Accession numbers: GSE50161, n = 34 and GSE50021, n = 32) (17, 20) and from our previously published dataset (n = 8) (6) were normalized to gene expression values of a housekeeping gene, hypoxanthine phosphoribosyltransferase 1 (HPRT1). This normalized WT1 gene expression values were compared with WT1 gene expression values of pediatric healthy brain specimens (Accession numbers: GSE50161, n = 13 and GSE50021, n = 9).
Quantitative RT-PCR
Total RNA of frozen tissue samples and cultured cells were extracted by homogenizing the samples in 1 mL of Trizol and 200 µL of chloroform with a mini-handheld homogenizer. The samples were centrifuged at 8000 rpm for 15 minutes. An equal volume of 70% ethanol was mixed with the supernatants and used to isolate RNA according to Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, Foster City, CA). The RNA was quantified using NanoDrop 2000 Spectrophotometer (ThermoScientific, Waltham, MA). Two mg of RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). 400 ng and 200 ng of cDNA from the tissues and cells were used, respectively. TaqMan Gene Expression Assay for WT1 (Assay ID Hs01103751_m1, ThermoFisher) and for HPRT1 (Assay ID Hs02800695_m1, ThermoFisher) was used to perform quantitative real-time polymerase chain reaction (qRT-PCR) on QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) was used. The Ct values of WT1 were normalized with Ct values of HPRT1.
Statistical Methods
Normality of all the data present in this study was assessed using the D’Agostino-Pearson omnibus normality test and visual inspection of histograms. Nonparametric tests were used to describe any outcome determined to be nonnormally distributed. WT1 protein intensity was compared between paired tumor and normal samples using a nonparametric Wilcoxon sign rank test. WT1 gene expression between pediatric glioblastomas (GBMs) and DIPGs and healthy brain samples was compared using a nonparametric Mann-Whitney U test. WT1 immunoreactivity score differences between mutation subgroups were assessed by the Fisher exact test. The error bars on the WT1 mRNA expression in tumors and H3.3K27M mutation subgroup samples are calculated with standard errors of the means. The level for significance was set at 0.05 and Prism was used for all analyses.
RESULTS
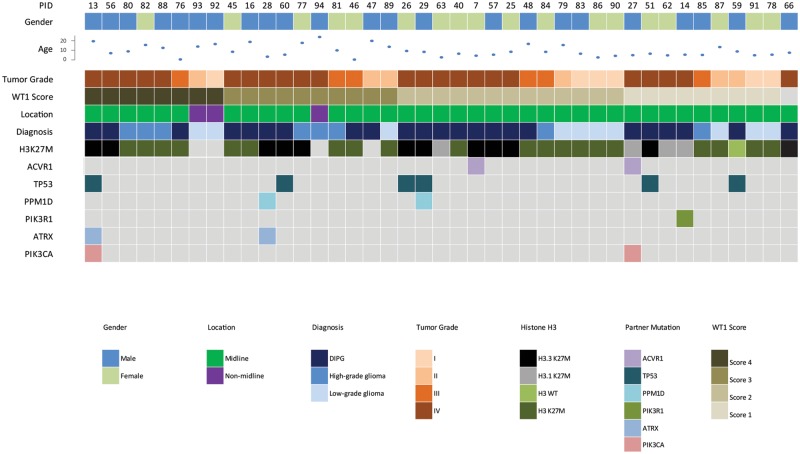
Given that WT1 has been associated with many types of cancers, including brain tumors (9), we investigated the expression of WT1 with specimens from a total of 41 pediatric patients diagnosed with CNS tumors (Table), 20 adjacent healthy brain specimens, and 1 healthy control brain specimen in this study. The median age of our patient cohort was 8.9 years old (range, 1–24 years old), and the tumor locations included: Pons (n = 23), thalamus (n = 6), and others (hypothalamus, posterior fossa, third ventricle, intramedullary cervical spine, n = 12).
TABLE.
Summary of Pediatric CNS Tumor Patient Samples Used in This Study
| PID | Oncohistone | Tumor Location | Sample Types | Histological Diagnosis | WHO Grade | WT1 Score | Gender | Age at Diagnosis (Yrs) |
|---|---|---|---|---|---|---|---|---|
| 13 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 4 | M | 19.7 |
| 56 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 4 | M | 7.5 |
| 80 | N/A | Thalamus, posterior fossa, pons | FFPE | Glioblastoma | IV | 4 | M | 9.4 |
| 82 | N/A | Thalamus, midbrain, third ventricle | FFPE | Glioblastoma | IV | 4 | F | 15.9 |
| 88 | N/A | Cerebellum | FFPE | Glioblastoma | IV | 4 | M | 12.9 |
| 76 | N/A | Pons | FFPE | DIPG | III | 4 | M | 1.2 |
| 93 | N/A | Temporal | FFPE | Low-grade astrocytoma | II | 4 | M | 14.3 |
| 92 | N/A | Cerebellum | FFPE | Pilocytic astrocytoma | I | 4 | M | 16.8 |
| 45 | N/A | Pons | FFPE | DIPG | IV | 3 | F | 9 |
| 16 | N/A | Pons | FFPE | DIPG | IV | 3 | M | 19 |
| 28 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 3 | M | 4 |
| 60 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 3 | M | 6 |
| 77 | H3.3 | Right lateral ventricle, corpus callosum | FFPE | Glioblastoma | IV | 3 | F | 18 |
| 94 | N/A | Frontal | FFPE | Glioblastoma | IV | 3 | M | 24 |
| 81 | N/A | Thalamus, third ventricle | FFPE | Anaplastic astrocytoma | III | 3 | F | 10.4 |
| 46 | N/A | Pons | FFPE | DIPG | III | 3 | F | 1 |
| 47 | N/A | Pons | FFPE | DIPG | II | 3 | M | 20 |
| 89 | N/A | Brainstem | FFPE | Low-grade astrocytoma | II | 3 | M | 14 |
| 26 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 2 | F | 9.8 |
| 29 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 2 | M | 8.9 |
| 63 | H3.1 | Pons | FFPE, Frozen | DIPG | IV | 2 | F | 3.3 |
| 40 | N/A | Pons | FFPE | DIPG | IV | 2 | F | 7 |
| 7 | H3.3 | Pons | FFPE | DIPG | IV | 2 | F | 5 |
| 57 | H3.3 | Pons | FFPE, Frozen | DIPG | IV | 2 | M | 6 |
| 84 | N/A | Brainstem | FFPE | Anaplastic astrocytoma | III | 2 | F | 8.9 |
| 48 | N/A | Pons | FFPE | DIPG | III | 2 | M | 17 |
| 25 | H3.3 | Pons | FFPE | DIPG | IV | 2 | F | 9 |
| 79 | N/A | Thalamus | FFPE | Low-grade astrocytoma | II | 2 | M | 15.8 |
| 83 | N/A | Thalamus | FFPE | Pilocytic astrocytoma | I | 2 | M | 7 |
| 86 | N/A | Hypothalamus | FFPE | Pilocytic astrocytoma | I | 2 | F | 3.2 |
| 90 | N/A | Hypothalamus | FFPE | Pilocytic astrocytoma | I | 2 | F | 4.8 |
| 27 | H3.1 | Pons | FFPE | DIPG | IV | 1 | M | 5.5 |
| 51 | H3.3 | Pons | FFPE | DIPG | IV | 1 | F | 7 |
| 62 | H3.1 | Pons | FFPE | DIPG | IV | 1 | F | 5.3 |
| 85 | N/A | Intramedullary c-spine | FFPE | Anaplastic astrocytoma | III | 1 | M | 5.8 |
| 14 | H3.1 | Pons | FFPE | DIPG | IV | 1 | M | 6 |
| 87 | N/A | Brainstem | FFPE | Low-grade astrocytoma | II | 1 | F | 13.8 |
| 59 | Wild-type | Pons | FFPE | DIPG | II | 2 | M | 9.3 |
| 91 | N/A | Posterior fossa | FFPE | Pilocytic astrocytoma | I | 1 | F | 5.4 |
| 78 | N/A | Thalamus | FFPE | Pilocytic astrocytoma | I | 1 | F | 6 |
| 66 | H3.3 | Pons | Frozen | DIPG | IV | N/A | M | 8 |
A total of 41 pediatric CNS tumor specimens were used in this study.
FFPE, formalin-fixed paraffin-embedded; WHO, World Health Organization; WT1, Wilms tumor protein.
WT1 Expression Is Robust in Pediatric High-Grade Gliomas
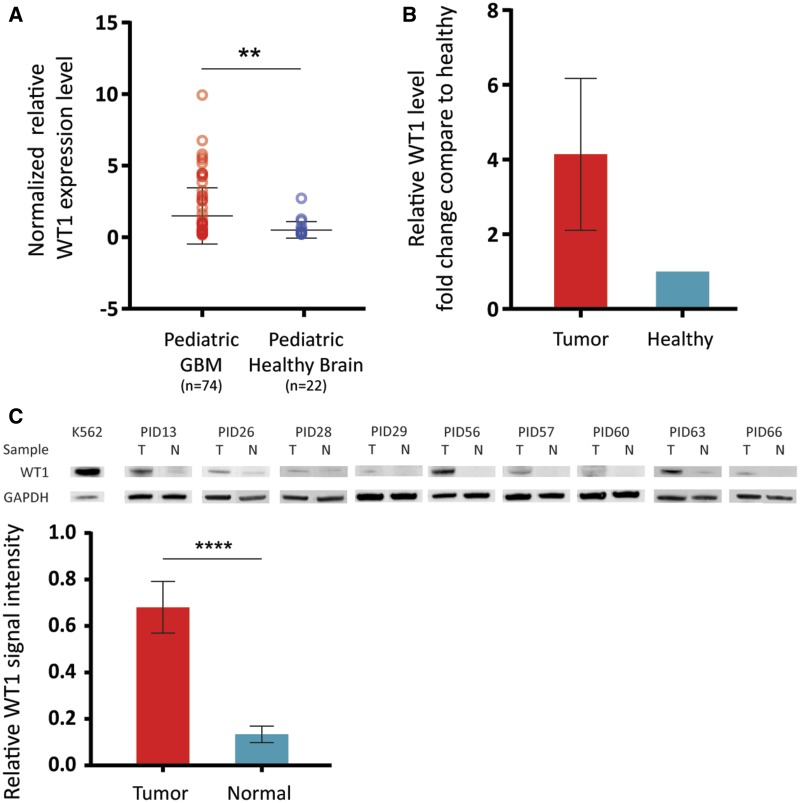
To assess the differential gene expression pattern in pediatric CNS cancers compared with pediatric healthy brain, we first analyzed publicly available gene expression data generated from 139 pediatric GBMs and compared with RNA expression obtained from 13 pediatric non-cancer brain specimens (Accession numbers: GSE34824, GSE50161, GSE19578, GSE36245) (4, 17–19) (Supplementary DataFig. S1). Then, WT1 expression specific comparative analysis was performed using the publicly available datasets (Accession numbers: GSE50161 and GSE50021) (17, 20). This analysis indicated a significant upregulation (2.9-fold, p = 0.023) of WT1 mRNA expression in GBM specimens, including DIPG (n = 74), compared with healthy control specimens (n = 22; Fig. 1A). Given that WT1 showed tumor specificity in the publicly available gene expression data, we performed qRT-PCR using cDNA from fresh frozen DIPG specimens, which confirmed the significantly higher expression (4.1-fold) of WT1 in tumor specimens (n = 5) compared with adjacent healthy brain samples (n = 5; Fig. 1B). We performed additional validation assays by Western blot using protein extracts from DIPG subjects (n = 9) for whom frozen tumor and adjacent normal tissue was available (Table). Western blot analysis showed significant overexpression (7.4-fold, p < 0.0001) of WT1 in DIPG specimens compared with adjacent normal samples (Fig. 1C).
FIGURE 1.
WT1 expression is associated with pediatric CNS cancers. (A) Analysis of publicly available gene expression profiles of a larger cohort of pediatric GBMs. This analysis showed that WT1 is highly expressed in pediatric GBMs (n = 74) compared with pediatric healthy brain specimens (n = 22) (2.9-fold, p = 0.023). (B) qRT-PCR using cDNA from DIPG specimens validated WT1 overexpression in tumor (n = 5) versus normal (n = 5) within each patient (4.1-fold). (C) WT1 overexpression in tumor tissues was validated by Western blot assays using DIPG tumor specimens (n = 9) and adjacent normal specimens (n = 9). Quantification of the Western blot assays of DIPG tumor versus normal showed significantly higher expression of WT1 in the tumors (7.4-fold, p < 0.0001).
WT1 Expression Is Associated with the Mutation Status of Oncohistone H3
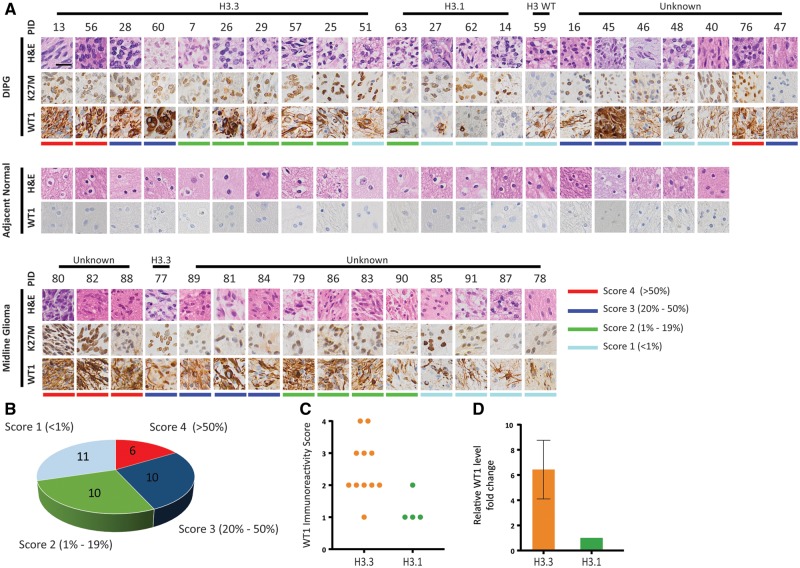
We observed that WT1 expression, at both the mRNA and protein level, was significantly higher in a number of DIPG specimens (Fig. 1B, C). This led us to expand our investigation to a cohort of 37 tumor specimens and 20 adjacent healthy specimens from 37 subjects diagnosed with midline glioma (Fig. 2) and probed for WT1 expression by IHC. H&E staining was analyzed by a neuropathologist indicating tumor grade for DIPGs (4 WHO grade II, 3 WHO grade III, 15 WHO grade IV) and non-DIPG midline gliomas (5 WHO grade I, 4 WHO grade II, 2 WHO grade III, 4 WHO grade IV; Fig. 3A). Using antibodies against H3K27M, we probed for mutant histones and found that 94.6% (35/37) of subjects harbored the H3K27M mutation (Fig. 3A). H3K27M antibody does not distinguish between H3.3 and H3.1 (K27M) mutant proteins thus genomic data (where available) were used to distinguish the H3.3K27M from the H3.1K27M patients. WT1 immunostaining was performed and histological stains were graded by a neuropathologist who was blinded to both diagnosis and histone mutation status (Fig. 3B). A close inspection of histological staining did not show any significant relationship between tumor grade and WT1 but revealed that differential expression of WT1 correlated with oncohistone mutation status. Specifically, for WT1 expression, 91% (10/11) of the H3.3 specimens scored greater than or equal to 2, while none of the H3.1 specimens scored >2. The distribution of WT1 immunoreactivity score was significantly different between H3.3 and H3.1 subtypes (Fisher exact test, p = 0.017; Fig. 3C). We validated this finding by performing qRT-PCR using fresh frozen H3.3K27M DIPG tumor specimen (n = 3) and H3.1K27M tumor specimens (n = 2). Our data showed higher expression (6.4-fold) in the H3.3-mutant samples (Fig. 3D).
FIGURE 2.
Demographic and clinical distribution of patients used for the study. Formalin-fixed paraffin-embedded (FFPE) specimens from a large cohort of pediatric CNS tumors (n = 41) were used in this study to examine the expression of WT1 in the tumors. The tumor grade of each specimen is represented by the orange shades. The driver and partner mutations were established using droplet PCR or WES. The specimens were checked for H3 mutation status by IHC only (n = 20; dark green boxes) or by either droplet PCR or WES (n = 16).
FIGURE 3.
Immunohistochemistry of a large cohort of DIPGs indicated high expression of WT1 in H3.3 subtype compared with H3.1 subtype DIPGs. (A) IHC of a large cohort of DIPGs and pediatric midline gliomas showed WT1 overexpression in tumor compared with adjacent healthy brain tissue was valid. Scale bar = 50 µm. (B) Each tumor specimen was reviewed by a blinded neuropathologist and given an immunoreactivity score based on the number of WT1-positive cells. The pie chart shows the WT1 scores distribution of the specimens. (C) Examination of WT1 immunoreactivity scores revealed that WT1 scores of the specimens are associated with histone H3 mutation status. The distribution of WT1 immunoreactivity score was significantly different between H3.3 and H3.1 subtypes (Fisher exact test, p = 0.017). (D) qRT-PCR using cDNA from H3.3K27M DIPG tumor tissue (n = 3) and H3.1K27M DIPG tumor tissue (n = 2) showed higher expression of WT1 in H3.3K27M tumor specimens compared with H3.1K27M tumor specimens (6.4-fold).
WT1 Expression Is Specific to Glial Tumor Cells with Cytosolic Localization
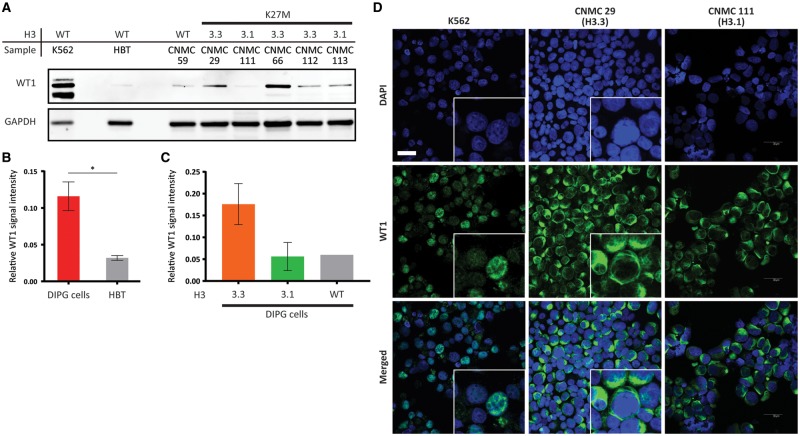
WT1 is known to be expressed in the endothelial cells of tumor vasculature. In order to show that the differential expression of WT1 in H3.3K27M tissues and H3.1K27M tissues was not derived from endothelial cells, we performed Western blot assays and IF assays to analyze human DIPG primary cells. Western blot assays (n = 6, 1 H3 wildtype, 2 H3.1K27M, and 3 H3.3K27M; Fig. 4A) and IF assays (n = 2, H3.3K27M; Fig. 4D) were performed using human DIPG primary cells. Western blot analysis using human DIPG primary cells showed significantly higher level of WT1 in DIPG tumor cells compared with healthy brain tissue specimens from pediatric patients without CNS disease (3.63-fold, p = 0.047; Fig. 4B). When we interrogated WT1 expression level in different subtypes of human DIPG primary cells, H3.3K27M subtype human DIPG cells showed higher WT1 expression compared with H3.1K27M cells, H3 wildtype cells and healthy brain tissue (2.83-fold, 2.86-fold, and 5.37-fold, respectively; Fig. 4C). This is a trend which supports our findings by IHC and qRT-PCR using DIPG tumor specimens. When we performed IF using human DIPG primary cells, WT1 was present in the cells but was localized to the cytosol in both human DIPG primary cells tested (Fig. 4D). Given the fact that WT1 is known to be a nuclear protein (21), this finding was surprising. A previous study had shown that WT1 expression was restricted to endothelial cells in normal brain and astrogliosis (16). This pattern of WT1 expression was also seen in our healthy pons specimen of a pediatric patient without CNS disease and adjacent normal brain specimens, serving as an internal control to confirm that WT1 detection by the antibody was specific (Supplementary DataFig. S3A, B). To further confirm that our antibody is detecting the correct protein, we used human chronic myelogenous leukemia K562 cells that are known to express high levels of WT1 protein as our positive control (Fig. 4D). We found that WT1 does, in fact, localize to the nucleus in K562 cells, suggesting the WT1 mislocalization in DIPGs. Healthy pediatric kidney specimens were also used to confirm our antibody specificity, which showed nuclear staining with the anti-WT1 antibody (Supplementary DataFig. S3C, D).
FIGURE 4.
Validation of WT1 differential expression in human DIPG primary cells. (A) A Western blot assay of WT1 using human DIPG primary cells (n = 6) and healthy brain tissue (HBT) lysates. (B) Quantification of the Western blot assays showed significantly higher WT1 in human DIPG primary cells compared with healthy brain tissue (3.63-fold, p = 0.047). (C) Human DIPG primary cells harboring H3.3K27M showed higher WT1 compared with human DIPG primary cells harboring H3.1K27M (2.83-fold) and histone H3 wildtype (2.86-fold). (D) Immunofluorescence staining of human DIPG primary cells (n = 2) showed WT1 expression in the tumor cells and localization of WT1, mainly in the cytoplasm of the tumor cells. The specificity of the antibody was validated by staining K562 cells, which showed WT1 localization in the nucleus. Scale bar = 30 µm.
DISCUSSION
There has been a rapid increase in studies investigating immunotherapy in CNS tumors. For instance, vaccines have been tested against the EGFR deletion mutant variant III in adults (22) and against EphA2, IL13Rα2, and BIRC5 (Survivin) in pediatrics (23). Checkpoint inhibitors and adoptive cellular therapy are also currently undergoing clinical investigation in childhood CNS cancers (24, 25). WT1-directed immunotherapeutic trials in adults with glioblastoma have indicated better survival of patients with higher WT1 expression levels receiving WT1-specific immunotherapy (26). In DIPG, a currently open phase I clinical trial investigating an H3.3K27M peptide vaccine highlights the interest in intracellular antigens similar to WT1.
In this study, we have used a large cohort of specimens obtained from children diagnosed with CNS cancers to show specific upregulation of WT1 at both protein and RNA levels in DIPGs. More specifically, our findings indicate that WT1 expression is significantly higher in DIPGs harboring H3.3K27M when compared with those with H3.1K27M alleles. Given the hypomethylated genome noted in both DIPGs (6, 27), this observed differential WT1 expression may be the result of the variation in the global methylation status of DIPG subtypes (H3.3K27M, vs H3.1K27M, vs H3WT). One archival FFPE sample (PID 47), for which histone status was not known, exhibited WT1 positivity but did not exhibit K27M positivity by IHC. Upon further investigation by performing H3K27 trimethylation IHC on the specimen, we noted that the specimen showed decreased H3K27 trimethylation compared with healthy brain control, suggesting that the specimen may be H3K27M (Supplementary DataFig. S2). However, there was some overlap in WT1 scores between the H3.1 and H3.3 specimens. The observed pattern of higher expression of WT1 in H3.3 specimens may become less marked in a larger cohort (Fig. 3C).
There have been multiple diseases, such as Denys-Drash syndrome (28) and Frasier syndrome (29), associated with germline WT1 mutation. WT1 has at least 36 putative isoforms that result from alternative splicing or coding by alternative start sites (10). Two major isoforms include –KTS and +KTS, which differ by 3 amino acids. The –KTS WT1 isoform is known to have a transcription factor function whereas the +KTS WT1 isoform is thought to have a RNA editing function. An analysis of the available genomic data of our sample cohort as well as the publicly available datasets on PedcBioportal (30–32) discovered infrequent genomic alterations associated with WT1 gene. Only 5 out of 1738 adult and pediatric GBM samples exhibited amplification, and 1 sample had WT1 gene deletion (data not shown). This observation suggests that WT1 upregulation is most likely a result of the global epigenetic changes or by tumor activated pathways such as cellular mitotic pathways rather than sequence mutation. Indeed, a study that examined the effects of WT1 knockdown in glioblastoma cell lines showed an upregulation of apoptosis-related genes such as PIK3CA and TP53 (33). Given the fact that most high-grade gliomas, including DIPGs, harbor mutations and thus upregulation of the PIK3CA and TP53 genes, we suspect that WT1 overexpression may be a compensatory mechanism for PIK3CA and p53 regulatory pathway upregulation.
Our cytosolic detection of WT1 is not surprising as previous studies have also shown cytosolic localization of WT1 in glioblastoma cells and lung cancer cells (34, 35); in fact, WT1 shuttles between the nucleus and cytoplasm in association with actively translating polysomes (36). Our finding, as well as the previous studies, suggests the possibility that WT1 has a role other than as a transcription factor in cancer, although it is possible that there exists overexpression of WT1 isoforms that are missing the nuclear localization signal. To fully understand the biological role of WT1 in cancer, including in pediatric high-grade gliomas, further molecular studies will be required.
There has been some discordance in tumor specificity of WT1 in the literature (37) and WT1 alone may not be an ideal marker to distinguish reactive astrocytes from neoplastic astrocytes. However, investigating the potential of WT1 as an immunotherapeutic target is still promising as even in cases of reactive gliosis, the vast majority of gliomas cases demonstrates stronger WT1 immunoreactivity. In summary, our study shows that WT1, which has been closely associated with prognosis in other solid cancers (38), is also highly expressed in one particular molecular phenotype of DIPG, H3.3K27M, and may serve as a strong immunotherapeutic target. Our study, together with the safety and efficacy demonstrated in WT1-based peptide vaccine clinical trial in GBM (13), and the prognostic value of WT1 protein level (26), advocates for further examination of WT1 as a potential immunotherapeutic target in children with DIPG.
Supplementary Material
ACKNOWLEDGMENTS
We would like to extend our gratitude to the patients and families who have generously donated samples. We would also like to thank Dr. Oren Becher of Northwestern University in Chicago for his expert advice in experimental design and manuscript preparation, and the Children’s National Health System Pathology Department and Children’s Brain Tumor Tissue Consortium (CBTTC) for their services.
Funding: Smashing Walnuts, DIPG Collaborative, The Goldwin Foundation, The Cure Starts Now Foundation, CNHS CTSI, Zickler Family Foundation, The Childhood Brain Tumor Foundation, Isabella Kerr Molina Foundation, Al Musella Foundation, Brain Tumor Foundation for Children, Kisses 4 Kayla, Matthew Larson Foundation, The Brad Kaminsky Foundation, iDance4aCURE, and Rally Foundation.
Footnotes
The authors have no duality or conflicts of interest to declare.
Supplementary Data can be found at academic.oup.com/jnen.
REFERENCES
- 1. Ramos A, Hilario A, Lagares A, et al. Brainstem gliomas. Semin Ultrasound CT MR 2013;34:104–12 [DOI] [PubMed] [Google Scholar]
- 2. Chassot A, Canale S, Varlet P, et al. Radiotherapy with concurrent and adjuvant temozolomide in children with newly diagnosed diffuse intrinsic pontine glioma. J Neurooncol 2012;106:399–407. [DOI] [PubMed] [Google Scholar]
- 3. Cohen K, Heideman R, Zhou T, et al. Temozolomide in the treatment of children with newly diagnosed diffuse intrinsic pontine gliomas: A report from the Children's Oncology Group. Neuro-oncology 2011;13:410–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartzentruber J, Korshunov A, Liu X, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31 [DOI] [PubMed] [Google Scholar]
- 5. Wu G, Broniscer A, McEachron T, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saratsis A, Kambhampati M, Snyder K, et al. Comparative multidimensional molecular analyses of pediatric diffuse intrinsic pontine glioma reveals distinct molecular subtypes. Acta Neuropathol 2014;127:881–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saratsis A, Yadavilli S, Magge S, et al. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol 2012;14:547–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yadavilli S, Scafidi J, Becher O, et al. The emerging role of NG2 in pediatric diffuse intrinsic pontine glioma. Oncotarget 2015;6:12141–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sugiyama H. WT1 (Wilms' Tumor Gene 1): Biology and cancer immunotherapy. Jpn J Clin Oncol 2010;40:377–87 [DOI] [PubMed] [Google Scholar]
- 10. Hastie N. Wilms' tumour 1 (WT1) in development, homeostasis and disease. Development 2017;144:2862–72 [DOI] [PubMed] [Google Scholar]
- 11. Haber D, Buckler A.. WT1: A novel tumor suppressor gene inactivated in Wilms' tumor. New Biol 1992;4:97–106 [PubMed] [Google Scholar]
- 12. Cheever M, Allison J, Ferris A, et al. The prioritization of cancer antigens: A National Cancer Institute Pilot Project for the Acceleration of Translational Research. Clin Cancer Res 2009;15:5323–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oji Y, Hashimoto N, Tsuboi A, et al. Association of WT1 IgG antibody against WT1 peptide with prolonged survival in glioblastoma multiforme patients vaccinated with WT1 peptide. Int J Cancer 2016;139:1391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kambhampati M, Perez J, Yadavilli S, et al. A standardized autopsy procurement allows for the comprehensive study of DIPG biology. Oncotarget 2015;6:12740–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikbakht H, Panditharatna E, Mikael L, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun 2016;7:11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schittenhelm J, Mittelbronn M, Nguyen T, et al. WT1 expression distinguishes astrocytic tumor cells from normal and reactive astrocytes. Brain Pathol 2008;18:344–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Griesinger A, Birks D, Donson A, et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J Immunol 2013;191:4880–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paugh B, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. JCO 2010;28:3061–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425–37 [DOI] [PubMed] [Google Scholar]
- 20. Buczkowicz P, Hoeman C, Rakopoulos P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 2014;46:451–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hohenstein P, Hastie N.. The many facets of the Wilms' tumour gene, WT1. Hum Mol Genet 2006;15:R196–201 [DOI] [PubMed] [Google Scholar]
- 22. O’Rourke D, Nasrallah M, Desai A, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 2017;9:eaaa0984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pollack I, Jakacki R, Butterfield L, et al. Antigen-specific immunoreactivity and clinical outcome following vaccination with glioma-associated antigen peptides in children with recurrent high-grade gliomas: Results of a pilot study. J Neurooncol 2016;130:517–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahmed N, Brawley V, Hegde M, et al. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma. JAMA Oncol 2017;3:1094–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wildes T, Grippin A, Dyson K, et al. Cross-talk between T cells and hematopoietic stem cells during adoptive cellular therapy for malignant glioma. Clin Cancer Res 2018;24:3955–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiba Y, Hashimoto N, Tsuboi A, et al. Prognostic value of WT1 protein expression level and MIB-1 staining index as predictor of response to WT1 immunotherapy in glioblastoma patients. Brain Tumor Pathol 2010;27:29–34 [DOI] [PubMed] [Google Scholar]
- 27. Bender S, Tang Y, Lindroth A, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 2013;24:660–72 [DOI] [PubMed] [Google Scholar]
- 28. Pelletier J, Bruening W, Kashtan C, et al. Germline mutations in the Wilms' tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell 1991;67:437–47 [DOI] [PubMed] [Google Scholar]
- 29. Barbaux S, Niaudet P, Gubler M, et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet 1997;17:467–70 [DOI] [PubMed] [Google Scholar]
- 30. Brennan C, Verhaak R, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 2017;32:520–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mackay A, Burford A, Molinari V, et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell 2018;33:829–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kijima N, Hosen N, Kagawa N, et al. Wilms' tumor 1 is involved in tumorigenicity of glioblastoma by regulating cell proliferation and apoptosis. Anticancer Res 2014;34:61–7 [PubMed] [Google Scholar]
- 34. Nakatsuka S, Oji Y, Horiuchi T, et al. Immunohistochemical detection of WT1 protein in a variety of cancer cells. Mod Pathol 2006;19:804–14 [DOI] [PubMed] [Google Scholar]
- 35. Oji Y, Suzuki T, Nakano Y, et al. Overexpression of the Wilms' tumor gene WT1 in primary astrocytic tumors. Cancer Sci 2004;95:822–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niksic M, Slight J, Sandford J, et al. The Wilms' tumour protein (WT1) shuttles between nucleus and cytoplasm and is present in functional polysomes. Hum Mol Genet 2003;13:463–71 [DOI] [PubMed] [Google Scholar]
- 37. Bourne T, Elias W, Lopes M, et al. WT1 is not a reliable marker to distinguish reactive from neoplastic astrocyte populations in the central nervous system. Brain Pathol 2010;20:1090–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qi X, Zhang F., Wu H, et al. Wilms' tumor 1 (WT1) expression and prognosis in solid cancer patients: A systematic review and meta-analysis. Sci Rep 2015;5:8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.